Back to Journals » Infection and Drug Resistance » Volume 16
Emerging and Fastidious Uropathogens Were Detected by M-PCR with Similar Prevalence and Cell Density in Catheter and Midstream Voided Urine Indicating the Importance of These Microbes in Causing UTIs
Authors Wang D, Haley E , Luke N , Mathur M, Festa RA, Zhao X, Anderson LA, Allison JL, Stebbins KL, Diaz MJ, Baunoch D
Received 5 August 2023
Accepted for publication 29 November 2023
Published 22 December 2023 Volume 2023:16 Pages 7775—7795
DOI https://doi.org/10.2147/IDR.S429990
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Dakun Wang,1 Emery Haley,2 Natalie Luke,2 Mohit Mathur,3 Richard A Festa,3 Xinhua Zhao,4 Lori A Anderson,5 Jennifer L Allison,6 Kristen L Stebbins,6 Manuel Jose Diaz,6 David Baunoch7
1Department of Writing, Stat4Ward, Pittsburgh, PA, USA; 2Department of Clinical Research, Pathnostics, Irvine, CA, USA; 3Department of Medical Affairs, Pathnostics, Irvine, CA, USA; 4Department of Statistical Analysis, Stat4Ward, Pittsburgh, PA, USA; 5L. Anderson Diagnostic Market Access Consulting, San Diego, CA, USA; 6DispatchHealth, Denver, CO, USA; 7Pathnostics, Irvine, CA, 92618, USA
Correspondence: David Baunoch, Pathnostics, 15545 Sand Canyon Suite 100, Irvine, CA, 92618, USA, Tel +1 714-966-1221, Fax +1 714-966-1231, Email [email protected]
Introduction: This study compared microbial compositions of midstream and catheter urine specimens from patients with suspected complicated urinary tract infections to determine if emerging and fastidious uropathogens are infecting the bladder or are contaminants.
Methods: Urine was collected by in-and-out catheter (n = 1000) or midstream voiding (n = 1000) from 2000 adult patients (≥ 60 years of age) at 17 DispatchHealth sites across 11 states. The two groups were matched by age (mean 81 years), sex (62.1% female, 37.9% male), and ICD-10-CM codes. Microbial detection was performed with multiplex polymerase chain reaction (M-PCR) with a threshold for “positive detection” ≥ 10,000 cells/mL for bacteria or any detection for yeast. Results were divided by sex.
Results: In females, 28 of 30 microorganisms/groups were found by both collection methods, while in males 26 of 30 were found by both. There were significant overlaps in the detection and densities of classical uropathogens including Escherichia coli, Enterococcus faecalis, and Klebsiella pneumoniae, as well as emerging uropathogens including Actinotignum schaalii and Aerococcus urinae. In females, detection rates were slightly higher in midstream voided compared to catheter-collected (p = 0.0005) urine samples, while males showed the opposite trend (p < 0.0001). More polymicrobial infections were detected in midstream voided compared to catheter-collected samples (64.4% vs 45.7%, p < 0.0001) in females but the opposite in males (35.6% vs 47.0%, p = 0.002).
Discussion: In-and-out catheter-collected and midstream voided urine specimens shared significant similarities in microbial detections by M-PCR, with some differences found for a small subset of organisms and between sexes.
Conclusion: Non-invasive midstream voided collection of urine specimens for microbial detection and identification in cases of presumed UTI does not result in significantly more contamination compared to in-and-out catheter-collected specimens. Additionally, organisms long regarded as contaminants should be reconsidered as potential uropathogens.
Keywords: urinary tract infection, standard urine culture, diagnostic testing, multiplex polymerase chain reaction, catheter, midstream voided
Introduction
Standard urine culture (SUC), used in combination with microscopy and urinalysis, is the current standard-of-care for UTI diagnosis.1 It is optimized for the detection of classical uropathogens, such as uropathogenic Escherichia coli (E. coli or UPEC), Citrobacter koseri (C. koseri), Citrobacter freundii (C. freundii), Enterococcus faecalis (E. faecalis), Klebsiella pneumoniae (K. pneumoniae), Proteus mirabilis (P. mirabilis), Pseudomonas aeruginosa (P. aeruginosa), Staphylococcus aureus (S. aureus), and Streptococcus agalactiae (S. agalactiae).2,3 Advances in enhanced culture techniques and culture-independent molecular methods, such as multiplex polymerase chain reaction (M-PCR), have helped to debunk the misconception that the bladder is sterile and have led to the discovery of a urinary microbiome, including emerging uropathogens, such as Actinotignum schaalii (A. schaalii) and Alloscardovia omnicolens (A. omnicolens). These organisms have been reported to also play a role in the pathogenesis of UTI.4–10 Additionally, Viridans group streptococci (VGS) and coagulase-negative staphylococci (CoNS) isolated from midstream voided urine by SUC are frequently reported as non-pathogenic specimen contaminants.11 However, recent studies have shown that using enhanced urine culture protocols, investigators were able to frequently detect species of VGS and CoNS, including Streptococcus anginosus (S. anginosus) and Staphylococcus lugdunesis (S. lugdunesis).2 The organisms were detected at densities from 1000 to over 100,000 CFU/mL in catheter-collected urine specimens, and the density of these microorganisms increased in symptomatic subjects, indicating that these bacteria may be pathogenic for UTI.2
Recent studies have also demonstrated that polymicrobial infections are common and that M-PCR is more sensitive than SUC for the detection of polymicrobial infections and microbes other than E. coli.12–15 Additionally, the more sensitive M-PCR-based diagnostic test in combination with phenotypic Pooled Antibiotic Susceptibility Testing (P-AST) has been associated with improved outcomes, including fewer hospitalizations, for older UTI patients, particularly those with polymicrobial or non-E. coli infections.11,16 Clinical management of UTI patients according to M-PCR/P-AST testing compared to SUC testing has also been shown to result in healthcare cost savings.17,18
However, both culture-dependent and independent UTI diagnostic tests use urine as a sample source, resulting in questions about the potential for detection of non-pathogenic contaminant organisms. Urine specimens collected via the non-invasive midstream voided method are most common in busy clinical practices but are presumed to be most vulnerable to contamination. When such non-invasive midstream voided self-collection is not possible or appropriate, the more invasive in-and-out transurethral catheterization (catheter-collected) method is used. Suprapubic needle aspiration, inserting a needle through the skin directly into the bladder, is considered the most sterile urine collection technique but is rarely used due to its highly invasive nature19 and was not included in this study.
Clinical guidelines citing SUC diagnostic microbial thresholds are inconsistent and recommended microbial thresholds differ for each urine specimen collection method, reflecting the presumed differences in contamination potential.20–22 Published guidance on thresholds for midstream voided urine vary from ≥10,000 to ≥100,000 CFU/mL and from any microbial detection to ≥10,000 CFU/mL for urine collected by in-and-out catheter.22 Our recent data on midstream voided specimens support the clinical validity of the ≥ 10,000 CFU/mL threshold.23 We have also demonstrated that microbial densities reported as cells/mL by M-PCR are equivalent to CFU/mL reported by SUC.24
Studies comparing differences in microbial identification in midstream voided and catheter-collected urine specimens from adult patients with symptoms of UTI are sparse. This study used an advanced M-PCR assay to compare the prevalence and cell density of microbes detected in catheter-collected versus midstream voided urine specimens from adult patients (≥60 years) symptomatic for UTI. The aim was to determine whether there was truly an absence of organisms typically considered “contaminants” (eg, VGS and CoNS) in the catheter-collected urine, and whether the two collection methods showed general agreement in the types of organisms and microbial densities detected.
Materials and Methods
Study Design
The study analyses were performed on specimens from consecutive patients with symptoms suggestive of a UTI who received medical services from 17 DispatchHealth sites across 11 states (https://www.dispatchhealth.com/). DispatchHealth provides comprehensive and trusted medical care in the comfort of home for serious health concerns, treating complex conditions that are commonly addressed in the emergency room or that often require a hospital stay. Urine specimens were collected through either midstream voided or in-and-out catheterization (catheter-collected) at the ordering physician’s discretion. Overall, 5971 subjects were enrolled between 07/01/22 and 04/10/23. Subjects ≥60 years of age were selected as the target population for the M-PCR test in this study’s analyses because these patients are more likely to have multiple comorbidities and/or immune-compromised states resulting in complications and adverse outcomes from a UTI.24–26 The selection for age resulted in a total of 5350 subjects, including 1158 subjects in the catheter-collected group and 4162 subjects in the midstream voided group. ICD-10-CM code (https://www.icd10data.com) information from these subjects was recorded. The M-PCR test was performed on all urine specimens upon receipt, as described below.
Data from the study was collected via database for which the subject cannot be readily identified or contacted. Therefore, the Western Institutional Review Board deemed the use of the data to be exempt under 45 CFR § 46.104(d)(4) as the information is used in a manner that the identity of the subject cannot be readily ascertained directly or through identifiers linked to the subjects, the subject is not contacted, and the investigator will not re-identify subjects.
One thousand subjects from each urine collection group were matched for age, sex, and the 5 most prevalent ICD-10-CM codes resulting in 2000 patients included in the study. The analyses focused on the comparison of microbes detected between the catheter-collected (n = 1000) and the matched midstream voided (n = 1000) cohorts (Figure 1) overall and between female (n = 621) and male (n = 379) subjects within each group.
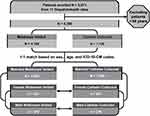 |
Figure 1 Study design. A flow chart of subject selection for analysis. |
Multiplex-Polymerase Chain Reaction (M-PCR) and Pooled Antibiotic Susceptibility Testing (P-AST)
The M-PCR/P-AST assay (Guidance® UTI, Pathnostics, Irvine, CA) provides susceptibility testing for 19 antibiotics, semi-quantification of 27 individual uropathogenic species and three bacterial groups, ESBL phenotype, and identification of 32 antibiotic-resistance genes. The assay was performed as described previously.25,26 Briefly, DNA was extracted from urine specimens with the KingFisher/MagMAX Automated DNA Extraction instrument and the MagMAX DNA Multi-Sample Ultra Kit (ThermoFisher, Carlsbad, CA). DNA specimens were vortexed with a universal PCR master mix and amplified with TaqMan technology on a Life Technologies 12K Flex Open Array System. DNA specimens were spotted in duplicate on 112-format Open Array chips. Plasmids for each organism in the panel were used as positive controls. Bacillus atrophaeus was used as an inhibition control. The microbial density of each organism was determined using the standard curve method. A proprietary bioinformatics tool developed by Pathnostics was used to report results.
Probes and primers were used for the following Classical uropathogens (see Supplementary Table 4): Candida albicans, Candida glabrata, Candida parapsilosis, Citrobacter freundii, Citrobacter koseri, Enterococcus faecalis, Enterococcus faecium, Escherichia coli, Klebsiella oxytoca, Klebsiella pneumoniae, Morganella morganii, Pantoea agglomerans, Proteus mirabilis, Providencia stuartii, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, Streptococcus agalactiae, and Enterobacter group [including Klebsiella aerogenes (formerly known as Enterobacter aerogenes) and Enterobacter cloacae].
Emerging uropathogens (see Supplementary Table 4): Acinetobacter baumannii, Actinotignum schaalii, Aerococcus urinae, Alloscardovia omnicolens, Candida auris, Corynebacterium riegelii, Gardnerella vaginalis, Mycoplasma hominis, Ureaplasma urealyticum, coagulase negative staphylococci group (CoNS) (including Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus lugdunensis, and Staphylococcus saprophyticus), and Viridans group streptococci (VGS) (including Streptococcus anginosus, Streptococcus oralis, and Streptococcus pasteurianus).
Results of the P-AST portion of the test, an antibiotic resistance and sensitivity assay which accounts for bacterial interactions, were not included in this analysis.
Statistical Analyses
Descriptive statistics were used to describe the patient demographics of age, sex, and ICD-10-CM codes, which were summarized for catheter-collected and midstream voided specimens and stratified by sex. The Wilcoxon rank sum test was used to compare microbial density distribution, and the median was used to compare microbial densities between the two collection-type groups.
Positive detection was defined as ≥ 10,000 cells/mL for bacteria, or any detection level for yeast in both types of urine specimens. The overall microbial positive detection rate, polymicrobial and monomicrobial positive detection rates, and positive detection rates for each individual organism were summarized and compared between the catheter-collected and midstream voided groups using the Chi-square test. Statistical comparisons for each collection method were also compared between male and female subjects. Based on the varying threshold guidelines, another definition of positive detection, ≥ 1000 cells/mL bacteria in catheter-collected urine, ≥ 10,000 cells/mL bacteria in midstream voided urine, or any detection of yeast in both types of urine specimens was also explored (Supplementary Data; Tables S1-S3). Monomicrobial positive detection was defined as only 1 microbe or microbe group identified over the threshold value and polymicrobial positive detection was defined as 2 or more microbes or microbe groups identified over the threshold criterion. P values < 0.05 from all statistical tests were considered statistically significant.
Results
Patient Demographics
Due to cohort matching, the two groups shared similar average age (81 years), ICD-10-CM codes, and sex distribution, with 621 female subjects (62.1%) and 379 male subjects (37.9%) in each group. All study subjects had a suspected diagnosis of UTI, with the majority (657, 65.7%) associated with an ICD-10-CM code of N39.0 (Urinary tract infection, site not specified) in each group (Table 1). The other four most prevalent ICD-10-CM codes included R30.0 (Dysuria), R35.0 (Frequency of micturition), R82.998 (Other abnormal findings in urine), and B99.9 (Unspecified infectious disease) with these top 5 ICD-10-CM codes accounting for over 70% of the total ICD-10-CM codes assigned to the matched cohorts. All other ICD-10-CM codes (“Other”) accounted for a total of 27%. Some cases were associated with more than one ICD-10-CM code.
 |
Table 1 Subject Demographics |
Microbial Densities in the Catheter-Collected and Midstream Voided Urine Specimens in Female and Male Subjects with Suspected UTI
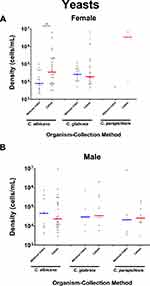
First, we compared microbial densities in catheter-collected vs midstream voided specimens for both male and female subjects (Table 2). Microbial densities of representative classical (Figure 2), emerging (Figure 3), CoNS and VGS (Figure 4), and yeast (Figure 5) uropathogens in matched catheter-collected and midstream voided urine specimens were compared for both male and female subjects.
 |
Table 2 Microbial Densities of Detected Organisms in the Catheter-Collected and Midstream Voided Urine Specimens in Female (n = 621) and Male (n = 379) Subjects |
The positive detection criterion accounts for total organisms detected at densities over 10,000 cells/mL; however, for this analysis, we included all detections to determine the median cell density. Microbial density comparisons revealed that the most frequent positively detected classical bacterial uropathogens (4 of 6 organisms in both females and males), emerging bacterial uropathogens (4 of 6 organisms in females and 3 of 6 organisms in males), and yeasts (3 of 4 yeasts in females and 4 of 4 yeasts in males), shared similar densities between the midstream voided and catheter-collected urine specimens in both female and male subjects. For example, in females, the median non-zero densities of E. coli were 23,892,818 cells/mL and 9,285,591 cells/mL (p = 0.08), in catheter-collected and midstream voided urine specimens, respectively, and in males, the median non-zero densities of E. coli were 20,827,707 cells/mL and 32,602,954 cells/mL (p = 0.78), in catheter-collected and midstream voided urine specimens, respectively (Table 2). The prevalence of E. coli and E. faecalis detected in this study, is consistent with those recently reported by others.27,28
For study subjects with non-zero CoNS detected, there were no statistically significant differences in median microbial densities between midstream voided and catheter-collected urine specimens for either female or male subjects (Figure 4). In females, the median densities of CoNS were 15,662 vs 53,334 cells/mL (p = 0.39), and in males, the median densities were 34,493 vs 116,200 cells/mL (p = 0.19), in midstream voided and catheter-collected specimens, respectively (Table 2). Similar findings were observed for VGS. In females, the median densities of VGS were 35,903 vs 39,650 cells/mL (p = 0.72), and in males, the median densities were 27,997 vs 341,368 cells/mL (p = 0.26), in midstream voided and catheter-collected specimens, respectively. When comparing the distribution of the non-zero densities, the two bacteria demonstrated similar densities between the catheter-collected and midstream voided cohorts, except for VGS being of overall higher densities in the catheter-collected than the midstream voided urine specimens from female subjects (p = 0.02).
Some bacteria or yeast demonstrated different densities between the two urine collection groups, most of which were higher in catheter-collected than midstream voided urines. For example, in males, the median non-zero densities of A. schaalii were higher in catheter-collected (2,799,497 cells/mL) than midstream voided urine (135,307 cells/mL) (p = 0.0002); in females, the median non-zero densities of E. faecalis were higher in catheter-collected (1,051,455 cells/mL) than midstream voided urine (27,454 cells/mL) (p < 0.0001) (Table 2).
Overall Positive Microbial Detection Rates in Catheter-Collected and Matched Midstream Voided Urine Specimens from Patients Suspected of UTI (n = 1000 in Each Group)
Using either positive detection (10,000 cells/mL or 100,000 cells/mL respectively), more than 80% of specimens had at least one organism positively detected and more than 40% of those positive specimens were polymicrobial (Supplementary Table 1). Due to the high similarity in detection densities between the two collection groups overall, the monomicrobial and polymicrobial (2 or more microbes detected) positive detection rates, and the most frequently detected organisms for each sex, we present the remaining analyses using only the criterion which defined positive detection using the same microbial thresholds (10,000 cells/mL for bacteria and any detection level for yeast) for both catheter-collected and midstream voided urine specimens (Table 3).
 |
Table 3 Positive Detection Rates of Organisms in Catheter-Collected and Midstream Voided Urine Specimens in Female Subjects, Using (≥ 10,000 Cells/mL for Bacteria or Any Detection Level for Yeast) |
Positive Rates of Top-Detected Organisms in Catheter-Collected and Matched Midstream Voided Urine Specimens in Female Subjects (n = 621 in Each Group)
In female subjects, slightly higher percentages of microbe-positive specimens were observed in midstream voided vs catheter-collected samples for any microbial detection (87.4% vs 80.2%, p = 0.0005) and for polymicrobial detection (65.5% vs 48.5%, p < 0.0001) (Table 3 and Supplementary Table 2). In the catheter-collected group, C. auris and P. agglomerans were not detected, making the total number of detected organisms in the catheter-collected group 28. In the midstream voided group, 28 organisms were detected, with C. auris and P. stuartii not detected (Table 3).
The top 5 classical uropathogens detected in catheter-collected urine specimens from female subjects were E. coli (217, 34.9%), E. faecalis (137, 22.1%), K. pneumoniae (89, 14.3%), P. mirabilis (71, 11.4%), and P. aeruginosa (59, 9.5%). There was significant overlap with the top 5 organisms in midstream voided urine specimens, were E. coli (278, 44.8%), E. faecalis (131, 21.1%), K. pneumoniae (80, 12.9%), P. mirabilis (33, 5.3%), and Enterobacter group (32, 5.2%).
For emerging bacterial uropathogens, the top 3 organisms detected in catheter-collected urine specimens from female subjects were A. schaalii (150, 24.2%), A. urinae (126, 20.3%), and A. omnicolens (15, 2.4%). The top 3 emerging bacterial uropathogens in midstream voided urine specimens, were A. urinae (254, 40.9%), A. schaalii (252, 40.6%), and G. vaginalis (50, 8.1%). These emerging uropathogens were detected more frequently in the midstream voided specimens than in the catheter-collected specimens (Table 3).
Positive Rates of Top-Detected Organisms in Catheter-Collected and Matched Midstream Voided Urine Specimens in Male Subjects (n = 379 in Each Group)
In contrast to the results for female subjects, in males, lower percentages of microbe-positive specimens were observed in midstream voided vs catheter-collected samples for any microbial detection (72.8% vs 85.5%, p < 0.0001) and for polymicrobial detection (36.9% vs 49.9%, p = 0.002) (Table 4 and Supplementary Table 3). In the catheter-collected group, C. auris, P. agglomerans, U. urealyticum, and G. vaginalis were not detected, making the total number of detected organisms in the catheter-collected group 26. In the midstream voided group, C. auris, S. marcescens, and A. baumannii were not detected, making the total number of detected organisms in the midstream voided group 27 (Table 4).
 |
Table 4 Positive Detection Rates of Organisms in Catheter-Collected and Midstream Voided Urine Specimens in Male Subjects, Using (≥ 10,000 Cells/mL for Bacteria or Any Detection Level for Yeast) |
The top 5 classical organisms detected as positive in catheter-collected urine specimens were E. faecalis (122, 32.2%), E. coli (93, 24.5%), P. aeruginosa (88, 23.2%), K. pneumoniae (48, 12.7%), and S. aureus (36, 9.5%). These significantly overlap with the top 5 organisms in midstream voided urine specimens, which were E. coli (90, 23.7%), E. faecalis (88, 23.2%), K. pneumoniae (29, 7.7%), P. aeruginosa (27, 7.1%), and Enterobacter group (19, 5.0%). Positive rates of these classical uropathogens were higher in catheter-collected urine specimens or similar between the two urine collection groups, depending on the organism (Table 4).
For emerging uropathogens, only two were detected in any appreciable amount, and they were the same between the catheter-collected and midstream voided urine specimen groups. These organisms are A. schaalii [55 (14.5%) vs 67 (17.7%), for catheter-collected and midstream voided, respectively, p = 0.24] and A. urinae [30 (7.9%) vs 74 (19.5%), for catheter-collected and midstream voided, respectively, p < 0.0001], with A. urinae more often detected in the midstream voided urine specimens (Table 4).
Discussion
This study compared the prevalence and cell density of 30 urinary microbes/microbe groups between catheter-collected and midstream voided samples, with the goal of determining if certain microbes were only present with any significance in voided samples, indicating that they could be contaminants that were not likely causative of UTIs.
A rigorous matching process between midstream voided and catheterized subject specimens was employed in order to minimize confounding factors and allow for a comparative analysis of prevalence and cell density between collection methods. Given that catheter-based specimen collection is less common, we matched each available subject with a catheter-collected specimen to a subject with a midstream voided specimen, based on ICD-10-CM codes, age, and sex. Out of all the successfully matched cases, 62.1% were female, which aligns with the clinical observation that females are more prone to UTIs compared to males.29,30
The data show that the two collection types were largely similar for both the prevalence and cell density of almost all microbes tested. Even with some differences, the vast majority of microbes had a higher prevalence in catheter-collected specimens than would be expected for microbes considered to be contaminants due to voiding. This is an important factor in understanding the value of detecting emerging and non-E. coli microbes with different diagnostic tests, including advanced technologies such as M-PCR. This finding indicates that all organisms detected in midstream voided urine specimens should be reconsidered as potential uropathogens when detected in the urine of patients with symptoms suggestive of UTI, rather than being presumed to be contaminants. Additionally, it supports the usefulness of the midstream voided urine collection method, which is the simplest and least invasive collection method and is commonly utilized in busy clinical settings.
All classical organisms were detected in both the midstream voided and catheter-collected specimens. For female subjects, both urine collection methods consistently detected commonly observed classical organisms, including E. coli, E. faecalis, K. pneumoniae, and P. mirabilis. Comparable densities were found among E. coli, K. pneumoniae, P. mirabilis, and P. aeruginosa. Similarly, in male subjects, E. coli, E. faecalis, K. pneumoniae, and P. aeruginosa were consistently detected in specimens from both collection methods. Comparable densities were found for E. coli, K. pneumoniae, Enterobacter group, and S. aureus. These findings highlight the convergence between the two collection methods regarding classical organism detection and density, indicating that these organisms are present directly in the bladder at high density and are not an artifact of voiding.
All emerging organisms targeted by the M-PCR assay were identified in both midstream voided and catheter-collected specimens. Among the frequently detected emerging organisms, A. schaalii and A. urinae were consistently identified as the predominant findings in both male and female subjects, regardless of the urine collection method utilized. This consistent identification of these two emerging uropathogens as the most prevalent organisms detected highlights their significant clinical importance. In yeast detection, three out of the four yeast organisms were detected in both midstream voided and catheter-collected methods with C. albicans and C. glabrata being the most frequently detected yeast species in both males and females. Notably, C. glabrata demonstrated similar densities and detection rates across both collection methods.
Some sex-based distinctions were found in prevalence and cell density. E. coli were more frequently identified in midstream voided urine samples in females, and the emerging uropathogens were more frequently identified in midstream voided for both males and females. The cell density of E. coli (in females only), A. schaalii, and A. urinae (in both males and females), however, were significantly lower in midstream voided samples compared to catheter-collected samples. K. pneumoniae had a higher detection rate in catheter-collected specimens from both males and females, but a higher average cell density in midstream voided specimens from females. In males, E. faecalis and P. aeruginosa exhibited a higher detection frequency and density in catheter-collected specimens than in midstream voided specimens. These microbes may be more prevalent in the catheterized population, or the catheter collection method may be more effective in capturing these organisms.11 In female subjects, P. mirabilis showed a higher detection rate in catheter-collected specimens, with similar densities between the collection methods.
G. vaginalis, VGS and CoNS, traditionally regarded as skin contaminants, displayed intriguing detection patterns in urine samples.11 Notably, G. vaginalis was present at densities ≥ 10,000 cells/mL in both midstream voided urine and catheter-collected samples in female subjects only, suggesting more clinical relevance in that group. However, the densities of G. vaginalis were higher in midstream voided samples in women supporting the notion that there is a skin niche for that organism. In male patients, while densities of G. vaginalis were below 10,000 cells/mL, there were similar densities between collection types. On the other hand, VGS and CoNS were detected at densities ≥ 10,000 cells/mL in both males’ and females’ urine, regardless of specimen type. The densities of VGS and CoNS in catheter-collected urine specimens were comparable to or higher than those found in midstream voided urine samples from both female and male subjects. These findings challenge the conventional belief that VGS and CoNS are merely contaminants, suggesting that they could act as colonizers or pathogenic organisms in the urinary tract. Consequently, identifying these organisms in either catheter-collected or midstream voided urine samples does not necessarily indicate contamination of the urine, emphasizing the necessity for further investigation into their clinical significance and implications in the diagnosis and treatment of urinary tract infections. Polymicrobial infections were frequently found in catheterized samples, indicating that they are not a result of contamination by voiding, and instead are relevant to diagnosing and managing UTIs. The incidence of polymicrobial detection, characterized by the identification of two or more organisms in the urine sample, was higher in midstream voided urine samples compared to catheter-collected specimens (65.5% vs 48.5%, respectively) for females, but lower for males (36.9% vs 49.9%, respectively). So, in both sexes, the fraction of all catheter-collected specimens that were polymicrobial was very high compared to traditional thought that presumed voiding is the cause of detecting polymicrobial infections.
The main limitation of this study was that the urine specimens were not collected by both methods from the same subjects. Instead, the two collection method cohorts consisted of specimens collected from subjects matched in age, sex, and ICD-10-CM codes. The two groups of subjects were not able to be matched by other clinical factors such as symptom severity, comorbidities, other clinical details of the UTI, and prior antibiotic usage. A future study comparing urine specimens which are collected by both in-and-out catheterization and midstream voided methods from the same subject may provide further clarity on potential similarities and differences in microbial detection and density between these 2 collection methods. Additionally, this study was focused on the population 60 years of age and older, which may limit the applicability of these findings to younger patients. However, since older populations are at a higher risk of adverse events from UTIs, these findings will be useful in managing UTIs in this high-risk group.
The strength of this study was the large sample size of 1000 subjects in each group, enhancing the statistical robustness of the findings. Samples were collected from patients in an urgent care setting diagnosed with elevated risk or complicated UTI, which is an important group for accurate diagnosis and management of UTIs to prevent serious adverse outcomes.
Conclusions
In this study of 2000 catheter and midstream voided urine specimens from matched patients symptomatic for UTI, we found that most of the 30 uropathogens (classical, emerging, yeasts, or traditionally regarded as contaminants) were detected in both urine specimen types, with similar detection frequencies and densities. There were a few sex-based differences in microbes detected and densities between the catheter-collected and midstream voided urine specimens. Overall, in-and-out catheter-collected and midstream voided urine specimens shared significant similarities in microbes detected by M-PCR, indicating that M-PCR identification of organisms in midstream voided urine specimens is largely representative of the bladder microbiome of these subjects. Therefore, non-invasive midstream voided collection of urine specimens is appropriate for microbial detection and identification in cases of presumed UTI and does not result in significantly more contamination compared to in-and-out catheter-collected specimens. Additionally, organisms long regarded as contaminants including Viridans Group streptococci (VGS) and coagulase-negative staphylococci (CoNS) should be reconsidered as potential uropathogens when detected in the urine of patients with symptoms suggestive of UTI.
Abbreviations
CFU, colony forming unit; CoNS, coagulase-negative Staphylococcus; P-AST, pooled-antibiotic susceptibility testing; M-PCR, multiplex-polymerase chain reaction; SUC, standard urine culture; UTI, urinary tract infection; VGS, Viridans group streptococcus.
Data Sharing Statement
All relevant data are within manuscript text, figures, and tables.
Ethics Approval and Informed Consent
Data from the study was collected via a database for which the the subject cannot be readily identified or contacted. The Western Institutional Review Board deemed the use of the data to be exempt under 45 CFR § 46.104(d)(4) as the information is used in a manner that the identity of the subject cannot be readily ascertained directly or through identifiers linked to the subjects, the subject is not contacted, and the investigator will not re-identify subjects.
Acknowledgment
The authors thank Laura Parnell from Precision Consulting and Michael Percaccio from Pathnostics for reviewing the manuscript.
Funding
Pathnostics funded the study.
Disclosure
D.B., N.L., E.H., R.A.F., and M.M. are employees of Pathnostics, and D.W. and X.Z. are paid consultants of Pathnostics. L.A.A. reports personal fees from Pathnostics, outside the submitted work. In addition, N.L. has patents (10,160,991, 11,053,532, 17/335,767, 63/493,416, AU2018254514 B2, BR112019021943-9 B1 and NZ 759292) issued to Pathnostics; pending patents (17/178,091 17/880,227 63/503,939) to Pathnostics. D.B. reports patents (US10160991, US11053532, US17178091, US17335767, AU2018254514B2, BR1120190219439B1, NZ759292) issued to PATHNOSTICS; pending patents (US17830227, US18351385, US18351286, US63493416, US63503393, US63514785, PCTUS2216816, PCTUS2277477, EP3612638, JP2022042545, CA3175879, CA3176586, CA3061015, HK620200143373, CN2018800399569, IL294577) to PATHNOSTICS. The authors report no other conflicts of interest in this work.
References
1. Schmiemann G, Kniehl E, Gebhardt K, Matejczyk MM, Hummers-Pradier E. The diagnosis of urinary tract infection: a systematic review. Dtsch Arztebl Int. 2010;107(21):361–367. doi:10.3238/arztebl.2010.0361
2. Price TK, Dune T, Hilt EE, et al. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol. 2016;54(5):1216–1222. doi:10.1128/jcm.00044-16
3. Brubaker L, Wolfe AJ. The female urinary microbiota, urinary health and common urinary disorders. Ann Transl Med. 2017;5(2):. doi:10.21037/13209
4. Wolfe AJ, Toh E, Shibata N, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol. 2012;50(4):1376–1383. doi:10.1128/jcm.05852-11
5. Lotte R, Lotte L, Ruimy R. Actinotignum schaalii (formerly Actinobaculum schaalii): a newly recognized pathogen—review of the literature. Clin Microbiol Infect. 2016;22(1):28–36. doi:10.1016/j.cmi.2015.10.038
6. Lotte L, Lotte R, Durand M, et al. Infections related to Actinotignum schaalii (formerly Actinobaculum schaalii): a 3-year prospective observational study on 50 cases. Clin Microbiol Infect. 2016;22(4):388–390. doi:10.1016/j.cmi.2015.10.030
7. Ogawa Y, Koizumi A, Kasahara K, et al. Bacteremia secondary to Alloscardovia omnicolens urinary tract infection. J Infect Chemother. 2016;22(6):424–425. doi:10.1016/j.jiac.2015.12.013
8. Higgins A, Garg T. Aerococcus urinae: an emerging cause of urinary tract infection in older adults with multimorbidity and urologic cancer. Urol Case Rep. 2017;13:24–25. doi:10.1016/j.eucr.2017.03.022
9. Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52(3):871–876. doi:10.1128/jcm.02876-13
10. Mueller ER, Wolfe AJ, Brubaker L. Female urinary microbiota. Curr Opin Urol. 2017;27(3):282–286. doi:10.1097/mou.0000000000000396
11. Korman HJ, Baunoch D, Luke N, et al. A diagnostic test combining molecular testing with phenotypic pooled antibiotic susceptibility improved the clinical outcomes of patients with non-E. coli or polymicrobial complicated urinary tract infections. Res Rep Urol. 2023;15:141–147. doi:10.2147/rru.s404260
12. Vollstedt A, Baunoch D, Wolfe A, et al. Bacterial interactions as detected by pooled antibiotic susceptibility testing(P-AST) in polymicrobial urine specimens. J Surg Urol. 2020;1:1.
13. Baunoch D, Luke N, Wang D, et al. Concordance between antibiotic resistance genes and susceptibility in symptomatic urinary tract infections. Infect Drug Resist. 2021;14:3275–3286. doi:10.2147/idr.s323095
14. Wojno KJ, Baunoch D, Luke N, et al. Multiplex PCR based urinary tract infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology. 2019;136:119–126. doi:10.1016/j.urology.2019.10.018
15. Vollstedt A, Baunoch D, Wojno K, et al. Multisite prospective comparison of multiplex polymerase chain reaction testing with urine culture for diagnosis of urinary tract infections in symptomatic patients. J Sur Urol. 2020:2020:JSU–102.
16. Haley E, Luke N, Korman H, et al. Improving patient outcomes while reducing empirical treatment with multiplex-polymerase-chain-reaction/pooled-antibiotic-susceptibility-testing assay for complicated and recurrent urinary tract infections. Diagnostics. 2023;13(19). doi:10.3390/diagnostics13193060
17. Ko DSC, Lukacz ES, Juster IA, et al. Real-world evidence that a novel diagnostic combining molecular testing with pooled antibiotic susceptibility testing is associated with reduced infection severity and lower cost compared with standard urine culture in patients with complicated or persistently recurrent urinary tract infections. JU Open Plus. 2023;1(5). doi:10.1097/ju9.0000000000000025
18. Daly A, Baunoch D, Rehling K, et al. Utilization of M-PCR and P-AST for diagnosis and management of urinary tract infections in home-based primary care. JOJ Uro Nephron. 2020;7(2):555707.
19. Plaza-Verduin MA, Lucas JK. Atlas of Emergency Medicine Procedures. Springer; 2022:641–643. doi:10.1007/978-3-030-85047-0_134
20. Markowitz MA, Wood LN, Raz S, Miller LG, Haake DA, Kim JH. Lack of uniformity among United States recommendations for diagnosis and management of acute, uncomplicated cystitis. Int Urogynecol J. 2019;30(7):1187–1194. doi:10.1007/s00192-018-3750-z
21. Kwok M, McGeorge S, Mayer‐Coverdale J, et al. Guideline of guidelines: management of recurrent urinary tract infections in women. Bju Int. 2022;130(Suppl 3):11–22. doi:10.1111/bju.15756
22. Hilt EE, Parnell LK, Wang D, Stapleton AE, Lukacz ES. Microbial threshold guidelines for UTI diagnosis: a scoping systematic review. Zhang DrP, ed. Pathol Lab Med Int. 2023;15:43–63. doi:10.2147/plmi.s409488
23. Parnell LKD, Luke N, Mathur M, et al. Elevated UTI biomarkers in symptomatic patients with urine microbial densities of 10,000 CFU/mL indicate a lower threshold for diagnosing UTIs. Baraniak A, ed. MDPI. 2023;13(16):1–15. doi:10.3390/diagnostics13162688
24. Festa RA, Opel M, Mathur M, et al. Quantitative multiplex PCR in copies mL–1 linearly correlates with standard urine culture in colonies mL−1 for urinary tract infection (UTI) pathogens. Lett Appl Microbiol. 2023;2023:1. doi:10.1093/lambio/ovad085
25. Wojno KJ, Baunoch D, Luke N, et al. Multiplex PCR based urinary tract infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology. 2020;136:119–126. doi:10.1016/j.urology.2019.10.018
26. Vollstedt A, Baunoch D, Wojno KJ, et al. Multisite prospective comparison of multiplex polymerase chain reaction testing with urine culture for diagnosis of urinary tract infections in symptomatic patients. J Surg Urol. 2020;2020:1.
27. Price TK, Hilt EE, Dune TJ, Mueller ER, Wolfe AJ, Brubaker L. Urine trouble: should we think differently about UTI? Int Urogynecol J. 2018;29(2):205–210. doi:10.1007/s00192-017-3528-8
28. Joyce C, Halverson T, Gonzalez C, Brubaker L, Wolfe AJ. The urobiomes of adult women with various lower urinary tract symptoms status differ: a re-analysis. Front Cell Infect Microbiol. 2022;12:860408. doi:10.3389/fcimb.2022.860408
29. Tan C, Chlebicki M. Urinary tract infections in adults. Singap Méd J. 2016;57(9):485–490. doi:10.11622/smedj.2016153
30. Medina M, Castillo-Pino E. An introduction to the epidemiology and burden of urinary tract infections. Ther Adv Urology. 2019;11:1756287219832172. doi:10.1177/1756287219832172
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.