Back to Journals » OncoTargets and Therapy » Volume 14
Expression and Diagnostic Value of tRNA-Derived Fragments Secreted by Extracellular Vesicles in Hypopharyngeal Carcinoma
Authors Xi J, Zeng Z, Li X, Zhang X, Xu J
Received 18 May 2021
Accepted for publication 29 June 2021
Published 14 July 2021 Volume 2021:14 Pages 4189—4199
DOI https://doi.org/10.2147/OTT.S320176
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Jian Xi,1– 3,* Zhiming Zeng,1,2,* Xin Li,1,2 Xin Zhang,1,2 Jing Xu1,2
1Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital of Central South University, Changsha, Hunan Province, 410008, People’s Republic of China; 2Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Changsha, 410008, Hunan, People’s Republic of China; 3Department of Neurosurgery, Xiangya Hospital of Central South University, Changsha, Hunan Province, 410008, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jing Xu
Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital of Central South University, Changsha, Hunan Province, 410008, People’s Republic of China
Email [email protected]
Objective: To detect the expression level of tRNA-derived fragments secreted by extracellular vesicles in hypopharyngeal cancer and explore the influence of tRNA-derived fragments on the occurrence of hypopharyngeal cancer and lung metastasis.
Methods: After high-speed centrifugation, tRNA, which was extracted from the extracellular vesicles of patients with hypopharyngeal cancer and healthy subjects, was sequenced using microarrays. The expression of three differentially expressed tRNAs in hypopharyngeal cancer, healthy subjects, human normal laryngeal epithelial cells and hypopharyngeal cancer line was detected by qRT-PCR. The correlation between the upregulated tRNA, as identified by qRT-PCR, and the clinicopathological features of the non-lung metastatic patients was further analyzed. Finally, the expressions of upregulated tRNA were compared between the non-lung metastatic and lung metastatic patients. The risk factors of hypopharyngeal cancer with lung metastatic were identified by the Cox regression analysis.
Results: By high-speed centrifugation, extracellular vesicles were extracted successfully. It was found that a variety of tRNAs in the extracellular vesicles from patients with hypopharyngeal cancer by sequencing. qRT-PCR validation indicated that tRF-1:30-Lys-CTT-1-M2 was significantly overexpressed in hypopharyngeal cancer patients and tumor cells, especially in lung metastatic patients. It was indicated that tRF-1:30-Lys-CTT-1-M2 overexpression was closely related to such pathological features as tumor staging, differentiation grade, smoking history and drinking history. According to the Cox regression analysis, stage III–IV, smoking history, drinking history and tRF-1:30-Lys-CTT-1-M2 overexpression were independent risk factors for metastasis of hypopharyngeal cancer.
Conclusion: tRF-1:30-Lys-CTT-1-M2 was overexpressed in patients with hypopharyngeal cancer and was identified as an independent risk factor for lung metastasis. It can be used as a novel biomarker for the diagnosis and lung metastasis monitoring of hypopharyngeal cancer.
Keywords: extracellular vesicles, tRNA-derived fragment, hypopharyngeal cancer, lung metastasis, cancer marker
Background
Hypopharyngeal cancer is not common in malignant tumors, accounting for about 2–6% of head and neck malignancies.1 Over 90% of hypopharyngeal cancer patients are squamous cell carcinoma. Hypopharyngeal cancer is one of the most malignant tumors in the head and neck. According to statistics, its 5-year survival rate is less than 50%.2 Most patients with hypopharyngeal cancer still have poor prognosis after comprehensive treatment, and the patients’ survival is not improved significantly. About one half of the patients relapse within one year.3 Therefore, understanding the molecular mechanism underlying the occurrence, development, and metastasis of hypopharyngeal cancer and looking for relevant biomarkers are of high importance for improving the therapeutic effect and prognosis of the patients.
Extracellular vesicles that can be released by most of the cells (including tumor cells) and have a diameter of about 30–150nm.4 Tumor cells can secrete a large amount of extracellular vesicles containing tumor-specific protective substances, so they have become a research object of particular interest to researchers in cancer biomarkers. Furthermore, extracellular vesicles regulate the behaviors of recipient cells and can be used as diagnostic biomarkers for human diseases.5 For example, Zhu et al found that glutaminase 2 and annexin A2 in the extracellular vesicles of human liver cancer cells were potential biomarkers for liver cancer.6 Other researchers have suggested that the tumor cell-derived extracellular vesicles can promote the proliferation, invasion, and drug resistance of tumor cells.7 However, the influence of extracellular vesicles secretions on hypopharyngeal cancer is not yet clarified and requires further investigation.
Along with the development of the high-throughput sequencing technology, an increasing number of transfer RNA (tRNA) have been discovered. In recent years, tRNAs have drawn growing attention in the field of tumors and other diseases due to their unique biological functions in gene transcription regulation.8 Moreover, tRNA-derived fragments (tRFs) are discovered in various organisms. A study has shown that tRFs play an important role in regulating the proliferation, invasion, metastasis, and gene expression of human cancers.9,10 In addition, the abnormal expressions of tRNAs and tRFs are closely related to tumor occurrence and development. tRNAs and tRFs may serve as candidate diagnostic biomarkers or therapeutic targets.11–13 However, the expressions of tRNAs and tRFs in hypopharyngeal cancer and their roles still remain largely unknown. In the present study, we attempted to investigate the expressions of tRFs in hypopharyngeal cancer and their influence on the occurrence and development of hypopharyngeal cancer.
Materials and Methods
Clinical Samples and Case Information
From January to December 2016, 30 patients newly diagnosed as hypopharyngeal cancer (treatment-naive and non-lung metastatic) who received surgery at the Department of Otolaryngology-Head and Neck Surgery of our hospital were recruited. In the meantime, 30 healthy subjects were chosen as controls. None of the patients had a history of radiotherapy and chemotherapy before the study. Neither did they have a history of other malignancies. All of them were pathologically confirmed as squamous cell carcinoma after surgery. First, plasma samples were collected from 5 newly diagnosed, treatment-naive, and non-lung metastatic patients and 5 volunteers. Extracellular vesicles were extracted for tRNA sequencing. The differentially expressed tRFs were chosen for qRT-PCR validation in the remaining newly diagnosed, treatment-naive, and non-lung metastatic patients, healthy subjects, and hypopharyngeal cancer cell line. Meanwhile, 30 hypopharyngeal cancer patients with lung metastatic were collected, and the expressions of relevant tRFs were detected in these patients. The patients’ clinicopathological parameters, such as age, gender, tumor staging, differentiation grade, and smoking and drinking history, were collected via the electronic medical record system. The present study was approved by the institutional ethics committee (approved number: 2015-S0127). All patients were given their consent before obtaining samples, and signed written informed consent. The informed consent form obtained from each subject was based on the Declaration of Helsinki.
Cell Sources and Cell Culture
Human normal laryngeal epithelial cell line and hypopharyngeal cancer cell line FaDu were purchased from the American Type Culture Collection (ATCC) and cultured in DMEM. The cell type and the treatment method were labeled. After disinfection with alcohol, the cells were incubated in an incubator (5% CO2, 37°C). After incubation for some time, the cell morphology was observed under an inverted microscope to determine whether the cells were adherent to the wall. Cells in the logarithmic phase of growth were harvested for subsequent experiments.
Extraction and Scanning Electron Microscopy of Extracellular Vesicles
The peripheral blood samples from the patients and healthy subjects were centrifuged at 500g for 5 min to obtain the supernatant, followed by ultra-high-speed centrifugation (The operation steps are detailed in Figure 1). The supernatant thus obtained was the extracellular vesicles, which were added with PBS, mixed well, and stored at −80°C. 10μL of the supernatant prepared was drawn and placed on the copper mesh. The extracellular vesicles supernatant was stained with 20μL of 2% uranyl acetate for 5 min. After staining, the samples were exposed to light for 10–15 min, observed under SEM (scanning electron microscopy), and photographed.
 |
Figure 1 The specific operation process of extracellular vesicles extraction. |
Extraction and Sequencing of Extracellular Vesicles RNA
Extracellular vesicles RNAs were extracted using the Trizol reagent (Invitrogen, USA). RNA concentration and purity of each sample were quantified using a NanoDrop ND-1000 spectrophotometer. For each sample, about 200ng of the extracted total RNA was taken for high-throughput sequencing. The total RNA was pretreated with the rtStar™ tRF and tiRNA Pretreatment Kit (Cat#AS-FS-005, Arraystar, MD, USA). The sequencing library was prepared from the pretreated total RNA by using the rtStar™ First-Strand cDNA Synthesis Kit (3ʹ and 5ʹ adapters) (Cat#AS-FS-005, Arraystar, MD, USA). The library was denatured with 0.1M NaOH to form single-stranded DNA molecules and diluted to a loading volume of 1.3mL and a loading concentration of 1.8pM. The diluted library was loaded to the kit according to the instruction manual. The sequencing was done by using the NextSeq 500 V2 sequencing reagent kit (#FC-404-2005, Illumina, California, USA) on the Illumina NextSeq 500 System. tRFs with fold change≥2 and P<0.05 were considered the differentially expressed ones.
Verification by Quantitative PCR
Extracellular vesicles RNAs were extracted using the Trizol reagent (Invitrogen, USA). The sequencing library was prepared from the pretreated total RNA using the rtStar™ First-Strand cDNA Synthesis Kit (3ʹ and 5ʹ adapters) (Cat#AS-FS-005, Arraystar, MD, USA). cDNA was diluted in nuclease-free water (dilution factor is 1:40), followed by qPCR for the nrStar™ Human tRNA Repertoire PCR Array (Arraystar) using the Applied Biosystems QuantStudio 7 Flex Real-Time PCR System (Thermo-Fisher) as per the manufacturer’s instructions. U6 was chosen as the control, and the tRNA expression level was determined by using the ΔΔCt method. The primer sequences for U6 were as follows: forward primer, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; reverse primer, 5ʹ-AACGCTTCACGAATTTGC GT-3ʹ. The primer sequences for other tRNAs were designed by using Primer 5.0. For more details, see Table 1.
 |
Table 1 The Basic Informations of tRNAs in Quantitative Real-Time PCR |
Statistical Analysis
Statistical analyses were performed using SSPS20.0. The data were expressed as mean±standard deviation. Relative expression levels of tRFs were analyzed by using a t-test in HNC patients vs healthy subjects and in non-metastatic vs metastatic HNC patients. A Chi-square test was performed to analyze the correlation between the tRF expression and clinicopathological features of the hypopharyngeal carcinoma patients. The receiver operating characteristic (ROC) curve was plotted to assess the diagnostic value of tRFs. P<0.05 indicated a significant difference.
Results
Identification of Extracellular Vesicles
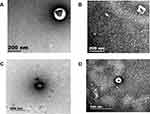
The extracellular vesicles extracted from the patients and the healthy subjects were dissolved in PBS. Under SEM, extracellular vesicles were microvesicles with a bilayer structure, with the diameter of about 200nm (Figure 2A and B) and 500nm (Figure 2C and D). Electron microscopy results demonstrated that extracellular vesicles had been successfully extracted from the subjects.
Expression of Extracellular Vesicles tRFs
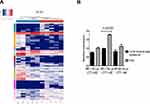
High-throughput sequencing was applied to detect the differential expressions of tRFs in the extracellular vesicles of patients with hypopharyngeal cancer and healthy subjects. All of the differentially expressed tRFs were compared by using the heat map (Figure 3A). There were 33 upregulated and 22 downregulated tRFs. Among them, 2 significantly upregulated and 1 significantly downregulated tRFs were chosen for qRT-PCR validation (Table 2). In addition, the percentages of each subtype of the differentially expressed tRFs were analyzed. It was found that over 75% of the subtypes were tRFs (primarily tRF-3a, tRF-3b, tRF-5a, tRF-5b, and tRF-5c); the remaining were tiRNAs (Figure 4A and B). The length distribution of each tRFs subtype in healthy individuals and hypopharyngeal cancer patients is shown in Figure 4C and D.
 |
Table 2 The Primer Sequence of tRNAs in Quantitative Real-Time PCR |
The Expression of tRFs in the Hypopharyngeal Cancer Patients and Cell Lines
The expressions of tRF-1:30-Lys-CTT-1-M2, tRF-1:30-Lys-CTT-2-M2 and tRF-60:76-Lys-CTT-1-M4 in the extracellular vesicles of the cancer patients and healthy subjects were verified by qRT-PCR. The results showed that all of these three tRNAs were expressed in the extracellular vesicles of the cancer patients and healthy subjects. As compared with the healthy subjects, tRF-1:30-Lys-CTT-1-M2 was significantly upregulated in the extracellular vesicles of the hypopharyngeal cancer patients (Figure 5A); however, there were no significant differences in the expressions of tRF-1:30-Lys-CTT-2-M2 and tRF-60:76-Lys-CTT-1-M4 between the two groups (Figure 5B and C). Later, the expressions of these three tRFs were detected in the human normal laryngeal epithelial cells and hypopharyngeal cancer cell line FaDu. The results showed that tRF-1:30-Lys-CTT-1-M2 was significantly upregulated in the FaDu cells than in the human normal laryngeal epithelial cells (Figure 3B). However, there were no significant differences in the expressions of tRF-1:30-Lys-CTT-2-M2 and tRF-60:76-Lys-CTT-1-M4 between the two cell lines (Figure 3B).
Correlation Between tRFs and Clinical Characteristics of Hypopharyngeal Cancer Patients
According to the qRT-PCR results, tRF-1:30-Lys-CTT-1-M2 was selected for the subsequent analysis of correlation with the clinical features. Based on the median expression level, the patients were divided into high expression group and low expression group. The subgroup analysis showed that tRF-1:30-Lys-CTT-1-M2 overexpression was closely related to the clinicopathological features of the patients, including tumor staging, differentiation grade, smoking history and drinking history. However, the tRF-1:30-Lys-CTT-1-M2 expression was not significantly correlated with other clinicopathological features (Table 3).
 |
Table 3 Correlation Analysis Between the Expression Level of tRNA Fragment and Its Clinicopathological Characteristics in 30 Newly Diagnosed and Newly Treated HNC Patients |
tRFs Were Closely Related to the Occurrence and Metastasis of Hypopharyngeal Cancer
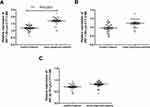
In order to investigate the relationship between tRFs and the occurrence of hypopharyngeal cancer, we plotted the ROC curve and evaluated the diagnostic value of tRFs in hypopharyngeal cancer. The results showed that tRF-1:30-Lys-CTT-1-M2 had a high diagnostic value in hypopharyngeal cancer (AUR=0.9172, P<0.0001. See Figure 6A). In contrast, the diagnostic value of tRF-1:30-Lys-CTT-2-M2 and tRF-60:76-Lys-CTT-1-M4 was relatively lower (See Figure 6B and C). The correlation between tRF-1:30-Lys-CTT-1-M2 expression and the lung metastasis of hypopharyngeal cancer was also analyzed. The results are shown in Figure 6D. As compared with the non-lung metastatic patients, the lung metastatic patients presented with a significant upregulation of tRF-1:30-Lys-CTT-1-M2. Finally, we analyzed the correlation between the tRF-1:30-Lys-CTT-1-M2 expression and the clinicopathological features of the lung metastatic patients. The results are presented in Table 4. It was found that stage III–IV, poor differentiation grade, drinking history, and smoking history were associated with a significant upregulation of tRF-1:30-Lys-CTT-1-M2 in the lung metastatic patients (P <0.05). However, the tRF-1:30-Lys-CTT-1-M2 expression was not significantly correlated with the patients’ age, gender, and site affected by cancer.
 |
Table 4 The Relationship Between the Expression Level of tRNA Fragments and the Clinicopathological Characteristics of 30 Patients with HNC Lung Metastasis |
Cox Regression Analysis of Risk Factors for the Metastasis of Hypopharyngeal Cancer
Univariate analysis and multivariate analysis were performed to determine the relationship between lung metastasis and clinicopathological features of hypopharyngeal cancer (Table 5). The results showed that stage III-IV, poor differentiation grade, drinking history, smoking history, and tRF-1:30-Lys-CTT-1-M2 overexpression were risk factors for the lung metastasis of hypopharyngeal cancer. According to the multivariate analysis, stage III-IV, drinking history, smoking history, and tRF-1:30-Lys-CTT-1-M2 overexpression were independent risk factors for the lung metastasis of hypopharyngeal cancer.
 |
Table 5 Univariate and Multivariate Cox Regression Analysis of Risk Factors for Patients with HNC Metastasis |
Discussion
Hypopharyngeal cancer most frequently occurs in the pyriform sinus, and very few cases are affected in the postcricoid region.14,15 Hypopharyngeal cancer is often found at a more advanced stage, where submucosal spread and neck lymph node metastasis are not uncommon.16 The five-year survival rate of hypopharyngeal cancer patients is only 20–40%.17 Therefore, Looking for ideal biomarkers for hypopharyngeal cancer has become a new research hotspot.18 In the present study, the expressions of extracellular vesicles tRFs in 30 treatment-naive non-lung metastatic patients and 30 lung metastatic hypopharyngeal cancer patients were analyzed. We found that tRF-1:30-Lys-CTT-1-M2 was significantly upregulated in the extracellular vesicles from the hypopharyngeal cancer patients. It could be used as a biomarker for the diagnosis and lung metastasis monitoring of hypopharyngeal cancer.
Extracellular vesicles can be extracted from a variety of cells (eg, endothelial cells and platelets) and body fluids (eg, blood and saliva).19,20 Extracellular vesicles contain a variety of substances, including lipids and different types of RNAs, such as miRNA, circRNA, lncRNA, and tRFs.21 It has been reported that miRNAs, circRNAs, and lncRNAs excreted by extracellular vesicles can be used as diagnostic or prognostic biomarkers for many tumors and other diseases.22,23 However, the use of extracellular vesicles-derived tRNAs as diagnostic biomarkers has been rarely discussed. We discovered a variety of tRFs in the extracellular vesicles from the hypopharyngeal cancer patients. Some tRFs were significantly upregulated in cancer patients as compared with the healthy subjects. These findings demonstrate the great potential of tRFs in cancer diagnosis.
tRFs are small non-coding RNAs derived from tRNA.9 tRFs are important components of the gene regulatory network and closely related to many diseases, including cancers, neurodegenerative disease, and inherited metabolic diseases.24 Some researchers have reported that tRFs are non-invasive diagnostic biomarkers for many diseases (especially cancers).25 In the present study, the extracellular vesicles- derived tRF-1:30-Lys-CTT-1-M2 was significantly upregulated in the treatment-naive non-lung metastatic patients as compared with the healthy subjects. ROC analysis also suggested that tRF-1:30-Lys-CTT-1-M2 had a high diagnostic value for hypopharyngeal cancer. These results proved that tRF-1:30-Lys-CTT-1-M2 was a candidate diagnostic marker for hypopharyngeal cancer. Our study also agreed with Wang, Kuang, and Zhao et al These researchers reported a high diagnostic value of tRFs in early breast cancer, lung cancer, and prostate cancer.26–28 We also found that the extracellular vesicles-derived tRF-1:30-Lys-CTT-1-M2 was significantly upregulated in the metastatic hypopharyngeal cancer patients as compared with the non-lung metastatic patients. That means tRF-1:30-Lys-CTT-1-M2 was able to differentiate between lung metastatic and non-lung metastatic hypopharyngeal cancer. If used as a biomarker, tRF-1:30-Lys-CTT-1-M2 has a bright prospect in the lung metastasis monitoring of hypopharyngeal cancer.
To conclude, we found that tRF-1:30-Lys-CTT-1-M2 was upregulated in treatment-naive non-lung metastatic hypopharyngeal cancer. More importantly, this tRF was more significantly upregulated in lung metastatic patients. Besides, tRF-1:30-Lys-CTT-1-M2 overexpression, tumor stage, smoking history and drinking history were independent risk factors for the lung metastasis of hypopharyngeal cancer. All of the above results demonstrated that tRFs might be closely associated with the occurrence and lung metastasis of hypopharyngeal cancer. Thus, tRF-1:30-Lys-CTT-1-M2 can be used as a novel biomarker for the diagnosis and lung metastasis monitoring of hypopharyngeal cancer. In addition, it was also found that tRFs are novel diagnostic and therapeutic targets for tumors.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Xiangya Hospital of Central South University. Informed consent was obtained from each subject in accordance with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no declarations of interest.
References
1. Takes RP, Strojan P, Silver CE, et al. Current trends in initial management of hypopharyngeal cancer: the declining use of open surgery. Head Neck. 2012;34(2):270–281. doi:10.1002/hed.21613
2. Hall SF, Groome PA, Irish J, et al. The natural history of patients with squamous cell carcinoma of the hypopharynx. Laryngoscope. 2010;118(8):1362–1371. doi:10.1097/MLG.0b013e318173dc4a
3. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
4. Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi:10.1016/j.semcdb.2015.02.010
5. Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83(11):1484–1494. doi:10.1016/j.bcp.2011.12.037
6. Zhu L, Qu XH, Sun YL, et al. Novel method for extracting exosomes of hepatocellular carcinoma cells. World J Gastroenterol. 2014;20(21):6651–6657. doi:10.3748/wjg.v20.i21.6651
7. Braicu C, Tomuleasa C, Monroig P, et al. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22(1):34–45. doi:10.1038/cdd.2014.130
8. Sun C, Fu Z, Wang S, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2017;4(14):16–25.
9. Lee YS, Shibata Y, Malhotra A, et al. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639–2649. doi:10.1101/gad.1837609
10. Huang B, Yang H, Cheng X, et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 2017;77(12):3194–3206. doi:10.1158/0008-5472.CAN-16-3146
11. Martens-Uzunova ES, Olvedy M, Jenster G. Beyond microRNA – novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 2013;340(2):201–211. doi:10.1016/j.canlet.2012.11.058
12. Mleczko AM, Celichowski P, Bkowska-Ywicka K. Ex-translational function of tRNAs and their fragments in cancer. Acta Biochim Pol. 2014;61(2):211–216. doi:10.18388/abp.2014_1888
13. Goodarzi H, Hoang CB, Zhang S, et al. Modulated expression of specific tRNAs drives gene expression and cancer pro-gression. Cell. 2016;165(6):1416–1427. doi:10.1016/j.cell.2016.05.046
14. Hamamoto T, Hayashi R, Miyazaki M, et al. A clinical study of post-cricoid carcinoma. Acta Otolaryngol. 2015;25(2):171–176.
15. Mura F, Bertino G, Occhini A, et al. Surgical treatment of hypopharyngeal cancer: a review of the literature and proposal for a decisional flow-chart. Acta Otorhinolaryngol Ital. 2013;33(5):299–306.
16. Jamali Z, Asl Aminabadi N, Attaran R, et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51(4):321–331. doi:10.1016/j.oraloncology.2015.01.008
17. Katsoulakis E, Riaz N, Hu M, et al. Hypopharyngeal squamous cell carcinoma: three-dimensional or Intensity-modulated radiotherapy? A single institution’s experience. Laryngoscope. 2016;126(3):620–626. doi:10.1002/lary.25509
18. Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2(11):698–704. doi:10.1016/S1470-2045(01)00560-5
19. van Niel G, Porto-Carreiro I, Simoes S, et al. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi:10.1093/jb/mvj128
20. Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi:10.3410/B3-15
21. Miranda KC, Bond DT, McKee M, et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78(2):191–199. doi:10.1038/ki.2010.106
22. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
23. Szabo G, Momen-Heravi F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2017;14(8):455–466. doi:10.1038/nrgastro.2017.71
24. Yin KJ, Hamblin M, Chen YE. Non-coding RNAs in cerebral endothelial pathophysiology: emerging roles in stroke. Neurochem Int. 2014;77:9–16. doi:10.1016/j.neuint.2014.03.013
25. Olvedy M, Scaravilli M, Hoogstrate Y, et al. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7(17):24766–24777. doi:10.18632/oncotarget.8293
26. Wang JY, Ma G, Li MH, et al. Plasma tRNA fragments derived from 5′ ends as novel diagnostic biomarkers for early-stage breast cancer. Mol Ther Nucleic Acids. 2020;21:954–964. doi:10.1016/j.omtn.2020.07.026
27. Kuang MY, Zheng DF, Tao XT, et al. tRNA-based prognostic score in predicting survival outcomes of lung adenocarcinomas. Int J Cancer. 2019;145(7):1982–1990. doi:10.1002/ijc.32250
28. Zhao CM, Tolkach Y, Schmidt D, et al. tRNA-halves are prognostic biomarkers for patients with prostate cancer. Urol Oncol. 2018;36(11):
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.