Back to Journals » OncoTargets and Therapy » Volume 14
DNASE1L3 as a Prognostic Biomarker Associated with Immune Cell Infiltration in Cancer
Authors Deng Z, Xiao M, Du D, Luo N, Liu D, Liu T, Lian D, Peng J
Received 2 December 2020
Accepted for publication 19 February 2021
Published 18 March 2021 Volume 2021:14 Pages 2003—2017
DOI https://doi.org/10.2147/OTT.S294332
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Zenghua Deng,1,2,* Mengmeng Xiao,3,4,* Dexiao Du,2 Nan Luo,1,2 Dongfang Liu,2 Tingting Liu,2 Dongbo Lian,1,2 Jirun Peng1,2
1Ninth School of Clinical Medicine, Peking University, Beijing, 100038, People’s Republic of China; 2Department of Surgery, Beijing Shijitan Hospital, Capital Medical University, Beijing, 100038, People’s Republic of China; 3Peking University International Hospital, Beijing, 102206, People’s Republic of China; 4Eighth School of Clinical Medicine, Peking University, Beijing, 102206, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jirun Peng Tel +86-10-63926721
Email [email protected]
Objectives: Deoxyribonuclease 1 like 3 (DNASE1L3) is critically involved in apoptosis and immune response, however, its role in cancer has yet to be deciphered. We aimed to explore the prognostic value of DNASE1L3 across a series of malignancies.
Methods: Based on Oncomine database and Tumor Immune Estimation Resource (TIMER), expression profiling of DNASE1L3 was detailed in malignancies. Using PrognoScan, Kaplan-Meier Plotter, GEPIA2, and bc-GenEcMiner v4.5, prognostic value of DNASE1L3 was estimated in diverse cancers. Based on TIMER, association between DNASEL13 expression and immune infiltration was examined in various cancers. Then, mRNA level of DNASE1L3 in hepatocellular carcinoma (HCC) samples (n=22) and stomach adenocarcinoma (STAD) samples (n=17) was measured with qRT-PCR. Immunohistochemistry was performed to confirm expression of DNASE1L3 in paraffin-embedded tissues of HCC (n=9) and lung adenocarcinoma (n=20).
Results: DNASE1L3 was downregulated in multiple cancers, including breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), liver hepatocellular carcinoma (LIHC), and lung adenocarcinoma (LUAD). A lower level of DNASE1L3 correlated with poorer prognosis in various cancers, especially in breast, liver, kidney, stomach, lung adenocarcinoma and sarcoma (SARC). Moreover, DNASE1L3 was positively related to immune cell infiltration in many cancers, including BRCA, LIHC, STAD, LUAD, and SARC. DNASE1L3 was significantly associated with CCR7/CCL19 in cancers. DNASE1L3 was downregulated in HCC and STAD tissues as demonstrated by qRT-PCR, as well as in HCC and LUAD samples, as shown by immunohistochemistry.
Conclusion: DNASE1L3 has potential to serve as a prognostic biomarker in cancer of the breast, kidney, liver, stomach, lung adenocarcinoma and sarcoma. Down-regulation of DNASE1L3 may participate in immune escape via CCR7/CCL19 axis.
Keywords: DNASE1L3, CCR7, CCL19, prognosis, immune infiltration, tumor
Introduction
Immunotherapy has shed light on treatment of cancer. Accumulating evidence has indicated that the immune system plays a key role in human cancer,1,2 so immunotherapy is one of the most promising directions in cancer treatment.3,4 Recently, immune checkpoint blockade has exhibited significant antitumor effects in several malignancies, such as melanoma, renal cell cancer, hepatocellular carcinoma (HCC), biliary tract carcinoma, gastric cancer, and head and neck cancer.5–7 Despite unprecedented efficacy of immune checkpoint blockade, majority of patients showed no response to immunotherapy, while some patients who had encouraging initial response, acquired resistance over time. Thus, a better understanding of underlying mechanisms is urgent for immunotherapy for cancer. For example, infiltrating lymphocytes in tumors may affect prognosis and response to immunotherapy.8–10 Collectively, elucidating various factors modulating immune infiltration and identifying new immunotherapeutic targets are essential to improve efficacy of cancer immunotherapy.
Deoxyribonuclease 1 like 3 (DNASE1L3) is a member of the deoxyribonuclease 1 family, with similar constituent to DNase,11,12 which was first identified from thymocytes of rat in 199413 and cloned as homolog of human DNASE1L3 in 1998.14 DNASE1L3 is important for DNA catabolism and apoptosis.15,16 Previous studies indicated that DNASE1L3 participated in signal transduction in breast cancer, including intracellular signaling cascade, receptor activity, and GTPase regulator.17 Moreover, DNASE1L3 is a prognostic indicator of HCC.18 Above all, DNASE1L3 probably plays a role in tumorigenesis.
In addition, DNASE1L3 secreted by dendritic cells (DCs), macrophages, and neutrophils16,19 can be detected in circulation. DNASE1L3 is involved in programmed cell death during development,20 immune response related to neutrophil activation, acute inflammatory response, and neutrophil mediated cytotoxicity.21,22 These findings indicate functions of DNASE1L3 in the immune system.
Considering interactions among cancer, immune system and DNASE1L3, we measured its expression, estimated its prognostic value, and explored its association with immune infiltration in human cancers comprehensively.
Materials and Methods
Genetic Analysis Using GEPIA2
Using GEPIA2 (http://gepia2.cancer-pku.cn/#index)23 the most differential survival and expression genes were explored in 33 different human cancers in TCGA and GTEx. DNASE1L3 was identified to be downregulated in almost all human cancers.
Genetic Analysis Using Oncomine
Using Oncomine gene expression array database (https://www.oncomine.org/resource/login.html),24 mRNA level of DNASE1L3 gene was assessed in human cancers. The threshold was defined as: P= 0.001, fold change =2.0, and gene ranking of all.
Prognostic Analysis Using PrognoScans
Using PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/),25 prognostic value of DNASE1L3 was evaluated in human cancers. The threshold was defined as Cox P<0.05, describing hazard ratio (HR) with 95% confidence intervals (CI).
Prognostic Analysis Using Kaplan-Meier Plotter
Using Kaplan-Meier plotter (http://www.kmplot.com/analysis/index.php?p=background),26,27 prognostic value of DNASE1L3 was estimated in human cancers. The HR with 95% CI was marked.
bc-GenEcMiner V4.5 Database Analysis
Using Breast Cancer Gene-Expression Miner v4.5 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1),28,29 the prognostic role of DNASE1L3 was explored in breast cancer. bc-GenExMiner v4.5 is a statistical mining tool of published annotated breast cancer transcriptomic data (DNA microarrays [n = 10,716] and RNA-seq [n = 4712]), which estimates prognostic values of specific genes in breast cancer.
TIMER Database Analysis
Using Tumor Immune Estimation Resource (TIMER) (https://cistrome.shinyapps.io/timer/),30,31 differential gene expression between multiple cancers and related normal tissues, as well as association between two molecules in cancers could be analyzed. The Cancer Genome Atlas (TCGA) was used to estimate immune infiltration. Moreover, gene expression against tumor purity can be normalized through TIMER.32 We analyzed DNASE1L3 expression across 32 types of human cancers as well as the relationship between DNASE1L3 and immune infiltration. Association between DNASE1L3 and multiple genetic markers of tumor-infiltrating immune cells was analyzed.33–36 Then, Spearman correlation and estimated statistical significance of a pair of defined genes in one cancer type were generated through correlation module. DNASE1L3 was applied to x-axis, and related markers were represented on y-axis. Gene expression level was displayed with log2 RSEM.
DNASE1L3 Expression in HCC, Stomach and Lung Adenocarcinoma
This study was approved by Ethical Committee of Beijing Shijitan Hospital and performed by complying with all relevant ethical regulations [No. sjtkyll-lx-2019(36)]. Written informed consent from each patient was obtained prior to study commencement, and all processes were conducted in accordance with the Declaration of Helsinki.
qRT-PCR was conducted to measure expression of DNASE1L3 in HCC samples and adjacent normal tissues (n=22), as well as the expression of DNASE1L3, CCR7, and CCL19 in STAD samples and adjacent normal tissues (n=17), which were collected in Beijing Shijitan Hospital. Briefly, total RNA was isolated using E.Z.N.A Total RNA Kit II (OMEGA R6934-02) according to the manufacturer’s protocol. ReverTra Ace qPCR RT Kit (TOYOBO FSQ-101) was used to synthesize the first-strand cDNA. Real-time qPCR was performed on Quant Studio 6 Real Time PCR System with SYBR (Thermo Fisher Scientific). 18S was used as reference gene. The sequences of primers were as follows: DNASE1L3 Forward: AGCCCTTTGTGGTCTGGTTC, Reverse: TCCTTAACGGATGTCTCTGGG; CCR7 Forward: TGAGGTCACGGACGATTACAT, Reverse: GTAGGCCCACGAAACAAATGAT. CCL19 Forward: CTGCTGGTTCTCTGGACTTCC, Reverse: AGGGATGGGTTTCTGGGTCA. 18S Forward: GTAACCCGTTGAACCCCATT, Reverse: CCATCCAATCGGTAGTAGCG.
Immunohistochemistry was used to explore protein expression of DNASE1L3 in formalin-fixed paraffin-embedded (FFPE) tissues collected at Beijing Shijitan Hospital. Briefly, 12 paired HCC and adjacent normal tissue FFPE samples, 20 paired lung adenocarcinoma and adjacent normal tissue FFPE samples were collected from the Department of Pathology. The primary and secondary antibodies were purchased from Abcam (ab203669, ab205718). Immunohistochemistry procedures followed the manufacturer’s instructions (https://www.abcam.com/protocols/immunostaining-paraffin-frozen-free-floating-protocol). The expression level of DNASE1L3 in each sample (collected from 3 random fields, 200×) was evaluated by AOD with Image J software. The clinical information was listed in Supplementary Tables 4 and 5.
Statistical Analysis
Data were analyzed by GraphPad Prism software (version 7.04; GraphPad Prism software Inc., San Diego, CA). Survival curves were generated by PrognoScan, Kaplan-Meier plot, GEPIA2 and bc-GenEcMiner v4.5. Data from Oncomine were represented as P-values, t-test values, fold changes, ranks and sample numbers. Results from Kaplan-Meier plots, PrognoScan, GEPIA2 and bc-GenEcMiner v4.5 were represented as HR, 95% CI and P or Cox P-values from Log rank tests. Association between two genes was evaluated by Spearman correlation and absolute value of correlation was defined as follows: 0.00–0.19: very weak; 0.20–0.39: weak; 0.40–0.59: moderate; 0.60–0.79: strong; 0.80–1.0: very strong. Paired t-test was used to compare DNASE1L3 expression levels between tumor and adjacent normal samples. A P <0.05 was considered as statistically significant.
Results
The mRNA Expression Profiling of DNASE1L3 in Various Cancers
The DNASE1L3 mRNA levels in human cancers and adjacent normal tissues were analyzed by Oncomine database. DNASE1L3 was downregulated in breast, cervical, colorectal, esophageal, gastric, head and neck, kidney, liver, lung, ovarian, pancreatic cancer, mesothelioma, leukemia, sarcoma, pleural malignant mesothelioma and pleural malignant fibrous histiocytoma. On the other hand, only lymphoma had higher expression of DNASE1L3 (Figure 1A). More information on DASE1L3 expression was collected in Supplementary Table 1.
Moreover, expression level of DNASE1L3 was also analyzed in TCGA database through TIMER. DNASE1L3 was downregulated in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) compared with adjacent normal tissues (Figure 1B).
Prognostic Value of DNASE1L3 in Human Cancers
Based on PrognoScan, DNASE1L3 might impact prognosis in blood cancer (follicular lymphoma and multiple myeloma), breast cancer, colorectal cancer, lung adenocarcinoma, head and neck cancer (squamous cell carcinoma), ovarian cancer, and skin cancer (melanoma). Lower expression of DNASE1L3 was related to worse prognosis in these cancers (Figure 2A-H, Supplementary Figure 1). In GSE31210 cohort, 204 lung adenocarcinoma patients were divided into DANSE1L3 higher expression group (n=108) and lower expression group (n=196), a lower level of DNASE1L3 was correlated with poorer overall survival (OS, HR=0.61, P=0.0212), HR=0.61 means the probability of OS in lower group/higher group=0.61 (Figure 2E).
Based on Kaplan-Meier plotter, DNASE1L3 might influence prognosis in bladder cancer, breast cancer, gastric carcinoma, liver cancer, lung adenocarcinoma, lung squamous cell carcinoma, ovarian cancer, cervical squamous cell carcinoma, head-neck squamous cell carcinoma, pancreatic adenocarcinoma, kidney renal clear cell carcinoma, sarcoma and mesothelioma. In all of these cancers/tumors except bladder cancer, lower expression of DNASE1L3 indicated worse prognosis (Figure 2I-P, Supplementary Figure 2).
In addition, prognostic value of DNASE1L3 was also estimated through GEPIA (Supplementary Figure 3). Interestingly, lower expression of DNASE1L3 in ACC, KICH, KIRC, LIHC, LUAD, PAAD, PCPG, and SARC correlated with poorer prognosis (reduced OS). By contrast, higher expression of DNASE1L3 in BCLA and UVM correlated with worse prognosis. As for relapse free survival (RFS), lower expression of DNASE1L3 in CESC, CHOL, KICH, KIRC, KIRP, LIHC, PAAD, and THCA correlated with poorer prognosis, whereas higher expression of DNASE1L3 in UVM correlated with worse prognosis (Supplementary Figure 3). Lastly, using bc-GenEcMiner v4.5 Database, prognostic value of DNASE1L3 was indicated in breast cancer. Lower expression of DNASE1L3 correlated with poorer OS and disease free survival (DFS) in breast cancer (Figure 3). Collectively, all these results indicated that DNASE1L3 might harbor significant prognostic value in various cancers.
DNASE1L3 Correlates with Immune Infiltration in Various Cancers
Tumor-infiltrating lymphocytes affect prognosis and response to immunotherapy of cancers.8–10 A previous study indicated that circulating DNASE1L3 could be secreted by dendritic cells (DC), macrophages, and neutrophils. Thus, DNASE1L3 may be connected with immune infiltration in various cancers. Then, TIMER was used to analyze relationship of DNASE1L3 expression with immune infiltration in multiple cancers. Interestingly, DNASE1L3 correlated with immune infiltration in diverse cancers. DNASE1L3 expression generally correlated with tumor purity in 31 human cancers, with B cells in 24 human cancers, with CD8+T cells in 19 human cancers, with macrophages in 13 human cancers, with neutrophils in 17 human cancers, as well as with dendritic cells in human cancers (Figure 4, Supplementary Figure 4). For 31 types of cancers with significant association between DNASE1L3 expression and tumor purity, relationship between immune infiltration and DNASE1L3 was defined when 4 or more types of immune cells correlated with DNASE1L3. In this way, 14 types of cancers were identified. Moreover, DNASE1L3 harbored prognostic value in 9 out of 14 cancers, including BRCA, HNSC, KIRP, LIHC, LUAD, SARC, SKCM, STAD, and UCEC (Figure 4). Taking BRCA as an example, DNASE1L3 expression correlated with tumor purity negatively, whereas with infiltration of CD8+ T cells, CD4+T cells, neutrophils and DCs (but not B cell and macrophage) positively. Similar results were observed in the other 8 types of cancers, except LIHC. Therefore, DNASE1L3 might participate in immune infiltration in these 9 types of cancers, especially for T cells (both CD4+ and CD8+) and DCs.
DNASE1L3 Correlates with Immune Markers in Various Cancers
To decipher the interaction of DNASE1L3 with immune infiltrating cells, association between DNASE1L3 expression and markers of various immune cells in BRCA, HNSC, KIRP, LIHC, LUAD, SARC, SKCM, STAD, and UCEC was evaluated through TIMER and GEPIA. As previously described,33 immune markers of CD8+ T cells, T cells (general), Th1 cells, Th2 cells, Tfh cells, Th17 cells, and regulatory T cells (Tregs), exhausted T cells, B cells, monocytes, tumor-associated macrophages (TAMs), M1 and M2 macrophages, neutrophils, NK cells and DCs were selected to analyze individual association with DNASE1L3 in the previously mentioned 9 cancers (Table 1, Supplementary Table 2). Interestingly, CD8+T cells (marked by CD8A and CD8B), general T cells (marked by CD3D, CD3E, or CD2), B cells (marked by CD19 or CD79A), monocytes (marked by CD86 or CD115) had a positive relationship with DNASE1L3 expression in all these 9 cancers, except LIHC. TAM (marked by CCL2, CD86, or IL10) and neutrophils (marked by CD66b, CD11b, or CCR7) widely correlated with DNASE1L3 expression in these 9 cancers, except HNSC. As to M1/M2 macrophages, DNASE1L3 correlated with both in BRCA, SARC and UCEC, whereas with either M1 or M2 in the other 6 cancers. DNASE1L3 expression correlated with natural killer cells (marked by KIR2DL1, KIR2DL3, KR2DL4, KIR3DL1, KIR3DL2, KR3DL3, or KIR2DS4) in these 9 cancers, except KIRP and LIHC. DNASE1L3 expression correlated with DCs (marked by HLA-DPB1, HTHELA-DQB1, HLA-DRA, HLA-DPA1, BDCA-1, BDCA-4, or CD11c) in all 9 cancers except LIHC. In addition, DNASE1L3 expression correlated with Tregs (marked by FOXP3, CCR8, STAT5B, or TGF-β) and exhausted T cells (marked by PD-1, CTLA4, LAG3, TIM-3, or GZMB) in all these 9 cancers, except LIHC. Thus, DNASE1L3 expression universally correlation with immune cells in tumor microenvironment (TME), especially with DCs and T cells.
 |
Table 1 Correlation Analysis Between DNASE1L3 and Markers of Immune Cells Through TIMER |
Then, the relationship between DNASE1L3 and DCs/exhausted T cells in tumor and normal tissue was explored from TCGA through GEPIA (Table 2, Supplementary Table 3). Interestingly, DNASE1L3 generally correlated with DCs/exhausted T cells in these 9 cancers. In addition, CCR7 (a marker of neutrophil) strongly correlated with DNASE1L3 in most of these cancers. Accordingly, DNASE1L3 strongly correlated with CCL19 (the ligand of CCR7) in these cancers (Figure 5). Collectively, DNASE1L3 correlated with immune infiltrating cells in diverse cancers, which may participate in tumorigenesis and affect prognosis of cancer patients.
 |
Table 2 Correlation Analysis Between DNASE1L3 and Immune Cells in Tumor/Normal Tissue of TCGA Through GEPIA2 |
DNASE1L3 is Downregulated in HCC, STAD, and LUAD
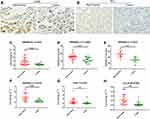
DNASE1L3 expression in HCC, STAD, and LUAD was analyzed. Firstly, mRNA level was measured by qRT-PCR in HCC and adjacent normal tissues. DNASE1L3 was downregulated in HCC (Figure 6C). DNASE1L3, CCR7, and CCL19 were downregulated in STAD (Figure 6F–H). Then, DNASE1L3 expression in lung adenocarcinoma (n=20) and HCC (n=9) was detailed by immunohistochemistry using FFPE tissues. DNASE1L3 was downregulated in LUAD and HCC (Figure 6A–B, D–E).
Discussion
In our present study, DNASE1L3 was markedly downregulated in various human cancers, and strongly correlated with poorer prognosis, especially in breast, kidney, liver cancer, stomach, lung adenocarcinoma and sarcoma. Furthermore, DNASE1L3 was related to immune infiltration, especially with CCR7/CCL19 axis. Our results suggest that DNASE1L3 could act as a prognostic marker and potential target for immune-related therapy in multiple human cancers.
DNASE1L3 (also termed DNase γ), an endonuclease, is a member of DNase I family. DNASE1L3, located on 3p14.3-p21.1, is recognized as a candidate tumor suppressor gene,11 since allelic loss of this region is common in cancers, including renal cell carcinoma and small cell lung cancers.37 Although a pathologic role of DNASE1L3 in cancer has yet to be extensively studied, DNASE1L3 might be involved in apoptosis.11,38,39 DNASE1L3 was overexpressed in the early stage of clear cell renal cancer, compared with late stage.40 In addition, DNASE1L3 was proposed as a prognostic factor of HCC.18 In our present study, DNASE1L3 had prognostic value in various types of cancers. Lower expression of DNASE1L3 correlated with poorer clinical outcomes in BRAD, STAD, LUAD, LIHC, ASRC, and KD; and probably in HNSC, melanoma, or UCEC. Moreover, DNASE1L3 correlated with immune infiltration and various immune cell markers in these cancers. Thus, our study provides supporting evidence about DNASE1L3 as a prognostic biomarker in cancers, as well as a potential target in immunotherapy of cancers.
DNASE1L3 is Downregulated in Various Tumors and Correlated with Poor Prognosis
In our study, DNASE1L3 expression in cancer tissue was compared with adjacent normal tissue through independent datasets in Oncomine and TCGA. In Oncomine, DNASE1L3 was downregulated in the following cancers: breast, cervical, colorectal, esophageal, gastric, head and neck, kidney, liver, lung, ovarian, pancreatic, mesothelioma, leukemia, sarcoma, pleural malignant mesothelioma and pleural malignant fibrous histiocytoma in one or more datasets. By contrast, higher expression of DNASE1L3 was observed only with lymphoma in two datasets (Figure 1A). Lower expression of DANSE1L3 was observed in various types of human cancers in TCGA database through TIMER (Figure 1B). Thus, DNASE1L3 was generally downregulated in human cancers. Moreover, lower expression of DNASE1L3 correlated with poorer prognosis in kidney cancer (Figure 2K-L, Supplementary Figure 2L-M, Supplementary Figure 3K-M), liver hepatocellular carcinoma (Figure 2M-N, Supplementary Figure 2E, Supplementary Figure 2N, Supplementary Figure 3P), lung adenocarcinoma (Figure 2E-F, Supplementary Figure 1M, Supplementary Figure 2O, Supplementary Figure 3Q), sarcoma (Figure 2O-P, Supplementary Figure 2U, Supplementary Figure 3Y) in PrognoScan, Kaplan-Meier Plotter, and TCGA databases. In addition, lower expression of DNASE1L3 correlated with poorer prognosis in breast cancer (Figure 2A-D, Supplementary Figure 2D, Supplementary Figure 2G, Supplementary Figure. 3C), HNSC (Supplementary Figure 1J, Supplementary Figure 2K, Supplementary Figure.3J), melanoma (Figure 2H, Supplementary Figure 3Z), STAD (Figure 2I-J, Supplementary Figure 2A, Supplementary Figure 2V, Supplementary Figure 3AA), and UCEC (Supplementary Figure 2Z, Supplementary Figure 3AE) in PrognoScan and Kaplan-Meier Plotter databases, but not in TCGA. Then, prognostic value of DNASE1L3 in breast cancer was identified in bc-GenEcMiner V4.5, which specifically collects data from breast cancer studies and is widely used in breast cancer research.28,29 Accordingly, lower expression of DNASE1L3 in breast cancer correlated with poorer probability of OS and DFS (Figure 3A). Furthermore, DNASE1L3 was downregulated in HCC at both transcriptional and translational levels (Figure 6B, C, E), as well as in STAD and LUAD at translational level (Figure 6A, D, F). Collectively, DNASE1L3 may serve as a prognostic marker in breast cancer, kidney cancer, liver hepatocellular carcinoma, lung adenocarcinoma and sarcoma. In addition, prognostic value of DNASE1L3 in HNSC, melanoma, STAD, and UCEC requires confirmation. As far as we know, this is the first report on prognostic value of DNASE1L3 in most of these types of cancers.
DNASE1L3 Correlates with Immune Infiltration in Various Tumors
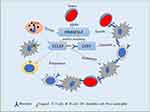
Mutation of DNASE1L3 was frequently connected to autoimmune diseases, like systemic lupus erythematosus, rheumatoid arthritis, and type 1 diabetes.41,42 As previously reported, DNASE1L3 was involved in immune response related neutrophil activation, and might regulate acute inflammatory response and neutrophil mediated cytotoxicity.21,22 To better understand functions of DNASE1L3 in cancers, we analyzed the relation between DNASE1L3 and immune infiltration in various cancers through TIMER. Interestingly, DNASE1L3 universally correlates with immune cells (Supplementary Figure 4). Specifically, DNASE1L3 correlates with prognosis in 9 cancers (breast invasive carcinoma, head and neck squamous cell carcinoma, kidney renal papillary cell carcinoma, liver hepatocellular carcinoma, lung adenocarcinoma, sarcoma, skin cutaneous melanoma, stomach adenocarcinoma, and uterine corpus endometrial carcinoma), and the strength of correlation varies from “moderate” (0.40<|R|<0.59) to “very week” (0.00<|R|<0.19) (Figure 4). Thus, DNASE1L3 may interact with immune infiltration in tumor microenvironment (TME), through which DNASE1L3 may affect prognosis of cancer patients. Moreover, DNASE1L3 correlates with multiple gene markers of tumor-infiltrating immune cells. For example, DNASE1L3 correlates with CD8+ T cell, general T cell and B cell in 8 of 9 cancers (except LIHC) (Table 1, Supplementary Table 2). DNASE1L3 universally correlates with monocytes, TAMs, M1/M2 macrophages, neutrophils, natural killer (NK) cells, dendritic cells (DCs), T-helper 1 (Th1) cells, T-helper 2 (Th2) cells, follicular helper T (Tfh) cells, T-helper 17 (Th17) cells, Tregs, and exhausted T cells, especially with CCR7 in neutrophils and BDCA-1 (CD1C) in dendritic cells. CCR7 (C-C motif chemokine receptor 7) is expressed in various lymphoid tissues, which activates B and T lymphocytes, as well as stimulating the maturation of dendritic cells. CCR7 acts in immune system and tumorigenesis when combined with its specific ligand CCL19 (C-C motif chemokine ligand 19).43–45 Accordingly, expression of DNASE1L3 significantly correlated with CCL19 (Figure 5). CCL19 promotes immune cells infiltrating into tumors to exert anti-tumor effects by inducing DCs into tumor tissue and by acting as a chemotactic factor on T cells through CCL19/CCR7 axis.46–48 Moreover, at least in lung adenocarcinoma and colorectal cancer cohorts, patients with CCL19-positive tumors had better prognosis.49,50 Combining findings from our present study and previous literature, we hypothesize a potential mechanism underlying the interaction of DNASE1L3 with tumor and TME (Figure 7): tumors could directly or indirectly inhibit CCL19-CCR7 axis by down-regulating DNASE1L3, which in turn produces a negative regulatory effect on the immune system, possibly by interfering with the maturation of DCs, inhibiting co-stimulatory activation of B/T cells, and undermining infiltration of multiple immune cells, ultimately facilitating in immune escape. Collectively, DNASE1L3 is widely associated with immune infiltration in TME of various cancers, and CCR7/CCL19 axis may play a key role in this process. Further experiments should be designed to confirm this hypothesis and explore molecular mechanisms in detail.
Conclusion
In our study, we comprehensively analyzed expression profiling and prognostic value of DNASE1L3, as well as its correlation with the immune system in multiple cancers. Notably, DNASE1L3 is widely downregulated in human cancers and associated with poorer prognosis in various cancers, especially in breast cancer, kidney cancer, LIHC, LUAD, and SARC. In addition, DNASE1L3 expression is universally related to immune cell infiltration in TME of various cancers, especially with CCR7-CCL19 axis. Thus, we infer that tumor cells may cause decrease in immune infiltration through down-regulating DNASE1L3 expression, followed by suppressing CCR7/CCL19 axis. This may represent a new pathway in which tumor cells escape from immune surveillance. Demonstrating a pathologic relationship of DNASE1L3 down-regulation with immune infiltration in TME, and deciphering molecular mechanisms by which CCR7/CCL19 axis can modify anti-tumor response, will facilitate improvement in cancer immunotherapy.
Data Sharing Statement
Correspondence and requests for materials should be addressed to Jirun Peng at [email protected] or [email protected].
Acknowledgments
We thank the Department of Pathology and the Central Lab of Beijing Shijitan Hospital, Capital Medical University, for providing technical support. This study was supported by National Natural Science Foundation of China (No. 82072601) and Beijing Natural Science Foundation (7182075).
Disclosure
All authors declare no conflicts of interest.
References
1. Role of immunity in therapy of human cancer. Discussion. Br J Cancer Suppl. 1973;1:299–308.
2. Sikora K, James N. Immune modulation and cancer. Br Med Bull. 1991;47(1):209–226.
3. Devaud C, John LB, Westwood JA, Darcy PK, Kershaw MH. Immune modulation of the tumor microenvironment for enhancing cancer immunotherapy. Oncoimmunology. 2013;2(8):e25961.
4. Sadreddini S, Baradaran B, Aghebati-Maleki A, et al. Immune checkpoint blockade opens a new way to cancer immunotherapy. J Cell Physiol. 2019;234(6):8541–8549.
5. Brahmer JR, Tykodi SS, Chow LQM, et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. 2012;366(26):2455–2465.
6. Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest. 2015;125(9):3384–3391.
7. Lin J, Yang X, Long J, et al. Pembrolizumab combined with lenvatinib as non-first-line therapy in patients with refractory biliary tract carcinoma. Hepatobiliary Surg Nutr. 2020;9(4):414–424.
8. Zhang H, Liu H, Shen Z, et al. Tumor-infiltrating neutrophils is prognostic and predictive for postoperative adjuvant chemotherapy benefit in patients with gastric cancer. Ann Surg. 2018;267(2):311–318.
9. Waniczek D, Lorenc Z, Snietura M, Wesecki M, Kopec A, Tumor-Associated Macrophages M-WM. Regulatory T cells infiltration and the clinical outcome in colorectal cancer. Arch Immunol Ther Exp (Warsz). 2017;65(5):445–454.
10. Kumpers C, Jokic M, Haase O, et al. Immune Cell Infiltration of the Primary Tumor, Not PD-L1 status, is associated with improved response to checkpoint inhibition in metastatic melanoma. Front Med (Lausanne). 2019;6:27.
11. Rodriguez AM, Rodin D, Nomura H, Morton CC, Weremowicz S, Schneider MC. Identification, localization, and expression of two novel human genes similar to deoxyribonuclease I. Genomics. 1997;42(3):507–513.
12. Shiokawa D, Tanuma S. Characterization of human DNase I family endonucleases and activation of DNase gamma during apoptosis. Biochemistry. 2001;40(1):143–152.
13. Shiokawa D, Ohyama H, Yamada T, Takahashi K, Tanuma S. Identification of an endonuclease responsible for apoptosis in rat thymocytes. Eur J Biochem. 1994;226(1):23–30.
14. Liu QY, Pandey S, Singh RK, et al. DNaseY: a rat DNaseI-like gene coding for a constitutively expressed chromatin-bound endonuclease. Biochemistry. 1998;37(28):10134–10143.
15. Errami Y, Naura AS, Kim H, et al. Apoptotic DNA fragmentation may be a cooperative activity between caspase-activated deoxyribonuclease and the poly(ADP-ribose) polymerase-regulated DNAS1L3, an endoplasmic reticulum-localized endonuclease that translocates to the nucleus during apoptosis. J Biol Chem. 2013;288(5):3460–3468.
16. Serpas L, Chan RWY, Jiang P, et al. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc Natl Acad Sci U S A. 2019;116(2):641–649.
17. Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274.
18. Xu B, Lv W, Li X, Zhang L, Lin J. Prognostic genes of hepatocellular carcinoma based on gene coexpression network analysis. J Cell Biochem. 2019.
19. Sisirak V, Sally B, D’Agati V, et al. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell. 2016;166(1):88–101.
20. Liu QY, Ribecco M, Pandey S, Walker PR, Sikorska M. Apoptosis-related functional features of the DNaseI-like family of nucleases. Ann N Y Acad Sci. 1999;887:60–76.
21. Jimenez-Alcazar M, Rangaswamy C, Panda R, et al. Host DNases prevent vascular occlusion by neutrophil extracellular traps. Science. 2017;358(6367):1202–1206.
22. Xie X, Liu PS, Percipalle P. Analysis of Global Transcriptome Change in Mouse Embryonic Fibroblasts After dsDNA and dsRNA Viral Mimic Stimulation. Front Immunol. 2019;10:836.
23. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019.
24. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180.
25. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18.
26. Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446.
27. Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227.
28. Jezequel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131(3):765–775.
29. Jezequel P, Frenel JS, Campion L, et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford). 2013;2013:bas060.
30. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110.
31. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174.
32. Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971.
33. Pan JH, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6.
34. Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One. 2017;12(7):e0179726.
35. Danaher P, Warren S, Dennis L, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer. 2017;5:18.
36. Sousa S, Maatta J. The role of tumour-associated macrophages in bone metastasis. J Bone Oncol. 2016;5(3):135–138.
37. Kovacs G, Erlandsson R, Boldog F, et al. Consistent chromosome 3p deletion and loss of heterozygosity in renal cell carcinoma. Proc Natl Acad Sci U S A. 1988;85(5):1571–1575.
38. Yakovlev AG, Wang G, Stoica BA, Simbulan-Rosenthal CM, Yoshihara K, Smulson ME. Role of DNAS1L3 in Ca2+- and Mg2+-dependent cleavage of DNA into oligonucleosomal and high molecular mass fragments. Nucleic Acids Res. 1999;27(9):1999–2005.
39. Mizuta R, Araki S, Furukawa M, et al. DNase gamma is the effector endonuclease for internucleosomal DNA fragmentation in necrosis. PLoS One. 2013;8(12):e80223.
40. Bhalla S, Chaudhary K, Kumar R, et al. Gene expression-based biomarkers for discriminating early and late stage of clear cell renal cancer. Sci Rep. 2017;7:44997.
41. Almlof JC, Nystedt S, Leonard D, et al. Whole-genome sequencing identifies complex contributions to genetic risk by variants in genes causing monogenic systemic lupus erythematosus. Hum Genet. 2019;138(2):141–150.
42. Westra HJ, Martinez-Bonet M, Onengut-Gumuscu S, et al. Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes. Nat Genet. 2018;50(10):1366–1374.
43. Diao R, Wei W, Zhao J, Tian F, Cai X, Duan YG. CCL19/CCR7 contributes to the pathogenesis of endometriosis via PI3K/Akt pathway by regulating the proliferation and invasion of ESCs. Am J Reprod Immunol. 2017;78:5.
44. Cheng S, Guo J, Yang Q, Yang X. Crk-like adapter protein regulates CCL19/CCR7-mediated epithelial-to-mesenchymal transition via ERK signaling pathway in epithelial ovarian carcinomas. Med Oncol. 2015;32(3):47.
45. Portero-Sainz I. Gomez-Garcia de Soria V, Cuesta-Mateos C, et al. A high migratory capacity of donor T-cells in response to the lymph node homing receptor CCR7 increases the incidence and severity of GvHD. Bone Marrow Transplant. 2017;52(5):745–752.
46. Braun SE, Chen K, Foster RG, et al. The CC chemokine CK beta-11/MIP-3 beta/ELC/Exodus 3 mediates tumor rejection of murine breast cancer cells through NK cells. J Immunol. 2000;164(8):4025–4031.
47. Hillinger S, Yang SC, Zhu L, et al. EBV-induced molecule 1 ligand chemokine (ELC/CCL19) promotes IFN-gamma-dependent antitumor responses in a lung cancer model. J Immunol. 2003;171(12):6457–6465.
48. Hillinger S, Yang SC, Batra RK, et al. CCL19 reduces tumour burden in a model of advanced lung cancer. Br J Cancer. 2006;94(7):1029–1034.
49. Lu J, Zhao J, Feng H, et al. Antitumor efficacy of CC motif chemokine ligand 19 in colorectal cancer. Dig Dis Sci. 2014;59(9):2153–2162.
50. Itakura M, Terashima Y, Shingyoji M, et al. High CC chemokine receptor 7 expression improves postoperative prognosis of lung adenocarcinoma patients. Br J Cancer. 2013;109(5):1100–1108.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.