Back to Journals » Infection and Drug Resistance » Volume 15
Distribution of mcr-1 Harboring Hypervirulent Klebsiella pneumoniae in Clinical Specimens and Lytic Activity of Bacteriophage KpnM Against Isolates
Authors Aslam B, Siddique MH, Siddique AB , Shafique M , Muzammil S, Khurshid M , Rasool MH, Ahmad M, Chaudhry TH, Amir A , Salman M, Baloch Z, Alturki NA, Alzamami A
Received 25 May 2022
Accepted for publication 23 September 2022
Published 1 October 2022 Volume 2022:15 Pages 5795—5811
DOI https://doi.org/10.2147/IDR.S374503
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Bilal Aslam,1 Muhammad Hussnain Siddique,2 Abu Baker Siddique,1 Muhammad Shafique,1 Saima Muzammil,1 Mohsin Khurshid,1 Muhammad Hidayat Rasool,1 Moeed Ahmad,1 Tamoor Hamid Chaudhry,3 Afreenish Amir,3 Muhammad Salman,3 Zulqarnain Baloch,4 Norah A Alturki,5 Ahmad Alzamami6
1Department of Microbiology, Government College University Faisalabad, Faisalabad, Pakistan; 2Department of Bioinformatics & Biotechnology, Government College University Faisalabad, Faisalabad, Pakistan; 3Public Health Laboratories Division, National Institute of Health, Islamabad, Pakistan; 4Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, People’s Republic of China; 5Clinical Laboratory Science Department, College of Applied Medical Science, King Saud University, Riyadh, Saudi Arabia; 6Clinical Laboratory Science Department, College of Applied Medical Science, Shaqra University, AlQuwayiyah, Saudi Arabia
Correspondence: Bilal Aslam, Department of Microbiology, Government College University Faisalabad, Faisalabad, Pakistan, Email [email protected] Ahmad Alzamami, Clinical Laboratory Science Department, College of Applied Medical Science, Shaqra University, AlQuwayiyah, Saudi Arabia, Email [email protected]
Background: The World Health Organization (WHO) has declared the multi-drug resistant (MDR) Klebsiella pneumoniae as one of the critical bacterial pathogens. The dearth of new antibiotics and inadequate therapeutic options necessitate finding alternative options. Bacteriophages are known as enemies of bacteria and are well-recognized to fight MDR pathogens.
Methods: A total of 150 samples were collected from different clinical specimens through a convenient sampling technique. Isolation, identification, and antibiotic susceptibility testing (AST) of K. pneumoniae were done by standard and validated microbiological procedures. Molecular identification of virulence factors and antibiotic resistance genes (ARGs) was carried out through polymerase chain reaction (PCR) by using specific primers. For bacteriophage isolation, hospital sewage samples were processed for phage enrichment, purification, and further characterization ie, transmission electron microscopy (TEM) and stability testing, etc. followed by evaluation of the lytic potential of the phage.
Results: Overall, a total of 41% of isolates of K. pneumoniae were observed as hypervirulent K. pneumoniae (hvKp). Among hvKp, a total of 12 (42%) were detected as MDR hvKp. A total of 37% of all MDR isolates were found resistant to colistin, and 66% of the colistin resistance isolates were recorded as mcr-1 positive. Isolated phage KpnM had shown lytic activity against 53 (79%) K. pneumoniae isolates. Remarkably, all 8 mcr-1 harboring MDR hvKp and non-hvKp isolates were susceptible to KpnM phage.
Conclusion: Significant distribution of mcr-1 harboring hypervirulent Klebsiella pneumoniae was observed in clinical specimens, which is worrisome for the health system of the country. Characterized phage KpnM exhibited encouraging results and showed the lytic activity against the mcr-1 harboring hvKp isolates, which may be used as a prospective alternative control strategy to fight this ominous bacterium.
Keywords: Klebsiella pneumoniae, bacteriophages, colistin, MDR, virulence
A Letter to the Editor has been published for this article.
A Response to Letter by Dr Panjaitan has been published for this article.
Introduction
At present, resistance to all classes of antibiotics is a serious health threat across the globe. The emergence of multidrug-resistant (MDR) bacterial pathogens is linked with different infections that are difficult to treat and pose a substantial economic effect on the healthcare settings of low-and middle-income countries (LMICs) like Pakistan. Moreover, various healthcare regulatory bodies of the world like the World Health Organization (WHO), Centers for Disease Control and Prevention USA, and the European Center for Disease Prevention and Control have listed and prioritized these MDR bacteria, which are considered significant health concerns worldwide.1
Klebsiella pneumoniae is one of these listed and prioritized bacteria, which is a frequent etiology of nosocomial infections. It is a member of Enterobacteriaceae, a non-motile, gram-negative bacterium with metabolic flexibility and a comparatively large genome that permits it to cause a variety of infections.2,3 In addition to a classical K. pneumoniae (cKp) recently emerged hypervirulent phenotype of K. pneumoniae is also considered a health challenge. The cKp is an important opportunistic nosocomial pathogen and is associated with hospital-acquired infections like wound infections, urinary tract infections (UTIs), and pneumonia. Various clinical manifestations linked with hypervirulent K. pneumoniae (hvKp) include eye or lung infection and liver abscess which may be developed into metastasis. Moreover, severe skin and soft tissue infections, epidural abscesses, and bone infections are considered some common clinical conditions of hvKp as well.4,5 Therefore, WHO declared that MDR K. pneumoniae has epidemic potential in health care systems.
In clinical settings, K. pneumoniae is a proven resistant pathogen against almost every antibiotic class by harnessing both intrinsic as well as acquired mechanisms of resistance. It has chromosomal mediated gene blaSHV which is responsible for intrinsic resistance, whereas, in the case of acquired resistance, K. pneumoniae use to produce extended-spectrum -lactamase (ESBLs) and carbapenemases that are associated with the resistance to different generations of cephalosporins and carbapenems.6,7 Moreover, K. pneumoniae displayed resistance against colistin due to the presence of plasmid-mediated mobilized colistin resistance (mcr) genes.8 Due to the emergence of resistant variants and a dearth of new antibiotics, treatment options for MDR K. pneumoniae are limited. This limited therapeutic regime warrants the exploitation of alternative approaches.
Bacteriophages are natural predators of bacteria with the self-replicating ability and rigorous specificity to spot and destroy the bacterial host by taking over the cellular machinery. Different structural components of bacteriophages responsible for the lysis of host bacterium include endolysins, holins, and spanins. These play a significant part in the assembly and release of the bacteriophages from host bacteria. The endolysins degrade the peptidoglycans, whereas holins and spanins disrupt the cell membrane. These specific particles efficiently kill the bacteria without doing any harm to the microbiota, so unlike antibiotics, they have no side effects eg, antibiotic-associated diarrhea. Additionally, bacteriophages neither produce any type of toxins nor disseminate antibiotic resistance genes (ARGs).9,10
Current incidence and inadequate treatment choices for MDR hvKp necessitate finding potential alternative approaches. The use of phages has been well reported and is a proven strategy across the globe to fight MDR pathogens. So, the present study was designed to estimate the distribution of mcr-1 harboring hypervirulent K. pneumoniae in clinical specimens and to evaluate the lytic potential of bacteriophages against the isolates, which may be a promising future therapeutic strategy against MDR K. pneumoniae.
Materials and Methods
Ethical Consideration
The study was approved by the Ethical Review Board (ERB), Government College University, Faisalabad Pakistan (letter No. GCUF/ERB/21/47). As the isolates in this study were a part of the routine laboratory procedure, the ERB exempted this research from informed consent.
K. pneumoniae Isolation and Identification
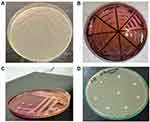
A total of 150 samples were collected through convenient sampling by employing standard microbiological sterile conditions. Sterile containers were used to collect the samples from different clinical sources including pus exudates, sputum, tracheal aspirates, catheters, and diabetic wound swabs. Initially, samples were inoculated on Petri dishes of MacConkey agar, plates were incubated for 24 hours at 37°C Figure 1A.11 Pink mucoid colonies that appeared on MacConkey agar were transferred on the selective K. pneumoniae agar (HiCromeTM Klebsiella selective agar M1573-500G) Petri plates, which were incubated for 24 hours at 37°C Figure 1B.12 Purified Purple colonies from the selective base were utilized for phage/host characterization. Biochemical characterization of K. pneumoniae was performed through API 20E kit API (BioMérieux, France).
Identification of Hypervirulent K. pneumoniae Isolates
Phenotypic identification of hypervirulent isolates was done through the string test method previously described.13 Briefly, overnight broth culture was streaked over MacConkey agar and incubated at 37°C for 24 hours. Fresh colonies were then touched and stretched gently over the surface by a sterilized platinum loop. The extended string was observed from the edge of the bacterial colony. For positive results, a string length of >5mm was observed for each isolate Figure 1C.
Virulence Genes Detection
For molecular detection, isolates were subjected to PCR. Different genes which were included in the study were wzy, rmpA, and fimH, details are given in the (Table 1). Subsequently, agarose gel electrophoresis, and documentation were performed for the analysis of PCR amplicons using a gel documentation system (BioRad®, USA).
 |
Table 1 Distribution of Virulence Genes and ARGs Among hvKp Isolates Along with the Details of the Primer Used in the Study |
Antimicrobial Susceptibility Testing (AST)
Kirby Bauer Disc diffusion test was used for the AST of the isolates.14 Briefly, suspensions were prepared from overnight grown bacterial culture. Exactly, a 0.5 McFarland standard (approximately 1–2×108 CFU/mL) was attained by mixing the bacterial colonies in sterile (0.85%) saline. The concentration was measured in HITACHI UH5300 UV-Vis/NIR Spectrophotometer according to the manufacturer’s instructions. The bacterial lawn was made over Muller–Hinton agar (Oxoid, UK) plates for further testing Figure 1D.15
Antibiotic susceptibility was determined against different classes of antibiotics. After the placement of antibiotic disks, the plates were incubated at 37 oC for 24 hours. Already characterized MDR K. pneumoniae isolates with GenBank no. MF953600 & MF953599 were kept as positive control. The zones of inhibition were observed for different antibiotics and analyzed according to CLSI 2020 criteria for interpretation of results as shown in Figure 1D.
Detection of ARGs
Extracted and quantified DNA (NanoDrop™) of K. pneumoniae was subjected to PCR and a number of ARGs were amplified by using specific primers. The PCR reaction mixture contained; 5 μL of sample DNA, 2 μL of 100 pM primers (F&R), 10 μL of 2X DreamTaq (Thermo-Scientific™), and 8 μL of Nuclease-free water (Ambion- AM9932) to adjust the final volume of 25 μL. The PCR reaction was carried out according to the specific annealing temperature for each of the ARGs listed in (Table 1). Afterward, agarose gel electrophoresis was performed to examine the PCR amplicons in a gel analyzer (BioRad, USA).
Rapid Polymyxin Test for Colistin-Resistant Isolates
The test was previously designed to detect colistin resistance associated with plasmid-acquired mcr-1 and mcr-2 genes.16 Briefly, 0.2 mg/mL colistin solution was prepared in Muller Hinton broth (MHB). The pH indicator phenol red at a concentration of 0.0125 g was in rapid polymyxin NP solution. Further, the pH of the rapid solution was maintained at 6.7 and confirmed by a litmus paper test. Filtered 10% anhydrous D-glucose was added in a rapid solution. Just before starting the procedure, colistin was added to the rapid polymyxin NP solution to attain a colistin-concentrated solution @ 5 μg/150μL. For this purpose, C1v1=C2V2 was applied to calculate the volume V1 of the stock solution having an initial concentration C1 of 0.2 mg/mL required to produce 5 μg /mL final colistin concentration C2 with a final volume V2 of 250 mL.
PCR Confirmation of mcr-1 Harboring of Klebsiella pneumoniae
Briefly, The PCR was performed in T3000 Thermocycler-48 (Biomerta, Germany) with the following conditions specific for mcr-1: 5 minutes of initial DNA denaturation at 94°C followed by 25 cycles of denaturation for 1 minute at 94°C. Primer Annealing for 90 secs at 61 °C followed by primer-specific extension for 1 minute at 72°C. The final extension was done for 10 minutes at 72°C.17
Bacteriophage Isolation
Sample Collection
To isolate the bacteriophages, hospital wastewater samples were collected from hospitals located in different localities of District Faisalabad Punjab Pakistan. Concisely, 500 mL of wastewater was collected in sterile containers and transported to the lab. Samples were placed to wait for 3 hours which facilitated the debris to settle down. Further, samples were treated with 1% chloroform, which destroy the unwanted bacterial contamination and facilitated the phage detachment from the contaminants.18
Bacteriophage Enrichment Assay
The bacteriophage enrichment assay was performed with several modifications as described previously.19 Briefly, 10 mL of already prepared 10X L.B broth media was first transferred to a 250 mL conical flask. 90 mL of wastewater containing potential bacteriophages was poured into each flask with 1X the final concentration of LB broth. A total of 50 µL of each bacterial broth grown at the early exponential phase was inoculated. The flasks were placed into a shaking incubator at 50–70 rpm with a temperature of 37 oC for 24 hours. After 24 hours of incubation 1.5 mL of broth from each flask was transferred to sterile Eppendorf tubes. The tubes were centrifuged at 12,000 rpm for 10 minutes. After centrifugation, the supernatant without filtration at this initial stage was utilized for spot test and plaque Assay explained further in later sections.
Host Range Determination
A spot test was performed to determine the host range of bacteriophages. Briefly, freshly grown overnight K. pneumoniae isolates were inoculated over LB agar plates. A total of 5µL of bacteriophage suspension was spotted over a freshly inoculated bacterial lawn. After drying the plates were placed in an incubator at 37 oC for 24 hours. The clear inhibition zones were observed after 8,16 and 24 hours.20 For the determination of inter-species hot range spot test was also performed against E. coli, P. aeruginosa, and S. Typhi.
Plaque Assay and Phage Purification
Plaque assay was performed for purification of bacteriophage and determination of average plaque formation time and plaque size through the double agar overlay method.21 Briefly, bacteriophage suspension from the enrichment assay was subjected to filtration through a 0.22 µm size cellulose filter. The filtrate was diluted by adding PBS till 10−11. A total of 900 μL of diluted phage suspension along with K. pneumoniae (100 μL) isolate was dispensed to LB soft 0.5% agar and poured onto the bottom 1.5% agar, this whole content was incubated for 24 hours at 37 oC. Afterward, the single plaques with different morphologies were picked with a sterile syringe and propagated with susceptible bacterial isolate for 24 hours at 37°C. After propagation centrifugation was done at 12,000 rpm for 10 minutes followed by filtration through 0.22 µm size filters for further characterization.22
Transmission Electron Microscopy (TEM)
To study the morphological characteristics of isolated bacteriophages TEM was performed. The TEM analysis was carried out at the National Institute of Biotechnology and Genetic Engineering (NIBGE), Punjab Pakistan. The filtered phage suspension was fixed over a formvar-carbon coated grid Cu Mesh 300, fixed with glutaraldehyde (1%), stained with the standard negative staining using uranyl acetate (2%), and examined by an EM 10C microscope (Zeiss, Germany) at 100 kV.23
Bacteriophage Stability
To check the bacteriophage stability various ranges of pH, temperature, and chloroform were used as previously described with minor adjustments.24 Different temperature ranges to observe the thermal stability include 0 °C, 4 °C, 15 °C, 37 °C, 48 °C, 55°C. Bacteriophage at 108 PFU/mL for 4 hours was incubated at these temperature ranges. Downstream, the double agar overlay method was performed to estimate the phage titer.25
The pH stability was determined at various pHs like 2, 4, 7, 9, and 11. The phage suspension 108 PFU/mL at these pH ranges was incubated for 4 hours. Likewise, the double agar overlay method was performed to determine the phage titer.
Bacteriophage stability was tested against various concentrations of chloroform as well which include 1%, 1. 5%, and 2%. A total of 100 µL of phage suspension (106 PFU/mL) was dispensed in 900 µL PBS along with these chloroform concentrations separately and survived phage titer was determined as PFU/mL.
Phage Adsorption Assay
The grown culture of phage susceptible K. pneumoniae was maintained at 108 CFU/mL in Mueller Hinton broth.24 This culture was mixed with an equal volume of phage (106 PFU/mL) suspension and incubated for 5- and 10-minutes intervals at 37°C. Subsequently, centrifugation at 10,000 × g for 5 min was done, followed by filtration (0.22 μm). Again double-layer agar method was performed to estimate the free phages. The reduced phage titer was taken as phage adsorption to the host bacterium.
One-Step Growth Curve
For this purpose, K. pneumoniae culture at exponential phase (~2× 108 CFU/mL) was used. At this point, phage with MOI of 0.1 was mixed with the culture and incubated for 10 minutes at 37°C. Afterward, centrifugation was performed to discard the unadsorbed phages and K. pneumoniae culture was resuspended in LB, then incubation was done in a shaking incubator @ 180 rpm at 37°C. The release of intracellular phages was facilitated by adding 1% chloroform, double-layer agar plate method was carried out to quantify the phage titer.26
Biofilm Inhibition and Eradication Assay
The phages isolated against K. pneumoniae were also tested for their ability to inhibit and eradicate biofilm production in vitro as previously described.27 Briefly, freshly grown bacterial culture at OD600 = 0.5 diluted with L.B broth at factor 10:100 was added to the first column of 96 well micro-titer plates followed by 100 µL of phage suspension at MOI of 0.01 to each well of the first column.28 The plates were incubated for 8 hours at 37°C. Biofilm eradication ability was determined by direct treatment of bacteriophage to mature biofilm. First, the planktonic cells were removed from the first column of 96 well plates after 8 hours of incubation. A total of 200 µL of phage suspension at 108 PFU/mL was added to each well of the first column and incubated again for 8 hours. After 8 hours the columns were washed three times with distilled water and air-dried for 10 minutes. A 2% crystal violet suspension was added to each well and incubated for 15 minutes followed by washing with distilled water. A total of 200 µL of ethanol was then added to each well to extract the absorbed CV onto biofilm. For positive control, bacterial suspension broth was added to column 2. The OD600 was then determined by using an ELISA plate reader (Multiskan™ FC Microplate Photometer Thermo Scientific™). The decrease in OD as compared to the positive control was analyzed.29
Results
Distribution of K. pneumoniae
Overall, K. pneumoniae was isolated from 67 (44%) samples. The highest incidence (48%) was estimated among catheter tips followed by pus samples and tracheal aspirates (47%) respectively. Whereas 42% of incidence was recorded from infectious wound swabs and 35% was detected in sputum samples, which was the lowest among various sample sources.
A total of 28 (41%) K. pneumoniae isolates were observed as hypervirulent K. pneumoniae (hvKp) Figure 1. Sample-wise distribution depicted that hvKp 9 (52%) isolates were from catheter tips followed by 8 (44%) from pus. Furthermore, 6 (35%) hvkp isolates were detected from infectious wound samples. Only 2 (28%) hvKp isolates were obtained from sputum and 3 (37%) were from tracheal aspirates (Table 2). Molecular detection of virulence factors was done through PCR; the incidence of virulence genes is given in Table 1.
 |
Table 2 Sample-Wise Distribution of Mcr-1 Harboring K. Pneumoniae (hvKp/Non-hvKp) |
Antibiotic Susceptibility Profiling
Among All 67 K. pneumoniae isolates 32 (47%) isolates were detected as MDR Klebsiella pneumoniae Figure 1D. Sample-wise distribution of MDR isolates revealed that a total of 10 (58%) MDR K. pneumoniae were detected from infectious wounds followed by 9 (50%) from pus and 8 (47%) from catheter tips. Overall, 2 (28%) isolates were detected as MDR from sputum, and 3 (37%) were from tracheal aspirates. Additionally, among 28 hvKp isolates total of 12 (42%) were detected as MDR hvKp. Whereas the sample-wise distribution of MDR hvKp isolates showed that a total of 3 (50%) were from infectious wounds and 4(50%) were from pus samples. A total of 4 (44%) MDR hvKp were detected from catheter tips and 1 (33%) MDR hvKp isolate was detected from sputum and tracheal aspirates (Table 2). All the isolates were subjected to the detection of ARGs, detailed distribution of ARGs is described in Table 1.
Among different classes of antibiotics used in this study maximum resistance was determined against ampicillin 92% and amoxicillin (88%). Furthermore, among cephalosporins, the maximum resistance was recorded against cefoxitin and cefotaxime which was (82%) and (81%) respectively. However, the minimum resistance was recorded against colistin (37%), imipenem, and gentamicin (48%) and (54%) respectively Figure 2A. Additionally, all 32 MDR isolates were subjected to a rapid polymyxin test. The color of the rapid polymyxin NP solution changed from orange to yellow after 5–6 hours of incubation. A total of 12 (37%) of all MDR isolates were resistant to colistin in the rapid polymyxin test Figure 2B.
Genotypic Characterization of Colistin Resistance
All 12 colistin resistance isolates were further characterized for the detection of the genotypic determinant of colistin resistance ie, the mcr-1 gene. A total of 8 (66%) of the colistin resistance isolates were positive for the mcr-1 gene Figure 2C. Among 12 MDR hvKp isolates 5 (41%) were detected as mcr-1 harboring MDR hvKp. Whereas a total of 3 (25%) mcr-1 harboring K. pneumoniae were among non-hvKp MDR isolates.
Bacteriophage Isolation and Host Range Determination
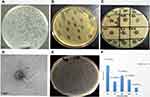
Overall, 15 (30%) wastewater samples were positive for K. pneumoniae lytic bacteriophage. All bacteriophages isolated from different samples were subjected to spot tests for the determination of the host range Figure 3B. The bacteriophage with maximum host range was then selected for further characterization Figure 3C and D. Phage KpnM had shown maximum host range against K. pneumoniae isolates. Among all 67 isolates, phage KpnM had shown lytic activity against 53 (79%) K. pneumoniae isolates. Furthermore, among 32 MDR K. pneumoniae, 26 (81%) were susceptible to KpnM phage while the other 33 phage-susceptible isolates were non-MDR K. pneumoniae. Remarkably, all 8 mcr-1 harboring MDR hvKp and non-hvKp isolates were susceptible to KpnM phage (Table 3). The comparative analysis of phage titer against colistin-resistant MDR Klebsiella pneumoniae isolates is shown in Figure 3F.
 |
Table 3 Comparative Analysis of Colistin Resistance with PCR Results of Mcr-1 Gene, Spot Test, and hvKp Isolates |
Plaque Assay
The susceptible isolates were also subjected to plaque assay by the double agar overlay method. The average plaque formation time was determined as 8–10 hours. The average plaque size was 0.5 to 3 mm Figure 3A. The Plaque Assay against the colistin-resistant MDR hvKp isolates is shown in Figure 3E.
Phage Stability Test
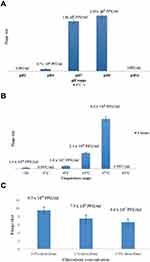
The stability of KpnM was determined under various environmental conditions including various ranges of pH, and temperature. In addition to that, chloroform stability was also determined. The bacteriophages were treated at various pH ranges 2, 4, 7, 9, and 11. After four hours of incubation at various pH ranges, bacteriophages were then directly subjected to plaque assay and the number of live phages was determined. At pH=2 No plaques were observed, while few plaques PFU/mL were observed at pH=4. At pH=7 highest titer was which is comparable with the titer at pH=9, whereas at pH=11 no plaques were developed Figure 4A.
Temperature
The KpnM were incubated at various ranges of temperature 0, 4 oC, 15 oC, 37 oC and 55 oC. After incubation, the titer of live phages was determined by using a plaque assay. No plaques were seen at 0 oC. Phages incubated at 4 °C resulted in a lower titer. At 15 oC for four hours a considerable number of plaques were produced. A higher phage titer was obtained when incubated at 37°C for four hours. Interestingly no plaques were seen at 48°C. Bacteriophages preserved at −20 oC after their attachment to a susceptible host with 50% glycerol produced high titer of bacteriophages in plaque assay after 6 months of preservation Figure 4B.
Chloroform
KpnM were also tested for their ability to tolerate Chloroform. After 4 hours of incubation (@ 0.5–1.5% chloroform conc.) bacteriophages were remain stable and plaques were shown in Figure 4C.
One-Step Growth Curve
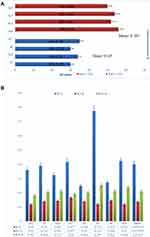
The bacteriophage infection activity was determined by a one-step or single-step growth curve using plaque assay Figure 5A. The method resulted in the determination of adsorption time, latent period, and burst size. The adsorption time for isolated bacteriophages was 6 minutes. The sudden early rise in phage titer at TN = 18 minutes after re-suspension, was determined as a latent period. The latent period of the phage was 24 minutes including 6 minutes of adsorption period. The burst size is defined as PFU/cell and is calculated as the ratio of PFU/mL at the plateau to PFU/mL at the start of the first rise. The burst size of the isolated phage was determined as 230 PFU/cell Figure 5B.
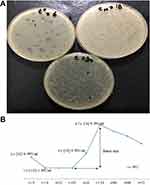 |
Figure 5 (A) Determination of one step growth curve through plaque assay. (B) Graphical analysis of phage titer at several time periods of one step growth curve. |
Biofilm Inhibition Assay
K. pneumoniae biofilm production was challenged by treatment with KpnM. The KpnM biofilm inhibition (B.I) and eradication (B.E) activity was determined against all MDR phage susceptible K. pneumoniae isolates. After 8 hours of incubation, the 96-well microtiter plate was examined through an ELISA plate reader at OD600, and values of biofilm inhibition and eradication assay were compared with positive control (ODs of biofilm assay B.A) Figure 6A. The mean value of OD600 obtained for the Biofilm assay was 0.401 while the mean value obtained for biofilm inhibition and eradication assay was 0.138 and 0.208. Interestingly, the comparison of means has shown strong biofilm inhibition and eradication ability of isolated bacteriophage KpnM Figure 6C.
Determination of Association of Phage Infection with mcr-1 Gene
The results of the biofilm inhibition assay were analyzed to determine the association between bacteriophage infection ability with the presence of the mcr-1 gene in susceptible isolates. The results showed a positive relation between phage infection and mcr-1 presence. All the isolates with mcr-1 genes were more efficiently inhibited by the KpnM than non-mcr-1 isolates. However, the presence or absence of the mcr-1 gene in isolates that already produced mature biofilm was observed as intermediately vulnerable to the same bacteriophages Figure 6B.
Discussion
Hypervirulent K. pneumoniae (hvKp) is a serious health challenge and difficult to treat. Recently, hvKp is disseminated globally as a community-acquired pathogen causing life-threatening infections. Furthermore, the emergence of colistin resistance hypervirulent K. pneumoniae is a major public health concern worldwide keeping in view that colistin is a last-resort antibiotic against carbapenem resistance hvKp.30 The purpose of our study was to estimate the occurrence of mcr-1 harboring hvKp and to find out an alternate approach to combat mcr-1 harboring MDR hvKp. The use of bacteriophages to control MDR bacteria may offer a non-antibiotic-based approach to treating infections caused by these pathogens. However, a detailed understanding of phage–host interactions is crucial to exploring the potential success of phage therapy for treatment.31
Overall, a significant incidence rate of MDR K. pneumoniae (Table 2) was estimated among different sample sources. In the study, maximum resistance rates were observed against ampicillin and amoxicillin as K. pneumoniae is considered naturally resistant to ampicillin and amoxicillin due to the intrinsic expression of the penicillinase enzyme. Similar findings have recently been documented for ampicillin amoxicillin resistance rates.32 The resistance ratio to imipenem, ciprofloxacin, and gentamicin was analyzed higher than reported in previous studies conducted in South Korea.33 As a previous intercontinental study demonstrates, higher resistance to carbapenems, fluoroquinolones, aminoglycosides, and colistin are directly associated with K. pneumoniae ability to cause HAIs and its dissemination.34 The resistance profiling of the present investigation is comparable with a recently executed study in Pakistan in which it was found that K. pnuemoniae showed maximum resistance against ceftriaxone ie 71%, then 40% against carbapenems. The least resistant 15% was observed in the case of colistin about 15%, however, is contrary to that in the case of colistin, we have observed a much high level of resistance ie 37% Figure 2A.35 Overall, for colistin limited data is available in the region, our study showed a significantly higher resistance rate than the previous investigation showing the resistance profiling of fosfomycin and colistin.36 Colistin resistance was determined using a rapid polymyxin NP test. Total 12 (37%) isolates were detected as colistin-resistant Figure 2B. The rapid polymyxin test had shown 98% sensitivity for the detection of resistant (positive control) isolates and 100% specificity for previously determined susceptible strains (negative control). Similar results with 100% specificity were obtained in a previously conducted study.37 In another study where carbapenem-resistant K. pneumoniae were subjected to various colistin resistance determination tests including the rapid NP polymyxin test, ColiSpot test, and the SuperPolymyxin medium. Aligned with our findings the results had shown 90% sensitivity and 94% specificity.16 Furthermore, genotypic analysis of colistin resistance was performed through the detection of the mcr-1 gene Figure 2C. Among 12 colistin resistance isolates 8 isolates were positive for the mcr-1 gene while 4 were not harboring mcr-1 gene. The more logical explanation of non-mcr-1 associated colistin resistance in 4 out of 12 isolates is probably due to some genetic mutation ie mgrB gene. A similar finding was previously documented for the mgrB-associated colistin resistance in K. pneumoniae isolates.38 Recent studies demonstrated that non-mcr-1 associated colistin resistance is particularly dependent on mgrB gene alterations, and variations in crrB, pmrB, phoQ, pmrA, and phoP genes.39 However, mcr-1 is not solely associated with colistin resistance other determinants including mcr-2 to 7 and more recently mcr-8 gene, located on a transferrable 95,983-bp IncFII-type plasmid may also be associated with colistin resistance in K. pneumoniae.40
The dearth of new antibiotics and the emergence of MDR strains warrant the search for potential alternative therapeutic approaches. Bacteriophages are considered one of the potential alternative strategies to fight MDR K. pneumoniae. The isolated and characterized Phage of the study ie KpnM had shown maximum host range (79%) against K. pneumoniae isolates. Our findings suggest that the phage isolation site may produce a great impact on the host range of bacteriophages because the isolation of bacteriophages from the environment where co-evolution of both phage and host occurs increases the chances of isolation of phages with a higher host range.41 As previously described exposure to multiple hosts may increase the host range of a particular bacteriophage.42 Keeping in view the aforementioned hypothesis the bacteriophage was first isolated from hospital waste where exposure to multiple capsular types of K. pneumoniae could have occurred. Additionally, the enrichment assay with multiple isolates of K. pneumoniae allowed more exposure. Also, phage-encoded depolymerases are directly associated with the phage host range. Previous studies have demonstrated that bacteriophages can be equipped with multiple depolymerases which individually augment the phage host range (polyvalency).43 As previously demonstrated, along with phage lytic activity degradation of capsular polysaccharides by depolymerases facilitates complement-mediated lysis and phagocytosis by macrophages.43 Therefore the broad host range with possibly multiple depolymerases makes our bacteriophage a potential candidate for in vivo phage therapy. However, sometimes the term broad host range is overestimated the lytic activity of other bacteria may be due to the production of lysins by nearby phages our lysis occurs due to media components.44
Furthermore, other key features of our isolated bacteriophage include an average plaque formation time of 8–10 hours with plaque size ranging from 0.5–3mm. One-step growth analysis had shown an adsorption period of 6 minutes followed by a latent period of 24 minutes with a burst size of 230 phage particles per bacterial cell. Similar results are also obtained in a recent study, phage vB_KpnM-VAC66 efficiently lysed K. pneumoniae host while aligned to our study phage vB_KpnM-VAC66 had a short adsorption time of 4 minutes and a latent period of 15 minutes.45 The short latent period and higher burst size are considered fundamental characteristics of bacteriophages suitable for phage therapy while the phage efficiently satisfies both these characteristics.46 In contrast, another investigation demonstrated that bacteriophage against Klebsiella had a 20 minutes latent period, which is aligned with this study whereas the burst size was different which was 80 PFU/cell and three times lower than the burst size obtained in our study. Additionally, our phage had shown stability under various broad ranges of environmental conditions. Phage KpnM remained stable at various concentrations of chloroform (0.5%–1.5%). In comparison to our study previously it has been demonstrated that the tail phages especially those belonging to the Siphoviridae family are chloroform-resistant.47 Similarly, maximum phage titer in PFU/mL was obtained at pH 7 while slightly lower titer was obtained at pH 9 and 11. Collectively the phage titer remained reasonably stable from pH 4–11 with maximum titer at pH 7. Similar results to our findings have previously been obtained for isolated bacteriophages which had shown stability to a wide range of pH 5–9 and retain residual activity at pH 3.48 Phage KpnM also showed thermal stability at various ranges of temperature while the maximum titer was obtained at 37 °C. However, in the present study phage genomics was not investigated which is a limitation of the study. Detailed genomics of the isolated phage may be conducted in the future to decipher the different molecular aspects and resistant or toxic genes exhibited in the phage genome.
The isolated bacteriophage KpnM was also tested for its ability to inhibit biofilms. The bacteriophage had successfully inhibited biofilm formation against all hvKp isolates. However, the ability of bacteriophages to eradicate mature biofilm was slightly moderate as compared to its activity for biofilm inhibition. The means of OD600 for biofilm inhibition assay and biofilm eradication assay were compared with the mean of O.D600 for biofilm formation assay. The mean O.D600 value of the biofilm formation assay was 0.401. The mean of O.D600 for biofilm inhibition assay was 0.138 with an S.D value. While the mean of O.D600 for biofilm eradication assay was 0.208, comparative analysis of means demonstrates effective biofilm inhibition and eradication ability of phage KpnM. The previous hypothesis about the association of the mcr-1 gene with the high susceptibility of bacteria towards bacteriophage was also analyzed by comparing the data of biofilm inhibition and eradication assay Figure 6C. The resulting plot demonstrates the association of the mcr-1 gene with higher susceptibility towards bacteriophage as reflected in more efficient clearance of biofilms by our isolated bacteriophage against mcr-1 positive K. pneumoniae isolates. Aligned to our findings the results from a previous study had shown the large plaque size in mcr-1 cultures.
Taking it together, it is concluded that the emergence of hvKp in the health setting is a serious health threat that needs to be addressed resourcefully. Due to limited antibiotic options especially the ineffectiveness of potent antibiotics like colistin, alternative approaches to control hvKp are indispensable. Characterized phage KpnM showed promising results and killed the mcr-1 harboring hvKp isolates, which may be used as a potential alternative therapeutic strategy against this pathogen. However, further studies are required to decipher the molecular basis of mechanisms opted by the phage to kill this ominous host bacterium.
Acknowledgment
The authors would like to thank the Deanship of Scientific Research at Shaqra University, Saudi Arabia for supporting this work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Aslam B, Khurshid M, Arshad MI., et al. Antibiotic resistance: one health one world outlook. Front Cell Infect Microbiol. 2021;11:771510. doi:10.3389/fcimb.2021.771510
2. Alvi RF, Aslam B, Rasool MH, et al. Transcriptional response of multidrug-resistant Klebsiella pneumoniae clinical isolates to ciprofloxacin stress. Can J Infect Dis Med Microbiol. 2021;2021:5570963. doi:10.1155/2021/5570963
3. Aslam B, Chaudhry TH, Arshad MI, et al. The First bla(KPC) Harboring Klebsiella pneumoniae ST258 Strain Isolated in Pakistan. Microb Drug Resist. 2020;26(7):783–786. doi:10.1089/mdr.2019.0420
4. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. doi:10.1111/joim.13007
5. Chaudhry TH, Aslam B, Arshad MI, et al. Emergence of bla (NDM-1) Harboring Klebsiella pneumoniae ST29 and ST11 in Veterinary Settings and Waste of Pakistan. Infect Drug Resist. 2020;13:3033–3043. doi:10.2147/IDR.S248091
6. Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. doi:10.2147/IDR.S173867
7. Seman A, Mihret A, Sebre S, et al. Prevalence and Molecular Characterization of Extended Spectrum β-Lactamase and Carbapenemase-Producing Enterobacteriaceae Isolates from Bloodstream Infection Suspected Patients in Addis Ababa, Ethiopia. Infect Drug Resist. 2022;15:1367–1382. doi:10.2147/IDR.S349566
8. Moosavian M, Emam N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect Drug Resist. 2019;12:1001–1010. doi:10.2147/IDR.S192597
9. Aslam B, Arshad MI, Aslam MA, et al. Bacteriophage Proteome: insights and Potentials of an Alternate to Antibiotics. Infect Dis Ther. 2021;10(3):1171–1193. doi:10.1007/s40121-021-00446-2
10. Rasool MH, Yousaf R, Siddique AB, Saqalein M, Khurshid M. Isolation, Characterization, and Antibacterial Activity of Bacteriophages Against Methicillin-Resistant Staphylococcus aureus in Pakistan. Jundishapur J Microbiol. 2016;9(10):e36135. doi:10.5812/jjm.36135
11. Procop GW, Church DL, Hall GS, Janda WM. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. Jones & Bartlett Publishers; 2020.
12. Rosenblueth M, Martínez L, Silva J, Martínez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol. 2004;27(1):27–35. doi:10.1078/0723-2020-00261
13. Li G, Shi J, Zhao Y, et al. Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur J Clin Microbiol Infect Dis. 2020;39(9):1673–1679. doi:10.1007/s10096-020-03890-z
14. Yao H, Liu J, Jiang X, Chen F, Lu X, Zhang J. Analysis of the Clinical Effect of Combined Drug Susceptibility to Guide Medication for Carbapenem-Resistant Klebsiella pneumoniae Patients Based on the Kirby–Bauer Disk Diffusion Method. Infect Drug Resist. 2021;14:79. doi:10.2147/IDR.S282386
15. Caneiras C, Lito L, Melo-Cristino J, Duarte A. Community-and hospital-acquired Klebsiella pneumoniae urinary tract infections in Portugal: virulence and antibiotic resistance. Microorganisms. 2019;7(5):138. doi:10.3390/microorganisms7050138
16. Conceição-Neto O, da Costa B, Pontes L, et al. Difficulty in detecting low levels of polymyxin resistance in clinical Klebsiella pneumoniae isolates: evaluation of Rapid Polymyxin NP test, Colispot Test and SuperPolymyxin medium. N Microbes New Infections. 2020;36:100722. doi:10.1016/j.nmni.2020.100722
17. Osei Sekyere J. Mcr colistin resistance gene: a systematic review of current diagnostics and detection methods. Microbiologyopen. 2019;8(4):e00682. doi:10.1002/mbo3.682
18. Uchiyama J, Rashel M, Maeda Y, et al. Isolation and characterization of a novel Enterococcus faecalis bacteriophage φEF24C as a therapeutic candidate. FEMS Microbiol Lett. 2008;278(2):200–206. doi:10.1111/j.1574-6968.2007.00996.x
19. Komijani M, Bouzari M, Rahimi F. Detection and characterization of a novel lytic bacteriophage (vB-KpneM-Isf48) against Klebsiella pneumoniae isolates from infected wounds carrying antibiotic-resistance genes (TEM, SHV, and CTX-M). Iranian Red Crescent Med J. 2017;19:548.
20. Manjunath N, Agsar D, Jagannath K, Rangaswamy B, Rao S, Anand S. Characterization and in vitro efficacy studies of wide host range lytic bacteriophage Φdmec-1 Infecting Escherichia coli isolated from pyogenic skin infections. DAMA Int. 2013;2(2):47–54.
21. Yazdi M, Bouzari M, Ghaemi EA. Isolation and characterization of a lytic bacteriophage (vB_PmiS-TH) and its application in combination with ampicillin against planktonic and biofilm forms of Proteus mirabilis isolated from urinary tract infection. J Mol Microbiol Biotechnol. 2018;28(1):37–46. doi:10.1159/000487137
22. Ghasemi SM, Bouzari M, Emtiazi G. Preliminary characterization of Lactococcus garvieae bacteriophage isolated from wastewater as a potential agent for biological control of lactococcosis in aquaculture. Aquaculture Int. 2014;22(4):1469–1480. doi:10.1007/s10499-014-9760-z
23. Stalin N, Srinivasan P. Characterization of Vibrio parahaemolyticus and its specific phage from shrimp pond in Palk Strait, South East coast of India. Biologicals. 2016;44(6):526–533. doi:10.1016/j.biologicals.2016.08.003
24. Jamal M, Hussain T, Das CR, Andleeb S. Characterization of Siphoviridae phage Z and studying its efficacy against multidrug-resistant Klebsiella pneumoniae planktonic cells and biofilm. J Med Microbiol. 2015;64(4):454–462. doi:10.1099/jmm.0.000040
25. Drulis-Kawa Z, Mackiewicz P, Kęsik-Szeloch A, et al. Isolation and characterisation of KP34—a novel φKMV-like bacteriophage for Klebsiella pneumoniae. Appl Microbiol Biotechnol. 2011;90(4):1333–1345. doi:10.1007/s00253-011-3149-y
26. Li L, Zhang Z. Isolation and characterization of a virulent bacteriophage SPW specific for Staphylococcus aureus isolated from bovine mastitis of lactating dairy cattle. Mol Biol Rep. 2014;41(9):5829–5838. doi:10.1007/s11033-014-3457-2
27. Pallavali RR, Degati VL, Durbaka VRP. Bacteriophages inhibit biofilms formed by multi-drug resistant bacteria isolated from septic wounds. bioRxiv. 2019;1:863076.
28. O’Toole GA. Microtiter dish biofilm formation assay. J Visualized Exp. 2011;2:47.
29. Fong SA, Drilling A, Morales S, et al. Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front Cell Infect Microbiol. 2017;7:418. doi:10.3389/fcimb.2017.00418
30. Olaitan AO, Diene SM, Kempf M, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44(6):500–507. doi:10.1016/j.ijantimicag.2014.07.020
31. Tan D, Zhang Y, Cheng M, et al. Characterization of Klebsiella pneumoniae ST11 isolates and their interactions with lytic phages. Viruses. 2019;11(11):1080. doi:10.3390/v11111080
32. Yang Y, Peng Y, Jiang J, et al. Isolation and characterization of multidrug‐resistant Klebsiella pneumoniae from raw cow milk in Jiangsu and Shandong provinces, China. Transbound Emerg Dis. 2021;68(3):1033–1039. doi:10.1111/tbed.13787
33. Kim D, Park BY, Choi MH, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrobial Chemother. 2019;74(1):190–199. doi:10.1093/jac/dky397
34. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nature Microbiol. 2019;4(11):1919–1929. doi:10.1038/s41564-019-0492-8
35. Imtiaz W, Syed Z, Rafaque Z, Andrews SC, Dasti JI. Analysis of Antibiotic Resistance and Virulence Traits (Genetic and Phenotypic) in Klebsiella pneumoniae Clinical Isolates from Pakistan: identification of Significant Levels of Carbapenem and Colistin Resistance. Infect Drug Resist. 2021;14:227. doi:10.2147/IDR.S293290
36. Gautam V, Thakur A, Sharma M, et al. Molecular characterization of extended-spectrum β-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. Indian J Med Res. 2019;149(2):208. doi:10.4103/ijmr.IJMR_172_18
37. Poirel L, Larpin Y, Dobias J, et al. Rapid Polymyxin NP test for the detection of polymyxin resistance mediated by the mcr-1/mcr-2 genes. Diagn Microbiol Infect Dis. 2018;90(1):7–10. doi:10.1016/j.diagmicrobio.2017.09.012
38. Malli E, Florou Z, Tsilipounidaki K, et al. Evaluation of rapid polymyxin NP test to detect colistin-resistant Klebsiella pneumoniae isolated in a tertiary Greek hospital. J Microbiol Methods. 2018;153:35–39. doi:10.1016/j.mimet.2018.08.010
39. Yang T-Y, Wang S-F, Lin J-E, et al. Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2020;55(3):105894. doi:10.1016/j.ijantimicag.2020.105894
40. Phetburom N, Boueroy P, Chopjitt P, et al. Klebsiella pneumoniae Complex Harboring mcr-1, mcr-7, and mcr-8 Isolates from Slaughtered Pigs in Thailand. Microorganisms. 2021;9(12):2436. doi:10.3390/microorganisms9122436
41. Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7(12):1225–1241. doi:10.1111/j.1461-0248.2004.00684.x
42. Yuan Y, Peng Q, Zhang S, et al. Phage reduce stability for regaining infectivity during antagonistic coevolution with host bacterium. Viruses. 2019;11(2):118. doi:10.3390/v11020118
43. Majkowska-Skrobek G, Latka A, Berisio R, et al. Phage-borne depolymerases decrease Klebsiella pneumoniae resistance to innate defense mechanisms. Front Microbiol. 2018;9:2517. doi:10.3389/fmicb.2018.02517
44. Ross A, Ward S, Hyman P. More is better: selecting for broad host range bacteriophages. Front Microbiol. 2016;7:1352. doi:10.3389/fmicb.2016.01352
45. Pacios O, Fernández-García L, Bleriot I, et al. Phenotypic and Genomic Comparison of Klebsiella pneumoniae Lytic Phages: vB_KpnM-VAC66 and vB_KpnM-VAC13. Viruses. 2022;14(1):6. doi:10.3390/v14010006
46. Merabishvili M, Pirnay J-P, De Vos D. Guidelines to Compose an Ideal Bacteriophage Cocktail. Bacteriophage therapy: Springer; 2018:99–110.
47. Kęsik-Szeloch A, Drulis-Kawa Z, Weber-Dąbrowska B, et al. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol J. 2013;10(1):1–12. doi:10.1186/1743-422X-10-100
48. Zhou S, Li L, Perseke M, Huang Y, Wei J, Qin Q. Isolation and characterization of a Klebsiella pneumoniae strain from mangrove sediment for efficient biosynthesis of 1, 3-propanediol. Sci Bulletin. 2015;60(5):511–521. doi:10.1007/s11434-015-0742-y
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.