Back to Journals » Infection and Drug Resistance » Volume 16
Epidemiological and Antimicrobial Resistant Patterns, and Molecular Mechanisms of Carbapenem-Resistant Klebsiella pneumoniae Infections in ICU Patients
Authors Lu F, Zhang L, Ji J, Xu Y , Wang B, Xia J
Received 1 March 2023
Accepted for publication 5 May 2023
Published 9 May 2023 Volume 2023:16 Pages 2813—2827
DOI https://doi.org/10.2147/IDR.S410657
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Fanbo Lu,1,* Luwen Zhang,1,* Juanjuan Ji,2 Yuanhong Xu,1 Bo Wang,1 Jinxing Xia1
1Department of Clinical Laboratory, First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 2Department of Respiratory and Critical Care Medicine, First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinxing Xia; Bo Wang, Department of Clinical Laboratory, First Affiliated Hospital of Anhui Medical University, No. 218 Jixi Road, Hefei, Anhui, People’s Republic of China, Email [email protected]; [email protected]
Objective: To study the epidemiological and antimicrobial resistant patterns, clinical characteristics and risk factors of critically ill patients infected with carbapenem-resistant Klebsiella pneumoniae (CRKP) from intensive care units (ICUs). The potential molecular mechanisms of antimicrobial resistance and virulence of CRKP were investigated through evaluation of associated genes.
Methods: Totally, 201 ICU patients infected with K. pneumoniae were recruited from January 2020 through January 2021. K. pneumoniae strains were collected from diverse clinical specimens and identified by microbial cultures and matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. Antimicrobial resistance was measured through broth micro-dilution or Kirby–Bauer assays. The carbapenemase-, virulence-, and capsular serotype-associated genes of CRKP were individually detected by PCR and sequencing. Demographic and clinical profiles were acquired from hospital databases to evaluate the correlation of CRKP infection incidence with clinical risk factors.
Results: Of the 201 K. pneumoniae strains, CRKP accounted for 41.29%. Seasonal bias existed in local prevalence of CRKP infections. CRKP strains mounted significantly strong resistance against major antimicrobial agents except ceftazidime–avibactam, tigecycline and minocycline. Recent exposure to certain antibiotics and prior treatment with invasive interventions were prone to increase CRKP infection risks with worsened infectious outcomes. The local top carbapenemase-encoding and virulence-associated genes of CRKP were blaKPC and irp 2, respectively. Nearly half of CRKP isolates harbored a capsular polysaccharide serotype of K14.K64 (wzi-64) which preferentially emerged in the cohort with worse outcomes of infection.
Conclusion: Featured epidemiology and typical clinical characteristics existed extensively in K. pneumoniae infections among ICU patients. The CRKP cohort exhibited substantially high antimicrobial resistance. Distinctive carbapenemase-, virulence-, and serotype-associated genes were intensively involved in the spread and pathogenesis of CRKP. These findings supported careful management of critically ill patients potentially infected with virulent CRKP in the ICUs.
Keywords: carbapenem-resistantKlebsiella pneumoniae, epidemiology, clinical features, antimicrobial resistance, virulence, ICU
Introduction
Klebsiella pneumoniae is classified as a common human- and animal-associated Gram-negative bacterium in the environment and has become a major cause of hospital-acquired infections worldwide.1 K. pneumoniae belongs to the commensal bacterial family with the potential to engage in a wide range of infectious conditions including soft tissue, wound, and respiratory infections, especially in patients with compromised immune systems.2 Numerous studies have reported K. pneumoniae infection frequencies can vary geographically and demographically, with a prevalence rate ranging from 1.30% to 16.00%.3–5 In China, K. pneumoniae was one of the three leading causative agents in patients with bacterium-associated acute respiratory infections between 2009 and 2019.6 The critically ill patients in intensive care units (ICUs) are at a high risk for K. pneumoniae infections, mainly due to impaired immune functions, severe underlying diseases, hospitalization time, and/or multiple invasive therapeutic interventions.
Carbapenems remain one of the last-line therapeutic drugs against K. pneumoniae in gravely ill patients. Though a majority of the patients were routinely infected with carbapenem-sensitive K. pneumoniae (CSKP), the emergence of carbapenem-resistant K. pneumoniae (CRKP) strains has been increasingly reported.7 Plasmids encoding carbapenemase can be transferred among bacteria by horizontal gene transfer and readily disseminated in hospitals. Notably, recent research groups have confirmed carbapenem resistance was partly attributed to increased length of hospital stay and led to an elevated mortality in patients.8 Therefore, the carbapenemase-mediated resistance poses a serious public health threat.9
K. pneumoniae pathogenicity is also associated with certain virulence factors that help it to evade host immune surveillance, including capsules, lipopolysaccharides, exopolysaccharides, adhesins, and iron uptake systems.10 Interestingly, continuous evolution of hypervirulence plasmids gives rise to a pathotype of hypervirulent K. pneumoniae. This type of K. pneumoniae has been increasingly reported in association with plasmid-mediated pathogenicity loci of iuc, iro, rmpA/rmpA2, etc.11,12 Of them, the iucABCD and iutA, and iroBCDN are involved in the biosynthesis of aerobactin, and salmochelin, respectively.13–15 While, the rmpA/rmpA2-encoded proteins regulate capsule productions in K. pneumoniae.16 In China, K. pneumoniae carrying virulence plasmids is often subject to excess morbidity and mortality.17,18 The presence and spread of hypervirulent K. pneumoniae is of great concern due to its virulence and lack of therapeutic options. In addition, multidrug resistance has been increased all over the world that is also considered a public health threat. Several recent investigations reported the emergence of virulent multidrug-resistant (MDR) bacterial pathogens from different origins that increase the necessity of the proper use of antibiotics as well as the application of rapid accurate diagnostic tools for screening the emerging virulent MDR strains including K. pneumoniae.19–24
In this study, we comprehensively investigated the epidemiology and drug sensitivities of CRKP and CSKP from ICU patients, and their clinical characteristics, multiple risk factors in combination with corresponding clinical data. In addition, typically important carbapenem-resistance genes, virulence-linked genes, as well as the wzi alleles that are conserved and used to predict capsular types of K. pneumoniae, were sequenced to explore the potential molecular mechanisms of CRKP.
Materials and Methods
Study Design, Bacterial Isolates and Patient Data Collection
The present study collected 201 cases infected with K. pneumoniae from the ICUs in the First Affiliated Hospital of Anhui Medical University, a Class A tertiary comprehensive hospital committed to delivering medical education and offering nearly 3000 beds for Anhui province in China, from January 2020 through January 2021. In cases with multiple detections of K. pneumoniae, the very first positive cultures we obtained were valid in this study. The relevant clinical data were accessed through the electronic medical database of the hospital. The information of all the cases generally included fundamental demographics, hospital stay length, concomitant diseases, invasive procedures or treatments, clinical medications and outcomes, as well as laboratory testing parameters.
Bacterial Isolation and Identification, and Antibiotic Susceptibility Testing
K. pneumoniae isolates were aseptically isolated and then identified to species level from blood, ascites, drainage fluid, cerebrospinal fluid, lower respiratory tract secretions and other materials of inpatients through an automatic microbial identification platform (BacT/AlerT 3D, BioMérieux, Marcy l’Etoile, France), blood agar cultures, and matrix-associated laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS, Vitek MS, BioMérieux, Marcy l’Etoile, France) within the hospital clinical laboratory according to the manufacturers’ instructions.
Antimicrobial susceptibility testing of the K. pneumoniae isolates to common clinically used antibiotics was performed via the broth micro-dilution method using an automated Vitek 2 Compact system with Advanced Expert System (BioMérieux, Marcy l’Etoile, France) or Phoenix 100 system (Becton Dickinson, Sparks, MD, USA), or by the Kirby–Bauer (Oxoid, Basingstoke, UK) approach according to the Clinical and Laboratory Standards Institute (CLSI) unified protocol.25 Specifically, the tested antibiotics included penicillins (ampicillin), β-lactam combination agents (ampicillin–sulbactam, piperacillin–tazobactam, cefoperazone–sulbactam, and ceftazidime–avibactam), cephalosporins (cefazolin, ceftazidime, ceftriaxone, cefotaxime, cefuroxime, and cefepime), cephamycins (cefotetan, cefmetazole), monobactams (aztreonam), carbapenems (ertapenem, imipenem, and meropenem), aminoglycosides (amikacin, gentamicin, and tobramycin), fluoroquinolones (ciprofloxacin, levofloxacin), tetracyclines (tetracycline, minocycline), and folate pathway antagonists (trimethoprim–sulfamethoxazole). Escherichia coli ATCC25922 and K. pneumoniae ATCC700603 were used as quality controls as previously described.26
Determination of Carbapenemase-Encoding Genes, Virulence-Associated Genes, and Capsular Polysaccharide Serotype-Associated Genes
Carbapenemase-encoding genes including blaKPC, blaNDM, blaOXA-23, blaOXA-24, blaOXA-48, blaOXA-50, blaOXA-55, blaOXA-58, blaOXA-60, blaOXA-69, blaVIM1, blaVIM2, blaIMP1, blaIMP2, blaSME, blaGES, blaGIM1, blaIMI, blaNMC, blaSIM1 and blaSPM1, and virulence-associated genes including iucA, iroB, irp2, terB, peg344, peg589, peg1631, rmpA, rmpA2 and crmpA were determined by PCR and sequencing assays.27,28 The PCR primers were commercially synthesized by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China), and the assays were performed in a DNA analyzer (Applied Biosystems 7500, Thermo Fisher Scientific, Waltham, MA, USA) with the PCR conditions as follows: (a) 5 minutes at 94 °C for initial denaturation, (b) 32 cycles of 30s at 94 °C for denaturation, 90s at a given temperature for annealing, and a given time period at 72 °C for extension), and (c) 10 minutes at 72 °C for last extension (Table 1). The resultant PCR products were confirmed by DNA agarose gel electrophoresis and quantified through a spectrophotometer (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA, USA), and then the typical carbapenem-resistant gene fragments underwent the process of sequencing service commercially provided by Sangon Biotech Co., Ltd. (Shanghai, China).
 |
Table 1 PCR Primer and Amplification Condition Information |
The capsular polysaccharide serotypes of CRKP strains were identified through wzi gene by PCR with the specific primers commercially available (Shanghai Sangon Biotech Co., Ltd., Shanghai, China) (Table 1), and followed by sequencing of the PCR products for wzi alleles, as previously described.29 The corresponding sequencing data then went through assignment analysis using an online database (http://bigsdb.pasteur.fr/klebsiella).
Statistical Analysis
The IBM Statistical Package for the Social Sciences (SPSS) (version 25.0, SPSS Inc., Chicago, IL, USA) and GraphPad Prism (version 5.0, GraphPad Software Inc., San Diego, CA, USA) were adopted to process statistically all the data collected. The χ2 or Fisher’s exact tests were applicable to the analysis regarding categorical variables when appropriate. The Student’s t-test was available for data with normal distribution, otherwise, Mann–Whitney U-test was applied. A P value of < 0.05 was regarded as statistical significance.
Results
Bacterial Identification and Distribution of K. pneumoniae
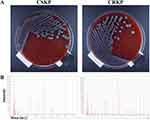
Throughout the entire study period, 201 of K. pneumoniae isolates from ICU patients were collected and identified by microbial cultures for macroscopic/microscopic examinations and MALDI-TOF MS (Figure 1). To be specific, 83 (41.29%) of these isolates were CRKP, and the remaining ones (n = 118, 58.71%) were determined to be CSKP. All the specimens involved were from diverse source origins. Lower respiratory tract secretions (n = 141, 70.15%) accounted for the majority of source types, followed by blood (n = 40, 19.90%), ascites (n = 5, 2.49%), drainage fluid (n = 3, 1.49%) and others (n = 12, 5.97%). Upon comparison of the isolation rates during the whole year, the data showed that CRKP was relatively more prevalent during the seasons with cold/hot weather switching, eg, spring (esp., March) and autumn (esp., October); while CSKP dominated in the entire winter (P < 0.01, Figure 2).
Clinical Features with K. pneumoniae Infections
The demographic and clinical profiles of patients infected with CSKP and CRKP are described in Table 2. Of these 201 patients, their average age was 58.25 ± 17.61 years, males outnumbered females by approximately 2.5-fold. The average time of hospital stay after CSKP and CRKP infections was 20.10 ± 23.35, and 24.41 ± 26.27 days, respectively. The predominant concomitant diseases in all the infected patients were primarily involved with cerebral diseases (59.70%), renal diseases (34.83%), hypertension (32.34%), cardiac diseases (31.34%) and others, among which renal diseases in ICU patients infected with CRKP were likely to concomitantly occur more often in comparison to those with CSKP in the present study.
 |
Table 2 Characteristics of the Critically Ill Patients Infected with K. pneumoniae |
As to such various risk factors related to CRKP infections, prior treatments/processes of urinary catheter, gastric cannula, blood product transfusion, mechanical ventilation, peripheral arterial catheter, central venous catheter, thoracic drainage, continuous renal replacement therapy (CRRT), as well as recent prior exposure to certain antibiotics of glycopeptides, carbapenems, quinolones, aminoglycosides, were found statistically different between CRKP and CSKP infection groups (P < 0.05). Whereas, the remaining risk factor variables did not make significant differences within both groups, mainly including most of the comorbidities, surgery, fiberoptic bronchoscopy, extracorporeal membrane oxygenation (ECMO), hemodialysis and plasma exchange, as well as prior administration of β-lactam/β-lactamase inhibitor, cephalosporins, linezolid, and antifungal agents. Furthermore, the clinical outcomes of CRKP-infected patients were worsened when compared to those infected with CSKP (P = 0.000), in terms of infection persistence (Table 2).
We next summarized and evaluated the laboratory testing parameters of the ICU patients infected with K. pneumoniae. As listed in Table 3, on average, white blood cell count (WBC, mainly neutrophils), prothrombin time (PT), activated partial thromboplastin time (APTT) and fibrinogen (FIB) levels in the entire infected population at admission were all beyond normal ranges, whereas, their mean concentrations of hemoglobin (Hb), hematocrit (Hct), total protein (TP, esp., albumin or ALB) were lower than normal levels. In comparison of CSKP and CRKP groups, we found significant differences in terms of red blood cell count (RBC), Hb and Hct (P < 0.001). We also observed a statistically significant difference in APTT (P < 0.05) rather than other coagulation-related indicators (eg, platelet or PLT, PT and FIB). The remaining laboratory parameters tested were not statistically different between the two groups.
 |
Table 3 Summary of Blood Test Data of the ICU Patients Infected with K. pneumoniae |
Antimicrobial Resistance Patterns of K. pneumoniae
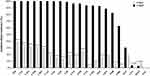
After classifying all the K. pneumoniae isolates into CRKP and CSKP groups, the overall results of in vitro susceptibility tests for these isolates were comprehensively analyzed. As shown in Figure 3, generally, CRKP from the ICU patients exhibited significantly lowered levels or even no any of susceptibility to all the tested antimicrobial agents except ceftazidime–avibactam (CZA), tigecycline and minocycline. Specifically, the data pointed out that the CRKP cohort presented remarkably higher levels of resistance against multiple antibiotics, particularly including the third generation of cephalosporin, carbapenem, the β-lactam/β-lactamase inhibitor combination, ampicillin, ciprofloxacin, gentamicin, amikacin, trimethoprim/sulfamethoxazole, when compared to the group of CSKP (P < 0.001). To be noted, both CRKP and CSKP showed comparable and nearly excellent in vitro susceptibility to CZA, tigecycline and minocycline.
Carbapenemase-Encoding Genes, Virulence-Associated Genes and Capsular Polysaccharide Serotypes Among CRKP
Twenty-one carbapenemase-encoding genes were selected for identification of the potential molecular mechanisms of the CRKP-infected cases. Among the 79 CRKP isolates that were successfully recovered from cryopreserved strains for subcultures, 77 isolates (97.47%) were positive for blaKPC, 6 (7.59%) for blaOXA-23, and 5 (6.33%) for blaNDM, and the remaining carbapenemase-encoding genes tested were absent in all the CRKP strains of this study. The typical carbapenemase-encoding gene fragments from Klebsiella pneumoniae strains were sequenced for validation of PCR products (data not shown). Notably, to our knowledge, the vast majority of the blaKPC-harboring CRKP strains (n = 69, 87.34%) simply carried blaKPC, while there were two strains (2.53%) of CRKP expressing blaNDM alone, in the present study. Whereas, there existed eight strains (10.13%) of CRKP possessing a combination of the carbapenemase-encoding genes for which the combined pattern of blaKPC and blaOXA-23 was the predominant (6.33%), followed by the pattern of blaKPC and blaNDM (2.53%), and the pattern of blaKPC, blaOXA-23 and blaNDM (1.27%). In comparison of the frequencies of carbapenemase-encoding genes in CRKP, there were no statistically significant differences among the three cohorts with different infection outcomes, despite the fact that nearly all the blaOXA-23 and blaNDM genes were barely found in the cohort with worse outcome of infection (Figure 4).
As for the virulence-associated genes presented in CRKP of the local area, the dominant candidates were irp2 (n = 77, 97.47%), rmpA2 (n = 57, 72.15%), peg589 (n = 55, 69.62%), iucA (n = 54, 68.35%), terB (n = 54, 68.35%) and peg1631 (n = 49, 62.03%). The presence of the other genes were all below 50.00% in this study. Likewise, no significant differences were observed among the three cohorts with different outcomes of infection when comparing the frequencies of CRKP virulence-associated genes, though terB and peg1631 exhibited considerably higher percentages in the cohort with worse outcome of infection than those in the other two cohorts (Table 4). Additionally, the capsular polysaccharide serotypes were further assessed for CRKP strains through PCR and sequencing of the wzi allele genes. Within these 79 CRKP isolates from the local region, three major capsular polysaccharide serotypes were identified as K14.K64 (wzi-64), KL47 (wzi-209) and K19 (wzi-19), accounting for 44.30% (n = 35), 27.85% (n = 22) and 16.46% (n = 13), respectively. The remaining serotypes involved were all less than 4.00% in proportion. Moreover, few significant differences were found in cohorts with different infection outcomes in comparison of the incidence of CRKP capsular polysaccharide serotypes, except the serotype of K28 (wzi-84) with P < 0.05. Interestingly, the KL47 (wzi-209) serotype appeared to prevail in the cohort with better outcome of infection, whereas the K14.K64 (wzi-64) dominated the cohort with worse one (Table 5). In addition, the overall data were summarized together to reveal potential correlation characteristics among the carbapenemase-encoding genes, polysaccharide serotyping (wzi allele) and virulence-associated genes from all the recovered CRKP strains, and meanwhile their MDR features were individually determined (>50.00% of CRKP strains were MDR) according to the criteria previously described (Supplementary Table S1).30
 |
Table 4 Virulence-Associated Genes in CRKP with Different Clinical Outcomes |
 |
Table 5 Capsular Polysaccharide Serotypes of CRKP with Different Clinical Outcomes |
Discussion
CRKP is considered as a public health threat worldwide as it is one of the leading causes of death among patients with hospital-acquired infections. The ICUs have been described as a factory for creating, disseminating, and amplifying antimicrobial resistance due to their extremely vulnerable population of critically ill patients, heavy use of invasive procedures, and frequent application of antimicrobials. Thus, regularly monitoring of CRKP would facilitate the choice and efficacy of empirical therapy, especially in the ICU settings.31 The present study assessed the local distribution features, antimicrobial resistance pattern, clinical risk factors, prevalence of the carbapenem-resistant genes, virulence-linked variant genes and capsular polysaccharide serotype-associated genes of CRKP strains isolated from clinical specimens of ICU patients in one of the largest tertiary comprehensive teaching hospitals in Central China. This study demonstrated a high prevalence of CRKP that carried a variety of carbapenem-resistant genes, virulence-linked variant genes and capsular polysaccharide serotypes.
CRKP infections threaten ICU patients because of the ability to contaminate environmental surfaces including hospital wards, increasing its transmission potentials among the environment, patients and medical staff.32 Previous researchers reported that the incidence of CRKP infections was connected with the switching of cold/hot seasons.33 Here we observed a relatively higher local prevalence of CRKP in spring and autumn. Although the underlying links between the seasons and the CRKP infection incidence remain unclear, understanding their potential relationship would contribute to the prophylaxis of such infections in future. Studies revealed that undergoing invasive procedures would increase such infection risks.34 Intubation or tracheotomy might disrupt normal body barriers and facilitate contact between the interior of body and the external environment, leading to easy invasion and attachment of opportunistic pathogens to the intubation lining, creating a biofilm cover that would be difficult to eradicate. As a result, pathogenic bacteria might enter deeper tissues of host and increase the chance of CRKP infections.35–37 Our ICU patients with multiple clinical invasive interventions had significant risks for CRKP infections. Moreover, prior treatment with certain antimicrobials substantially increased the emergence of CRKP. Some investigators suggested that frequent exposure to one antimicrobial class would augment the impact of exposure to another antimicrobial class on the CRKP infection risks.38,39 Hence, antimicrobial combination therapy and long-term high-dose treatment with carbapenems likely increase antimicrobial selection pressure and allow carbapenem-resistant bacteria to further develop novel resistance mechanisms. It is worth noting that MDR bacteria have been increasingly emerging worldwide.20 Our study indicated over half of the CRKP population with MDR phenotype. The control and prevention of MDR pathogens, particularly including MDR K. pneumoniae strains from the ICU settings, urgently need rational utilization of antimicrobials and rapid screening strategies.19–24 Based on our laboratory data, we speculated that CRKP-infected ICU patients were more likely accompanied by symptoms of anemia and bleeding, due to relatively lowered RBC/Hb/Hct and extended APTT, which might aggravate the severity of diseases and K. pneumoniae infections.
K. pneumoniae is able to resist antimicrobial actions through multiple mechanisms including alteration of targets, inactivation of drugs, enhanced activity of efflux pumps, and attenuated permeability of cells.19 K. pneumoniae resistance to carbapenems is mainly attributable to the presence of carbapenemases, often assisted by other resistance pathways such as extended-spectrum β-lactamases (ESBLs).40 Our findings illustrated that CRKP in ICU patients exhibited significantly lower levels of susceptibility to all antibiotics except CZA, tigecycline and minocycline. It reported that administration of tigecycline in patients with CRKP infections contributed to rapid development of in vitro resistance.41 The novel FDA-approved CZA has been verified to mount a good activity against CRKP, especially against the KPC-producing, but not NDM-producing CRKP.42,43 We also noticed that merely 3.45% of CRKP were resistant to CZA. Therefore, to prevent the development of new CZA-resistant strains in the local area of China, identification of resistance mechanisms ought to be an essential prerequisite for development of a rational antibiotic regimen.
Recent CRKP prevalence rates of Chinese major provinces ranged from 0.80% to 28.10%, with an average rate of 11.30%, according to China Antimicrobial Resistance Surveillance System (CARSS, http://www.carss.cn), and the dominant carbapenem-resistant genotype of CRKP was blaKPC, accounting for approximately 65.00%.44 Based on the findings in the present study, the local incidence of CRKP infections among the ICU patients was higher than the average level of China, which should draw special attention for our clinicians to focusing on the current situation. With the first KPC-producing CRKP isolate emerging in Hangzhou City, KPC-type enzymes have constantly been the predominant carbapenemases of CRKP in China.45,46 Similarly, such type of enzymes were the most prevalent amidst our tested CRKP isolates, followed by OXA-23 and NDM. Interestingly, eight of these isolates were carrying combinations of blaKPC, blaOXA-23 and/or blaNDM genes. Further studies on the combined carbapenemase-associated genes might help to understand the evolution of CRKP. Previous studies reported several defining factors in pathogenic agents for virulence and toxicity, mainly including capsules, pili and siderophores, which fundamentally affect both classic and virulent strains of K. pneumoniae causing severe bacteremia, pneumonia and even meningitis in vulnerable population with compromised immunity such as the ICU patients.10,47,48 Acquisition of virulence-associated genes in CRKP is one of the main strategies to produce highly virulent CRKP.49 It is proposed that MDR clones would be more likely to subsequently acquire virulence genes than highly pathogenic clones that, however, would be prone to get access to resistance genes, suggesting that CRKP strains with pre-existing resistance genes might take an advantage of subsequent acquisition of virulence-associated genes, when compared to CSKP strains.50 In the present study, nearly all of CRKP strains from ICU patients possessed a variety of virulence-associated genes tested, indicating that these gene members might be ubiquitously spread among CRKP isolates in the local region. Finally, according to previous epidemiology studies, the capsular polysaccharide serotype of K14.K64 (wzi-64) in K. pneumoniae was identified as a common type within CRKP strains in China as well as other countries.51,52 Here, our observation pointed out that such serotype of K14.K64 (wzi-64) was certainly the main type for local CRKP strains. In addition, the serotypes of KL47 (wzi-209) and K19 (wzi-19) also occupied relatively high proportions in the local CRKP population. Further investigations will be needed to consolidate the understanding of potential connections between the capsular polysaccharide serotypes and treatment of K. pneumonia infections.
Conclusion
The current study reported epidemiological and antimicrobial resistance patterns, clinical characteristics and risk factors of CRKP in critically ill patients. Importantly, carbapenemase-encoding genes, virulence- and capsular polysaccharide serotype-associated genes of CRKP were well documented and evaluated. Our data revealed seasonal bias in the prevalence of CRKP infections for ICU patients. The CRKP strains showed significantly attenuated susceptibility to most of commonly used antimicrobials and were subject to MDR phenotype. Recent exposure to certain antibiotics and prior treatment with invasive interventions were prone to increase CRKP infection risk with worsened infectious outcomes. In the local area, the top three carbapenemase-encoding genes of CRKP were blaKPC, blaOXA-23 and blaNDM. The virulence-associated gene irp2 dominated the regional CRKP population. Moreover, nearly half of the CRKP isolates presented the capsular polysaccharide serotype of K14.K64 (wzi-64) which preferentially emerged in the cohort with worse infectious outcome at the bedside. In short, our findings supported the strict policies for antibiotic administration, cautious implementation of invasive procedures, and careful management of critically ill patients infected with virulent CRKP in the ICUs.
Abbreviations
K. pneumoniae, Klebsiella pneumoniae; ICU, intensive care unit; CSKP, carbapenem-sensitive Klebsiella pneumoniae; CRKP, carbapenem-resistant Klebsiella pneumoniae; MDR, multidrug-resistant; MALDI-TOF MS, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry; CLSI, Clinical and Laboratory Standards Institute; PCR, polymerase chain reaction; CRRT, continuous renal replacement therapy; ECMO, extracorporeal membrane oxygenation; WBC, white blood cell; PT, prothrombin time; APTT, activated partial thromboplastin time; FIB, fibrinogen; Hb, hemoglobin; Hct, hematocrit; TP, total protein; ALB, albumin; RBC, red blood cell; PLT, platelet; CZA, ceftazidime–avibactam; ESBL, extended-spectrum β-lactamase; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillinase.
Ethics Approval
The study protocol was approved by the Ethics Committee of Clinical Medicine Research of the First Affiliated Hospital of Anhui Medical University. All the subjects provided written informed consents according to the Helsinki statement.
Author Contributions
Fanbo Lu and Luwen Zhang share first authorship. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (21604079) (JX), and the University Scientific Research Collaborative Key Project of Anhui Province (2022AH040162) (YX).
Disclosure
All the authors declared no competing interests for this work.
References
1. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
2. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi:10.1128/CMR.11.4.589
3. Chang D, Sharma L, Dela Cruz CS, Zhang D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front Microbiol. 2021;12:750662. doi:10.3389/fmicb.2021.750662
4. Martin RM, Cao J, Brisse S, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella Pneumoniae. MSphere. 2016;1(5):e00261–16.
5. Gorrie CL, Mirceta M, Wick RR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65(2):208–215. doi:10.1093/cid/cix270
6. Li ZJ, Zhang HY, Ren LL, et al. Etiological and epidemiological features of acute respiratory infections in China. Nat Commun. 2021;12(1):5026. doi:10.1038/s41467-021-25120-6
7. Tängdén T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med. 2015;277(5):501–512. doi:10.1111/joim.12342
8. Schwaber MJ, Klarfeld-Lidji S, Navon-Venezia S, Schwartz D, Leavitt A, Carmeli Y. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother. 2008;52(3):1028–1033. doi:10.1128/AAC.01020-07
9. Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707. doi:10.1128/CMR.05035-11
10. Riwu KHP, Effendi MH, Rantam FA, Khairullah AR, Widodo A. A review: virulence factors of Klebsiella pneumonia as emerging infection on the food chain. Vet World. 2022;15(9):2172–2179. doi:10.14202/vetworld.2022.2172-2179
11. Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi:10.1016/j.gene.2004.05.008
12. Lam MMC, Wyres KL, Duchêne S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. doi:10.1038/s41467-018-05114-7
13. de Lorenzo V, Martinez JL. Aerobactin production as a virulence factor: a reevaluation. Eur J Clin Microbiol Infect Dis. 1988;7(5):621–629. doi:10.1007/BF01964239
14. Russo TA, Gulick AM. Aerobactin synthesis proteins as antivirulence targets in hypervirulent Klebsiella pneumoniae. ACS Infect Dis. 2019;5(7):1052–1054. doi:10.1021/acsinfecdis.9b00117
15. Crouch MLV, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2008;67(5):971–983. doi:10.1111/j.1365-2958.2007.06089.x
16. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
17. Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin Infect Dis. 2014;58(2):225–232. doi:10.1093/cid/cit675
18. Chen Y, Chen Y. Clinical challenges with hypervirulent Klebsiella pneumoniae (hvKP) in China. J Transl Intern Med. 2021;9(2):71–75. doi:10.2478/jtim-2021-0004
19. Kareem SM, Al-Kadmy IMS, Kazaal SS, et al. Detection of gyrA and parC mutations and prevalence of plasmid-mediated quinolone resistance genes in Klebsiella pneumoniae. Infect Drug Resist. 2021;14:555–563. doi:10.2147/IDR.S275852
20. Algammal AM, Hetta HF, Mabrok M, Behzadi P. Editorial: emerging multidrug-resistant bacterial pathogens “superbugs”: a rising public health threat. Front Microbiol. 2023;14:1135614. doi:10.3389/fmicb.2023.1135614
21. Algammal AM, Abo Hashem ME, Alfifi KJ, et al. Sequence analysis, antibiogram profile, virulence and antibiotic resistance genes of XDR and MDR gallibacterium anatis isolated from layer chickens in Egypt. Infect Drug Resist. 2022;15:4321–4334. doi:10.2147/IDR.S377797
22. Algammal AM, Ibrahim RA, Alfifi KJ, et al. A first report of molecular typing, virulence traits, and phenotypic and genotypic resistance patterns of newly emerging XDR and MDR Aeromonas veronii in Mugil seheli. Pathogens. 2022;11(11):1262. doi:10.3390/pathogens11111262
23. Algammal AM, Hashem HR, Al-Otaibi AS, et al. Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 2021;21(1):237. doi:10.1186/s12866-021-02287-y
24. Algammal AM, Eidaroos NH, Alfifi KJ, et al. oprL gene sequencing, resistance patterns, virulence genes, quorum sensing and antibiotic resistance genes of XDR Pseudomonas aeruginosa isolated from broiler chickens. Infect Drug Resist. 2023;16:853–867. doi:10.2147/IDR.S401473
25. Weinstein Melvin P, Lewis James S, Clinical T. Laboratory Standards Institute Subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes. J Clin Microbiol. 2020;58(3):e01864–01819. doi:10.1128/JCM.01864-19
26. Zhang Y, Xu Y, Huang Y. Virulence genotype and correlation of clinical severeness with presence of the type VI secretion system in Klebsiella pneumoniae isolates causing bloodstream infections. Infect Drug Resist. 2022;15:1487–1497. doi:10.2147/IDR.S353858
27. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
28. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–18. doi:10.1128/JCM.00776-18
29. Liao W, Huang N, Zhang Y, et al. Comparison of carbapenem-resistant Klebsiella pneumoniae strains causing intestinal colonization and extraintestinal infections: clinical, virulence, and molecular epidemiological characteristics. Front Public Health. 2021;9:783124. doi:10.3389/fpubh.2021.783124
30. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
31. Brusselaers N, Vogelaers D, Blot S. The rising problem of antimicrobial resistance in the intensive care unit. Ann Intensive Care. 2011;1:47. doi:10.1186/2110-5820-1-47
32. Yan Z, Zhou Y, Du M, et al. Prospective investigation of carbapenem-resistant Klebsiella pneumonia transmission among the staff, environment and patients in five major intensive care units, Beijing. J Hosp Infect. 2019;101(2):150–157. doi:10.1016/j.jhin.2018.11.019
33. Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi:10.1080/22221751.2020.1799721
34. Li J, Li Y, Song N, Chen Y. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection: a meta-analysis. J Glob Antimicrob Resist. 2020;21:306–313. doi:10.1016/j.jgar.2019.09.006
35. Jiao Y, Qin Y, Liu J, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection/colonization and predictors of mortality: a retrospective study. Pathog Glob Health. 2015;109(2):68–74. doi:10.1179/2047773215Y.0000000004
36. Tuon FF, Rocha JL, Toledo P, et al. Risk factors for KPC-producing Klebsiella pneumoniae bacteremia. Braz J Infect Dis. 2012;16(5):416–419. doi:10.1016/j.bjid.2012.08.006
37. Li Y, Shen H, Zhu C, Yu Y. Carbapenem-resistant Klebsiella pneumoniae infections among ICU admission patients in central China: prevalence and prediction model. Biomed Res Int. 2019;2019:9767313. doi:10.1155/2019/9767313
38. Del Mar Tomas M, Cartelle M, Pertega S, et al. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin Microbiol Infect. 2005;11(7):540–546. doi:10.1111/j.1469-0691.2005.01184.x
39. Kritsotakis EI, Tsioutis C, Roumbelaki M, Christidou A, Gikas A. Antibiotic use and the risk of carbapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae infection in hospitalized patients: results of a double case-control study. J Antimicrob Chemother. 2011;66(6):1383–1391. doi:10.1093/jac/dkr116
40. Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi:10.1128/AAC.01019-15
41. van Duin D, Cober ED, Richter SS, et al. Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin Microbiol Infect. 2014;20(12):O1117–O1120. doi:10.1111/1469-0691.12714
42. Ehmann DE, Jahić H, Ross PL, et al. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A. 2012;109(29):11663–11668. doi:10.1073/pnas.1205073109
43. van Duin D. Bonomo. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis. 2016;63(2):234–241. doi:10.1093/cid/ciw243
44. Han R, Shi Q, Wu S, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
45. Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi:10.1016/j.jgar.2020.09.004
46. Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51(2):763–765. doi:10.1128/AAC.01053-06
47. Parrott AM, Shi J, Aaron J, Green DA, Whittier S, Wu F. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York City hospital through screening of virulence genes. Clin Microbiol Infect. 2021;27(4):583–589. doi:10.1016/j.cmi.2020.05.012
48. Meatherall BL, Gregson D, Ross T, Pitout JDD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med. 2009;122(9):866–873. doi:10.1016/j.amjmed.2009.03.034
49. Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. doi:10.1016/j.jgar.2021.02.020
50. Wyres KL, Wick RR, Judd LM, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15(4):e1008114. doi:10.1371/journal.pgen.1008114
51. Koh TH, Cao D, Shan QY, Bacon A, Hsu L-Y, Ooi EE. Acquired carbapenemases in Enterobactericeae in Singapore, 1996–2012. Pathology. 2013;45(6):600–603. doi:10.1097/PAT.0b013e3283650b1e
52. Zhou K, Xiao T, David S, et al. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi:10.3201/eid2602.190594
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.