Back to Journals » Infection and Drug Resistance » Volume 12
Diversity of Virulence Genes in Multidrug Resistant Escherichia coli from a Hospital in Western China
Authors Li X, Luo Q, Yu X, Zhang Y, Cao X, Li D
Received 5 August 2019
Accepted for publication 16 November 2019
Published 5 December 2019 Volume 2019:12 Pages 3817—3826
DOI https://doi.org/10.2147/IDR.S226072
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eric Nulens
This paper has been retracted
Xue Li, 1,* Qi Luo, 1,* Xinyu Yu, 1 Yanling Zhang, 1 Xiaoyue Cao, 1 Dan Li 2, 3
1Non-Coding RNA and Drug Discovery Key Laboratory of Sichuan Province, Chengdu Medical College, Chengdu, Sichuan 610500, People’s Republic of China; 2School of Medical Laboratory Science, Chengdu Medical College, Chengdu, Sichuan 610500, People’s Republic of China; 3Sichuan Provincial Engineering Laboratory for Prevention and Control Technology of Veterinary Drug Residue in Animal-Origin Food, Chengdu Medical College, Chengdu, Sichuan 610500, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dan Li
School of Medical Laboratory Science, Chengdu Medical College, Chengdu, Sichuan 610500, People’s Republic of China
Tel/Fax +86 28 62739520
Email [email protected]
Background: Escherichia coli strains are the most commonly isolated bacteria in hospitals. The normally harmless commensal E. coli can become a highly adapted pathogen, capable of causing various diseases both in healthy and immunocompromised individuals, by acquiring a combination of mobile genetic elements. Our aim was to characterize E. coli strains from a hospital in western China to determine their virulence and antimicrobial resistance potential.
Methods: A total of 97 E. coli clinical isolates were collected from the First Affiliated Hospital of Chengdu Medical College from 2015 to 2016. Microbiological methods, PCR, and antimicrobial susceptibility tests were used in this study.
Results: The frequency of occurrence of the virulence genes fimC, irp2, fimH, fyuA, lpfA, hlyA, sat, and cnf1 in the E. coli isolates was 93.81, 92.78, 91.75, 84.54, 41.24, 32.99, 28.86, and 7.22%, respectively. Ninety-five (97.9%) isolates carried two or more different virulence genes. Of these, 44 (45.4%) isolates simultaneously harbored five virulence genes, 24 (24.7%) isolates harbored four virulence genes, and 17 (17.5%) isolates harbored six virulence genes. In addition, all E. coli isolates were multidrug resistant and had a high degree of antimicrobial resistance.
Conclusion: These results indicate a high frequency of occurrence and heterogeneity of virulence gene profiles among clinical multidrug resistant E. coli isolates. Therefore, appropriate surveillance and control measures are essential to prevent the further spread of these isolates in hospitals.
Keywords: Escherichia coli, clinical isolates, virulence genes, antimicrobial resistance, MDR
Introduction
Most Escherichia coli strains that colonize the human intestines rarely cause illness in healthy individuals. However, a number of pathogenic strains can cause intestinal or other diseases in healthy, as well as immunocompromised individuals.1 Commensal E. coli strains can evolve into highly adapted pathogens capable of inducing diseases following the acquisition of a combination of mobile genetic elements, including virulence genes.1–3
The occurrence of multidrug resistant (MDR) E. coli strains has increased in recent years, leading to a severe problem in healthcare settings, especially in developing countries.4–6 MDR E. coli strains complicate treatment, as they require prolonged hospitalization and antibiotic treatment and increase the need of surgery, which eventually increase mortality.7,8
E. coli strains have been well documented in healthcare settings in western China; however, their characterization has often been limited to phenotypic tests and the identification of resistance genes,9–13 with limited information regarding their virulence factors. Previously, we examined the virulence gene profiles of 13 diarrheagenic E. coli (DEC) strains isolated from a hospital in western China, as well as the molecular characteristics of their genes.14 Here, we characterized E. coli strains from a hospital in western China and determined their virulence and antimicrobial resistance potential, to better understand the prevalence of virulence genes and antimicrobial resistance in clinical E.coli isolates. This study emphasizes the importance of preventing the spread of E.coli isolates that harbor both antimicrobial resistance and virulence genes in the clinical setting.
Methods
Bacterial Isolates
A total of 97 non-duplicated clinical E. coli isolates were collected from 97 different patients in various departments (gastroenterology, urology, endocrinology, neurosurgery, and other wards) of the First Affiliated Hospital of Chengdu Medical College, Chengdu, Sichuan, China from 2015 to 2016. The isolates were identified using standard laboratory methods and the ATB New system (bioMérieux, Lyons, France). Patients who satisfied the following three criteria were included in the analysis: 1) age >18 years; 2) suspected of having an infection, based on their clinical symptoms (e.g. fever, abdominal pain, nausea, vomiting, dehydration and tenesmus); and 3) their bacterial culture yielded E. coli. The E. coli isolates were collected from biofluid samples including blood, urine, sputum, wound exudates and abscesses. Each isolate was further verified by PCR amplification of a 369-bp internal control region from the E. coli marker gene, alr.15 All bacterial strains were stored at −80 °C and were grown on MacConkey Agar (Oxoid, Hampshire, UK).
The study protocol was approved by the Ethics Committee of Chengdu Medical College, in accordance with the Helsinki declaration. In all cases, the patients or their family members were informed and their written consents was obtained.
Detection of Adherence and Virulence Genes
All E. coli isolates were subjected to PCR to detect 12 adherence (bfp, daaD, daaE, fimC, fimH, aggA, aafA, agg3A, agg4A, lpfA, sfa, and pap) and 27 virulence (aggR, pic, astA, stx1, stx2, eae, ipaH, est, elt, irp2, fyuA, escJ, escN, escV, espP, nleB, nleE, ent/espL2, cnf1, cnf2, cdt-I, cdt-II, invE, hlyA, pet, sat, and subAB) genes. The primers used to amplify these genes are listed in Table 1.
 |
Table 1 Gene Primers Used in This Study |
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of 24 antimicrobial agents against the E. coli isolates were determined by agar dilution methods, according to the 2019 Clinical and Laboratory Standards Institute guidelines.16 The following 24 antimicrobial agents were tested: sulfonamide, doxycycline, tetracycline, cefotaxime, ampicillin, ticarcillin, nalidixic acid, cefoperazone, piperacillin, gentamicin, ciprofloxacin, levofloxacin, ofloxacin, tobramycin, cefoxitin, ceftazidime, minocycline, aztreonam, kanamycin, amikacin, chloramphenicol, meropenem, imipenem, and ertapenem. The results were used to classify the isolates as resistant or susceptible to a particular antibiotic using standard reference values.16
Results
Detection of E. coli Adherence and Virulence Genes
The presence of 12 adherence genes and 27 toxin-encoding genes was examined in all E. coli strains by PCR. As shown in Figure 1, the detection rate of fimC, irp2, fimH, fyuA, lpfA, hlyA, sat, and cnf1 in the isolated E. coli strains was 93.81, 92.78, 91.75, 84.54, 41.24, 32.99, 28.86, and 7.22%, respectively. All isolates were negative for the other genes tested (bfp, daaD, daaE, aggA, aafA, agg3A, agg4A, sfa, pap, aggR, pic, astA, stx1, stx2, eae, ipaH, est, elt, escJ, escN, escV, espP, nleB, nleE, ent/espL2, cnf2, cdt-I, cdt-II, invE, pet, and subAB).
 |
Figure 1 Frequency of virulence genes among E.coli isolates. |
Different combinations of multiple virulence genes were detected in the E. coli isolates. The number of virulence genes in each isolate and the specific virulence gene combinations are shown in Table 2. Two or more different virulence genes were identified in ninety-five (97.94%) isolates. Of these, 44 (45.37%) isolates simultaneously harbored five virulence genes, 24 (24.74%) isolates harbored four virulence genes, 17 (17.53%) isolates harbored six virulence genes, five (5.15%) isolates harbored three virulence genes, two (2.06%) isolates harbored two virulence genes, two (2.06%) isolates harbored seven virulence genes, and only one (1.03%) isolate harbored eight virulence genes.
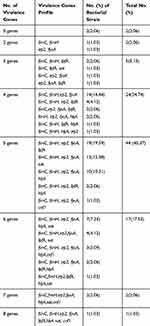 |
Table 2 Distribution of Virulence Genes Among E. coli Isolates |
Resistance to Antimicrobial Agents
The 24 most commonly used antimicrobials in Chinese practice clinical were used in this study to test the antibiotic resistance of the 97 E. coli isolates,14,18–20 including penicillin (ampicillin, ticarcillin, piperacillin), cephems (cefoxitin, cefoperazone, cefotaxime, ceftazidime), monobactams (aztreonam), carbapenems (meropenem, imipenem, ertapenem), aminoglycosides (tobramycin, kanamycin, gentamicin, amikacin, chloramphenicol), tetracyclines (deoxycycline, minocycline, tetracycline), quinolones (levofloxacin, ofloxacin, nalidixic acid, ciprofloxacin).16 The resistance profiles of the E. coli isolates against these 24 antibiotics are detailed in Table 3. The isolates exhibited a high degree of resistance, especially against sulfonamide (97.94%), ampicillin (94.85%), ticarcillin (90.72%), nalidixic acid (90.72%), tetracycline (81.44%), doxycycline (78.49%), ciprofloxacin (70.10%), ofloxacin (68.04%), cefotaxime (68.04%), and levofloxacin (60.82%). Furthermore, all E. coli isolates were susceptible to meropenem and imipenem. The sensitivity rate of the E. coli strains to ertapenem, amikacin, cefoxitin, ceftazidime, aztreonam, and chloramphenicol was 92.79, 88.66, 74.22, 67.01, 67.01, and 64.95%, respectively.
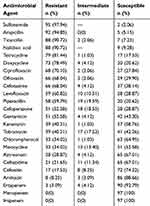 |
Table 3 Antimicrobial Susceptibility of E. coli Clinical Isolates |
Importantly, all isolates were resistant to at least three different classes of antimicrobial agents and were considered as multidrug resistant.17 Of the 97 MDR E. coli isolates, five (5.16%), one (1.03%), one (1.03%), three (3.09%), three (3.09%), six (6.19%), nine (9.28%), six (6.19%), nine (9.28%), twelve (12.37%), nine (9.28%), eight (8.25%), eight (8.25%), four (4.12%), three (3.09%), two (2.06%), three (3.09%), two (2.06%), and three (3.09%) isolates exhibited resistance to 3–21 types of antibiotics, respectively, as shown in Table 4 and Figure 2.
 |
Table 4 Number of E. coli Isolates Resistant to Different Classes of Antibiotics |
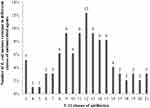 |
Figure 2 Number of E. coli isolates resistant to different classes of antimicrobial agents. |
Frequency of Virulence Gene Occurrence in Isolated E. coli Strains Exhibiting Antimicrobial Resistance
The frequencies of virulence gene occurrence in isolated E. coli strains exhibiting antimicrobial resistance are detailed in Table 5. The frequencies for fimC, irp2, and fimH among the resistant E. coli isolates were nearly > 90%, whereas that of fyuA was > 80%. Moreover, the frequencies of lpfA, hlyA, sat, and cnf1 in the resistant isolates were higher than 40, 30, 20, and 5%, respectively.
 |
Table 5 Frequency of Virulence Genes Among Antibiotic Resistant E. coli Isolates |
Discussion
E. coli strains are the most commonly isolated bacteria in hospitals.18–20 Although these strains have been frequently reported in hospitals in western China, data regarding the virulence genes present in these strains are limited.9–13 Thus, in this study, we investigated the presence of virulence genes and antimicrobial resistance in E. coli strains at a hospital in the western region of China in order to further expand our knowledge of the characteristics of E. coli strains prevalent in China.
We first detected 12 adherence and 27 virulence genes in 97 clinical E. coli isolates. Our results showed that most of the E. coli isolates contained multiple and heterogeneous virulence genes (Table 2). Type 1 fimbriae is an E. coli adhesion factor encoded by the fimC and fimH genes. It enables E. coli to bind to intestinal epithelial cells by attaching on mannose-containing receptors. In our study, fimC and fimH were identified in 93.81 and 91.75% of the strains, respectively. Nuesch-Inderbinen et al21 detected the presence of fimC and fimH in all human E. coli strains isolated in Switzerland, while Malekzadegan and Khashei22 found fimH in all isolates from Iranian patients. These reports are in agreement with our findings; the high frequency of occurrence of fimC and fimH among E. coli strains points to their importance in E. coli adhesion.
Some E. coli strains, contain another type of fimbria, long polar fimbriae (LPF), encoded by the conserved gene lpfA.23,24 We found that 41.24% of the E. coli isolates carried lpfA, which is similar to the frequency (50%) reported in Mexico.25 Initial studies conducted on human biopsy samples have suggested that adherence and the attaching and effacing lesion caused by E. coli do not require LPF.24 Therefore, it is possible that LPF are not necessary for E. coli pathogenicity.
The High-Pathogenicity Island (HPI) marker genes, irp2 and fyuA, were detected in 92.78 and 84.54%, respectively, of E. coli isolates in this study. The irp2 and fyuA genes have been detected in a number of studies examining pathogenic E. coli isolated from humans,26–28 similar to the results of the present study. The iron-uptake system of highly pathogenic strains is mediated via yersiniabactin, which is encoded by irp2 and fyuA and is associated with strain virulence.29,30 A considerable number of bacteria isolated from food harbor irp2 and fyuA (involved in iron capture systems).31,32 This could be the reason for the frequent detection of irp2 and fyuA in pathogenic E. coli isolated from humans.
The hlyA gene was detected in 32.99% of the E. coli isolates. In Iran, Malekzadegan and Khashei22 reported that 28.6% of the E. coli strains were positive for hlyA, whereas Dale et al33 found that 26% of E. coli strains in the UK carried hlyA, and Bozcal et al34 identified this gene in 15.4% of E. coli strains in Turkey. The percentage of E. coli harboring hlyA in our study was higher than detected in the above-mentioned studies. α-hemolysin (HlyA) belongs to a group of pore-forming leukotoxins containing RTX repeats, and is thus consider a virulence factor in E. coli.35–37 Depending on its concentration and the type of cell affected, HlyA either displays cytolytic activity or hijacks innate immune signaling pathways.37–39 The high percent of hlyA in this study suggests that HlyA is involved in the mechanisms underlying E. coli pathogenicity in 32 (32.99%) patients.
The sat gene was detected in 28.86% of E. coli isolates. Sat is frequently detected in pathogenic E. coli strains.5,40,41 As demonstrated by Guignot et al,42 Sat can cause tight junction lesions between epithelial cells, which may lead to an increase in their permeability. These findings indicate that Sat probably plays a role in E. coli pathogenesis in 28 (28.86%) of the patients.
The cnf1 gene was found in seven (7.22%) E. coli isolates, similar to the 7.2% reported in Turkey.34 Moreover, Bouzari et al43 reported that 29.4% of E. coli strains harbor cnf1 genes. Cytotoxic necrotizing factor type 1 (CNF1) is a monomeric protein previously shown to effect rabbit skin cell necrosis and multinucleation of various cultured eukaryotic cells.44–46 Our results are in agreement with the low occurrence of cnf1 in E. coli strains.
We next examined the antimicrobial resistance of the 97 E. coli strains. The E. coli isolates were insensitive to first-line antibiotics such as nalidixic acid, sulfonamide, ticarcillin, ampicillin, tetracycline, doxycycline, ofloxacin, cefotaxime, ciprofloxacin, and levofloxacin (Table 3). The antibiotic resistance rates of the E. coli isolates exceeded those reported in developing countries such as Brazil, Turkey, and Ghana.5,34,47 Moreover, the resistance rates observed in our study were higher than noted in the CHINET project.18–20 Unexpectedly, we found that all E. coli isolates were MDR and over half of them were resistant to > 12 classes of antibiotics (Table 4 and Figure 2). These results highlight the increasing severity of antibiotic misuse in clinical practice in western China.
Carbapenem-resistant Enterobacteriaceae (CRE) are highly prevalent in China, the United States, Italy, Israel, Colombia, Greece, the Indian subcontinent, North Africa, and Turkey.48,49 China (especially the regions of Beijing, Changsha, Chongqing, Fuzhou, Guangzhou, Hangzhou, Hebei, Hong Kong, and Zhengzhou) is thought to be one of main endemic regions of these bacteria around the world.50,51 In our study, we found that three (3.09%) CRE among the 97 E. coli isolates were resistant to ertapenem (Table 3). Carbapenem-resistant E. coli have been frequently reported in western China in recent years;52–54 most probably owing to the use of carbapenems as antimicrobial agents in this region.
Lastly, but most importantly, we found that the E. coli strains harbor a high rate of virulence genes in addition to high antimicrobial resistance (Table 5). These findings explain how the E. coli isolates are able to successfully invade the human body and evade antibiotic treatment. Our findings indicate that clinical MDR E. coli isolates harbor a high frequency of virulence genes and that their virulence gene profiles are highly heterogeneous. Therefore, surveillance and control measures need to be enhanced to prevent these isolates from spreading further in hospitals.
Conclusions
This study demonstrates a high frequency of occurrence and heterogeneity of virulence gene profiles among clinical multidrug resistant E. coli isolates. We conclude that appropriate surveillance and control measures are essential to prevent the further spread of these isolates in hospitals. However, further investigations are needed including additional hospitals in western China and a greater number of E.coli isolates to better understand the prevalence of virulence genes and antimicrobial resistance of the E.coli in western China.
Acknowledgments
This work was supported by grants from the Natural Science Foundation of Sichuan Provincial Education Department (grant 15ZB0239) and the Natural Science Foundation of Chengdu Medical College (grant CYZ11-008). We also thank Professor Xu Jia at Chengdu Medical College for providing technical support and Professor Ying Xu at the First Affiliated Hospital of Chengdu Medical College for supplying E. coli clinical isolates.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi:10.1038/nrmicro818
2. Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi:10.1128/CMR.11.1.142
3. Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–880. doi:10.1128/cmr.00022-13
4. Pathak A, Chandran SP, Mahadik K, et al. Frequency and factors associated with carriage of multi-drug resistant commensal Escherichia coli among women attending antenatal clinics in central India. BMC Infect Dis. 2013;13:2–9. doi:10.1186/1471-2334-13-199
5. Spano LC, da Cunha KF, Monfardini MV, et al. High prevalence of diarrheagenic Escherichia coli carrying toxin-encoding genes isolated from children and adults in southeastern Brazi. BMC Infect Dis. 2017;17:773. doi:10.1186/s12879-017-2872-0
6. Zheng S, Yu F, Chen X, et al. Enteropathogens in children less than 5 years of age with acute diarrhea: a 5-year surveillance study in the Southeast Coast of China. BMC Infect Dis. 2016;16:434. doi:10.1186/s12879-016-1760-3
7. Pop-Vicas A, Tacconelli E, Gravenstein S, et al. Influx of multidrug-resistant, gram-negative bacteria in the hospital setting and the role of elderly patients with bacterial bloodstream infection. Infect Control Hosp Epidemiol. 2009;30:325–331. doi:10.1086/596608
8. Kollef MH. Introduction: update on the appropriate use of meropenem for the treatment of serious bacterial infections. Clin Infect Dis. 2008;47:1–2. doi:10.1086/590060
9. Zhang L, Lu X, Zong Z. The emergence of blaCTX-M-15-carrying Escherichia coli of ST131 and new sequence types in Western China. Ann Clin Microbiol Antimicrob. 2013;12:35. doi:10.1186/1476-0711-12-35
10. Long H, Feng Y. The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci Rep. 2019;9:696. doi:10.1038/s41598-018-37125-1
11. Liu L, Feng Y, McNally A, et al. blaNDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J Antimicrob Chemother. 2018;73:2336–2339. doi:10.1093/jac/dky226
12. Ma K, Feng Y, Zong Z. Fitness cost of a mcr-1-carrying IncHI2 plasmid. Front Microbiol. 2018;13:331. doi:10.1371/journal.pone.0209706
13. Wu W, Feng Y. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32:115–118. doi:10.1128/cmr.00115-18
14. Li D, Shen M, Xu Y, et al. Virulence gene profiles and molecular genetic characteristics of diarrheagenic Escherichia coli from a hospital in western China. Gut Pathog. 2018;10:35. doi:10.1186/s13099-018-0262-9
15. Preethirani PL, Isloor S, Sundareshan S, et al. Isolation, biochemical and molecular identification, and in-vitro antimicrobial resistance patterns of bacteria isolated from bubaline subclinical mastitis in South India. PLoS ONE. 2015;10:e0142717. doi:10.1371/journal.pone.0142717
16. CLSI. Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. 2019.
17. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi:10.1111/j.1469-0691.2011.03570.x
18. Fupin HU, Zhu D, Wang F, et al. CHINET 2014 surveillance of bacterial resistance in China. Chin J Infect Chemother. 2015;15:401–410.
19. Fupin HU, Zhu D, Wang F, et al. CHINET 2015 surveillance of bacterial resistance in China. Chin J Infect Chemother. 2016;16:685–694. doi:10.16718/j.1009-7708.2016.06.003
20. Hu F, Guo Y, Zhu D, et al. CHINET surveillance of bacterial resistance across China: report of the results in 2016. Chin J Infect Chemother. 2017;17:481–491. doi: 10.16718/j.1009-7708.2017.05.001.
21. Nuesch-Inderbinen M, Cernela N, Wuthrich D, et al. Genetic characterization of Shiga toxin producing Escherichia coli belonging to the emerging hybrid pathotype O80: h2isolated from humans 2010–2017 in Switzerland. Int J Med Microbiol. 2018;308:534–538. doi:10.1016/j.ijmm.2018.05.007
22. Malekzadegan Y, Khashei R. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infect Dis. 2018;18:572. doi:10.1186/s12879-018-3467-0
23. Torres AG, Kanack KJ, Tutt CB, et al. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157: h7and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol Lett. 2004;238:333–344. doi:10.1016/j.femsle.2004.07.053
24. Tatsuno I, Mundy R, Frankel G, et al. The lpf gene cluster for long polar fimbriae is not involved in adherence of enteropathogenic Escherichia coli or virulence of Citrobacter rodentium. Infect Immun. 2006;74:265–272. doi:10.1128/iai.74.1.265-272.2006
25. Munhoz DD, Nara JM, Freitas NC, et al. Distribution of major pilin subunit genes among atypical enteropathogenic Escherichia coli and influence of growth media on expression of the ecp operon. Front Microbiol. 2018;9:942. doi:10.3389/fmicb.2018.00942
26. Schubert S, Cuenca S, Fischer D, et al. High-pathogenicity island of Yersinia pestis in enterobacteriaceae isolated from blood cultures and urine samples: prevalence and functional expression. J Infect Dis. 2000;182:1268–1271. doi:10.1086/315831
27. Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–272. doi:10.1086/315217
28. Cespedes S, Saitz W, Del Canto F, et al. Genetic diversity and virulence determinants of Escherichia coli strains isolated from patients with Crohn’s disease in Spain and Chile. Front Microbiol. 2017;8:639. doi:10.3389/fmicb.2017.00639
29. Schubert S, Rakin A, Karch H, et al. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect Immun. 1998;66:480–485.
30. Carniel E. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 2001;3:561–569. doi:10.1016/S1286-4579(01)01412-5
31. Johnson JR, Delavari P, O’Bryan TT, et al. Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2005;2:38–49. doi:10.1089/fpd.2005.2.38
32. Johnson JR, McCabe JS, White DG, et al. Molecular analysis of Escherichia coli from retail meats (2002–2004) from the United States National antimicrobial resistance monitoring system. Clin Infect Dis. 2009;49:195–201. doi:10.1086/599830
33. Dale AP, Pandey AK, Hesp RJ, et al. Genomes of Escherichia coli bacteraemia isolates originating from urinary tract foci contain more virulence-associated genes than those from non-urinary foci and neutropaenic hosts. J Infect. 2018;77:534–543. doi:10.1016/j.jinf.2018.10.011
34. Bozcal E, Eldem V, Aydemir S, et al. The relationship between phylogenetic classification, virulence and antibiotic resistance of extraintestinal pathogenic Escherichia coli in Izmir province, Turkey. Peer J. 2018;6:e5470. doi:10.7717/peerj.5470
35. Welch RA. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi:10.1111/mmi.1991.5.issue-3
36. Bhakdi S, Bayley H, Valeva A, et al. Staphylococcal alpha-toxin, streptolysin-O, and Escherichia coli hemolysin: prototypes of pore-forming bacterial cytolysins. Arch Microbiol. 1996;165:73–79. doi:10.1007/s002030050300
37. Linhartova I, Bumba L, Masin J, et al. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010;34:1076–1112. doi:10.1111/j.1574-6976.2010.00231.x
38. Gur C, Coppenhagen-Glazer S, Rosenberg S, et al. Natural killer cell-mediated host defense against uropathogenic E. coli is counteracted by bacterial hemolysinA-dependent killing of NK cells. Cell Host Microbe. 2013;14:664–674. doi:10.1111/j.1574-6976
39. Wiles TJ, Mulvey MA. The RTX pore-forming toxin alpha-hemolysin of uropathogenic Escherichia coli: progress and perspectives. Future Microbiol. 2013;8:73–84. doi:10.2217/fmb.12.131
40. Mansan-Almeida R, Pereira AL, Giugliano LG. Diffusely adherent Escherichia coli strains isolated from children and adults constitute two different populations. BMC Microbiol. 2013;13:22. doi:10.1186/1471-2180-13-22
41. Lima IF, Boisen N, Quetz Jda S, et al. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern Brazil. J Med Microbiol. 2013;62:683–693. doi:10.1099/jmm.0.054262-0
42. Guignot J, Chaplais C, Coconnier-Polter MH, et al. The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell Microbiol. 2007;9:204–221. doi:10.1111/j.1462-5822.2006.00782.x
43. Bouzari S, Oloomi M, Oswald E. Detection of the cytolethal distending toxin locus cdtB among diarrheagenic Escherichia coli isolates from humans in Iran. Res Microbiol. 2005;156:137–144. doi:10.1016/j.resmic.2004.09.011
44. Caprioli A, Falbo V, Roda LG, et al. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect Immun. 1983;39:1300–1306.
45. Caprioli A, Donelli G, Falbo V, et al. A cell division-active protein from E. coli. Biochem Biophys Res Commun. 1984;118:587–593. doi:10.1016/0006-291X(84)91343-3
46. De Rycke J, Gonzalez EA, Blanco J, et al. Evidence for two types of cytotoxic necrotizing factor in human and animal clinical isolates of Escherichia coli. J Clin Microbiol. 1990;28:694–699.
47. Forson AO, Tsidi WB, Nana-Adjei D, et al. Correction to: Escherichia coli bacteriuria in pregnant women in Ghana: antibiotic resistance patterns and virulence factors. BMC Res Notes. 2019;12:29. doi:10.1186/s13104-019-4057-y
48. Gupta N, Limbago BM, Patel JB, et al. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. doi:10.1093/cid/cir202
49. Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821–830. doi:10.1111/1469-0691.12719
50. Qin S, Fu Y, Zhang Q, et al. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother. 2014;58:4275–4282. doi:10.1128/aac.02813-13
51. Berrazeg M, Diene S, Medjahed L, et al. New Delhi Metallo-beta-lactamase around the world: an eReview using Google Maps. Euro Surveill. 2014;19:
52. Zong Z, Fenn S, Connor C, et al. Complete genomic characterization of two Escherichia coli lineages responsible for a cluster of carbapenem-resistant infections in a Chinese hospital. J Antimicrob Chemother. 2018;73:2340–2346. doi:10.1093/jac/dky210
53. Hu Y, Liu L, Zhang X, et al. In vitro activity of neomycin, streptomycin, paromomycin and apramycin against carbapenem-resistant enterobacteriaceae clinical strains. Front Microbiol. 2017;8:2275. doi:10.3389/fmicb.2017.02275
54. Liu L, Feng Y, Zhang X, et al. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother. 2017;61:e01757–17. doi:10.1128/AAC.01757-17
55. Muller D, Greune L, Heusipp G, et al. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol. 2007;73:3380–3390. doi:10.1128/AEM.02855-06
56. Antikainen J, Tarkka E, Haukka K, et al. New 16-plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. Eur J Clin Microbiol Infect Dis. 2009;28:899–908. doi:10.1007/s10096-009-0720-x
57. Chandra M, Cheng P, Rondeau G, et al. A single step multiplex PCR for identification of six diarrheagenic E. coli pathotypes and Salmonella. Int J Med Microbiol. 2013;303:210–216. doi:10.1016/j.ijmm
58. J Y O, Kang MS, Yoon H, et al. The embryo lethality of Escherichia coli isolates and its relationship to the presence of virulence-associated genes. Poult Sci. 2012;91:370–375. doi:10.3382/ps.2011-01807
59. Lopes LM, Fabbricotti SH, Ferreira AJ, et al. Heterogeneity among strains of diffusely adherent Escherichia coli isolated in Brazil. J Clin Microbiol. 2005;43:1968–1972. doi:10.1128/JCM.43.4.1968-1972.2005
60. Boisen N, Struve C, Scheutz F, et al. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76:3281–3292. doi:10.1128/IAI.01646-07
61. Prorok-Hamon M, Friswell MK, Alswied A, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761–770. doi:10.1136/gutjnl-2013-304739
62. Bai X, Zhao A, Lan R, et al. Shiga toxin-producing Escherichia coli in yaks (Bos grunniens) from the Qinghai-Tibetan Plateau, China. PLoS ONE. 2013;8:e65537. doi:10.1371/journal.pone.0065537
63. Pass MA, Odedra R, Batt RM. Multiplex PCRs for identification of Escherichia coli virulence genes. J Clin Microbiol. 2000;38:2001–2004.
64. Barletta F, Ochoa TJ, Cleary TG. Multiplex real-time PCR (MRT-PCR) for diarrheagenic. Methods Mol Biol. 2013;307–314. doi:10.1007/978-1-60327-353-4_21
65. Chakraborty S, Deokule JS, Garg P, et al. Concomitant infection of enterotoxigenic Escherichia coli in an outbreak of cholera caused by Vibrio cholerae O1 and O139 in Ahmedabad, India. J Clin Microbiol. 2001;39:3241–3246. doi:10.1128/JCM.39.9.3241-3246.2001
66. Arikawa K, Meraz IM, Nishikawa Y, et al. Interleukin-8 secretion by epithelial cells infected with diffusely adherent Escherichia coli possessing Afa adhesin-coding genes. Microbiol Immunol. 2005;49:493–503. doi:10.1111/j.1348-0421.2005.tb03754.x
67. Kyaw CM, De Araujo CR, Lima MR, et al. Evidence for the presence of a type III secretion system in diffusely adhering Escherichia coli (DAEC). Infect Genet Evol. 2003;3:111–117. doi:10.1016/S1567-1348(03)00008-X
68. Coombes BK, Wickham ME, Mascarenhas M, et al. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl Environ Microbiol. 2008;74:2153–2160. doi:10.1128/AEM.02566-07
69. Patzi-Vargas S, Zaidi MB, Perez-Martinez I, et al. Diarrheagenic Escherichia coli carrying supplementary virulence genes are an important cause of moderate to severe diarrhoeal disease in Mexico. PLoS Negl Trop Dis. 2015;9:e0003510. doi:10.1371/journal.pntd.0003510
70. Arikawa K, Nishikawa Y. Interleukin-8 induction due to diffusely adherent Escherichia coli possessing Afa/Dr genes depends on flagella and epithelial Toll-like receptor 5. Microbiol Immunol. 2010;54:491–501. doi:10.1111/j.1348-0421.2010.00244.x
71. Paton AW, Paton JC. Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli strains producing the novel subtilase cytotoxin. J Clin Microbiol. 2005;43:2944–2947. doi:10.1128/JCM.43.6.2944-2947.2005
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
