Back to Journals » OncoTargets and Therapy » Volume 10
Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non-small cell lung cancer
Authors Wan L, Zhang L, Fan K, Wang JJ
Received 17 August 2017
Accepted for publication 11 October 2017
Published 28 November 2017 Volume 2017:10 Pages 5695—5702
DOI https://doi.org/10.2147/OTT.S149314
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Ingrid Espinoza
Li Wan, Lin Zhang, Kai Fan, Jian-Jun Wang
Department of Thoracic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China
Aim: We have previously shown that the long noncoding RNA prostate cancer-associated transcript 6 (PCAT6) promoted the proliferation and invasion of lung adenocarcinoma (LUAD) cells. In this study, the diagnostic significance of tissue and serum PCAT6 was evaluated in non-small cell lung cancer (NSCLC).
Materials and methods: Tissue expression of PCAT6 was systematically evaluated in five Gene Expression Omnibus datasets (GSE19804, GSE18842, GSE30219, GSE19188, and GSE27262). Circulating and tissue expressions of PCAT6 were detected by quantitative reverse-transcriptase polymerase chain reaction in NSCLC patients from Union Hospital.
Results: PCAT6 was significantly increased in lung cancer tissues and could be used to distinguish LUAD from adjacent normal tissues with an area under the receiver operating characteristic curve (AUC) of 0.9210 (p<0.0001; sensitivity, 98.82%; specificity, 78.57%) in GSE30219, 0.9333 (p<0.0001; sensitivity, 86.67%; specificity, 90.77%) in GSE19188, 0.9584 (p<0.0001; sensitivity, 92.00%; specificity, 96.00%) in GSE27262, and 0.9574 (p<0.0001; sensitivity, 95.89%; specificity, 87.67%) in patients from Union Hospital. As for lung squamous cell carcinoma (LUSC), the AUC of PCAT6 was 0.9567 (p<0.0001; sensitivity, 100%; specificity, 85.71%) in GSE30219, 0.9795 (p<0.0001; sensitivity, 96.30%; specificity, 92.31%) in GSE19188, and 0.9942 (p<0.0001; sensitivity, 100%; specificity, 98.04%) in patients from Union Hospital. We further noticed that the plasma levels of PCAT6 were significantly increased in 73 LUAD and 51 LUSC patients compared with 39 healthy controls (p<0.0001). The AUC of circulating PCAT6 was 0.9213 (p<0.0001; sensitivity, 87.67%; specificity, 97.44%) in LUAD and 0.9583 (p<0.0001; sensitivity, 94.12%; specificity, 100%) in LUSC.
Conclusion: Together with our previous findings, our results suggest that PCAT6 could be used as a potential diagnostic and prognostic biomarker in NSCLC.
Keywords: lncRNAs, PCAT6, diagnosis, lung adenocarcinoma, lung squamous cell carcinoma
Introduction
Non-small cell lung cancer (NSCLC) has become the main cause of cancer-related deaths in China, and the mortality of this disease has increased for more than four times during the past three decades.1,2 Furthermore, NSCLC could remain a major health problem for at least the next 50 years.3 Although the development of new diagnostic and treatment strategies has promoted the survival of NSCLC patients, the overall 5-year survival rate is still <20%, mainly because it is usually detected at an advanced stage.4 Diagnosis of NSCLC at an early stage is one way that can improve a patient’s survival;5 however, early diagnosis remains a challenge due to the lack of specific biomarkers.
In recent years, methods for lung cancer diagnosis include the use of imaging techniques and blood-fluid tests for tumor markers. For example, the largest lung cancer screening trial showed that screening high-risk group with computerized tomography (CT) or low-dose CT relatively reduced mortality by 20% compared to chest radiography.6 However, it often leads to overdiagnosis and unnecessary surgeries due to increased false-positive results, and the cumulative exposure to radiation resulted by annual examinations also represents a considerable health risk.7,8 Thus, developing blood-fluid tests for the diagnosis of early stage NSCLC is clinically important, as blood samples are easily acquired in a relatively noninvasive manner compared with biopsy or surgery. The most widely used serum biomarkers of lung cancer screening include carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CYFRA21-1), and neuron-specific enolase (NSE). However, the sensitivity and specificity of these biomarkers is commonly ranged between 50% and 90%, and false-negative rate is commonly >50% for the diagnosis of NSCLC at an early stage.9 Given these limitations, identification of novel serum biomarkers with high sensitivity and specificity for the diagnosis of NSCLC is of immediate need.
During the past decade, mounting evidence has confirmed that dysregulation of lncRNAs, acting either as oncogenes or as tumor suppressors, is an important cause of certain cancers.10–12 Increasing evidence have shown that long noncoding RNAs (lncRNAs) has led to the development of a new field of molecular diagnosis of cancer. Plasma level of long intergenic non-protein-coding RNA 152 (LINC00152) is significantly elevated and has the potential to be used as a blood-based biomarker for the diagnosis of gastric cancer in patients.13 lncRNA16 (ENST00000539303) is significantly elevated in plasma samples of lung cancer patients, and GAS5 expression is decreased in NSCLC plasma samples, thus making lncRNAs a potential biomarker of lung cancer diagnosis.5,14
lncRNA prostate cancer-associated transcript 6 (PCAT6) was first identified in keratinocyte-enhanced cellular proliferation and colony formation of prostate cancer cells in an androgen-independent way.15 In lung cancer, PCAT6 was also found to be upregulated using Affymetrix HG-U133 plus 2.0 array with an lncRNA classification pipeline.16 Previously, we have confirmed that PCAT6 is significantly upregulated in cancer tissues compared with adjacent normal tissues and positively correlated with metastasis of lung adenocarcinoma (LUAD) patients.17 In addition, PCAT6 was found to be negatively correlated with overall survival of lung cancer patients with retrospective analysis.17 Therefore, we set out to investigate whether serum PCAT6 was increased in LUAD and lung squamous cell carcinoma (LUSC), two major types of NSCLC, and has the potential to be used as a noninvasive diagnostic biomarker of NSCLC.
Materials and methods
GEO lung cancer gene expression data
To identify PCAT6 expression in lung cancer, we searched relevant Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/geo/). As a result, five panels of lung cancer gene expression datasets, including GSE27262, GSE19804, GSE19188, GSE30219, and GSE18842, were selected to compare PCAT6 expression between lung cancer tissues and normal tissues. Table 1 summarizes the details of these five datasets.
Participants and tissue samples
Seventy-three plasma samples, biopsy specimens of LUAD tissues and adjacent normal tissues, 51 plasma samples and biopsy specimens of LUSC tissues and adjacent normal tissues were collected from the department of thoracic surgery of Union Hospital (Wuhan, People’s Republic of China). Tissue biopsy specimens were collected and immediately snap-frozen in liquid nitrogen and stored at −80°C until to be used. Thirty-nine control plasma samples were collected from healthy donors without cancer. Approximately 4 mL of venous blood was collected from each participant, and plasma was separated within 2 h by centrifugation at 1,200 ×g for 10 min at 4°C to spin down blood cells, followed by centrifugation at 12,000 ×g for 10 min at 4°C to completely remove cellular components or cell debris. The supernatant plasma was then carefully collected and stored at −80°C until to be used. The study was approved by the Ethical Review Board for Research of Union Hospital, affiliated to Tongji Medical College of Huazhong University of Science and Technology. All the participants had signed the written informed consent.
RNA extraction and cDNA synthesis
Total RNA was extracted from tissue specimens using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and plasma RNA was extracted using TRIzol LS reagent (Invitrogen). The isolated RNA concentration was calculated and normalized with RNase-free water and then reverse-transcribed into cDNA using PrimeScript™ RT reagent kit with gDNA Eraser (RR047A; Takara, Dalian, People’s Republic of China). Reverse transcription conditions were performed as follows: 42°C for 2 min, and then 37°C for 15 min, 85°C for 5 s, followed by storage at 4°C. All cDNA samples were stored at −80°C until use.
Quantitative reverse-transcriptase polymerase chain reaction
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was conducted using SYBR Premix Ex Taq (Takara), according to the manufacturer’s instructions. Briefly, all the reactions were carried out on an ABI7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA), and 2 μL cDNA was used as template. The qRT-PCR amplification was performed as follows: an initial denaturation at 95°C for 5 min, followed by 45 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The 2−ΔΔCt method was used to quantify the fold change of PCAT6 expression in tumor samples versus normal control samples as we previously described.17 GAPDH was used as an internal control, and all reactions were performed in triplicate. The primer sequences were as follows: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse); PCAT6, 5′-CAGGAACCCCCTCCTTACTC-3′ (forward) and 5′-CTAGGGATGTGTCCGA AGGA-3′ (reverse).15
Statistical analysis
All data are presented as mean ± SD from at least three separate experiments and analyzed by using the GraphPad Prism V.6.00 software (GraphPad Software, La Jolla, CA, USA). Comparison between two groups for statistical significance was performed with two-tailed Student’s t-test. For more groups, one-way ANOVA followed by Newman–Keuls post hoc test was used. Receiver operating characteristic (ROC) curves were established to evaluate the diagnostic value of PCAT6 for differentiating tumors from controls. A p-value of <0.05 was considered to be statistically significant.
Results
Tissue PCAT6 is upregulated and has a significant diagnostic value in LUAD and LUSC.
In a previous study, we have confirmed that PCAT6 expression is increased in patients with LUAD and predicts a poor overall survival.17 Then, we further evaluated PCAT6 expression in lung cancer patients from five GEO datasets (GSE19804,18 GSE18842,19 GSE30219,20 GSE19188,21 and GSE2726222). As shown in Table 1, these five GEO datasets include patients with NSCLC, that is, LUAD and LUSC. PCAT6 expression was significantly increased in tissues of LUAD and LUSC compared with normal noncancerous tissues in these five datasets (Figure 1). Then, we further evaluated the diagnostic value of PCAT6 in these five datasets using ROC analyses. The results showed that the value of the area under the ROC curve (AUC) was 0.9578 (95% CI 0.9209–0.9947) in GSE19804 (Figure 2A), 0.9584 (95% CI 0.8915–1) in GSE27262 (Figure 2B), 0.9210 (95% CI 0.8326–1) in GSE30219 (Figure 2C), and 0.9333 (95% CI 0.8864–9802) in GSE19188 (Figure 2D) for LUAD and normal controls. With regard to LUSC, the AUC value was 0.9567 (95% CI 0.8935–1) in GSE30219 (Figure 2E) and 0.9795 (95% CI 0.9548–1) in GSE19188 (Figure 2F) versus normal controls.
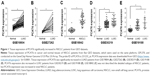  | Figure 1 Tissue expression of PCAT6 significantly increased in NSCLC patients from GEO datasets. |
Next, we further validated the expression of PCAT6 in 73 paired LUAD tissues and normal counterparts, and 51 paired LUSC tissues and normal counterparts from our local hospital. As shown in Figure 3A and C, PCAT6 was significantly increased in LUAD tissues (p<0.0001, Figure 3A) as well as in LUSC tissues (p<0.0001, Figure 3C) compared with normal counterparts. The diagnostic value of PCAT6 for LUAD tissues and normal counterparts was 0.9574 (95% CI 0.9232–0.9916; Figure 3B), and for LUSC tissues, it was 0.9942 (95% CI 0.9825–1; Figure 3D). The sensitivity, specificity, accuracy, and Yoden index of PCAT6 for distinguishing LUAD and LUSC from normal controls in five GEO datasets and our local hospital are summarized in Table 3. The results showed that PCAT6 had a significant higher level of sensitivity and specificity than CEA, which had a 51.1% sensitivity and a 84.8% specificity in NSCLC.14 Together, these results indicate that PCAT6 has a potential significance with respect to sensitivity and specificity in the diagnosis of NSCLC.
Plasma PCAT6 has a diagnostic potential significance in the diagnosis of NSCLC
Plasma samples are easily acquired in a relatively noninvasive manner compared with biopsy or surgery. Thus, developing noninvasive techniques for the diagnosis of early stage NSCLC is clinically important. To examine whether plasma PCAT6 had diagnostic potential, plasma from the abovementioned 73 LUAD patients, 51 LUSC patients, and 39 normal healthy donors was collected. Plasma PCAT6 expression was significantly increased in LUAD and LUSC patients compared with normal healthy donors (p<0.0001, Figure 4A). Then, we further evaluated the correlation between plasma PCAT6 and the patients’ clinicopathological characteristics (Table 2). Plasma expression of PCAT6 was positively correlated with TNM stage (p<0.0001) and metastasis status (p<0.0001) of LUAD and LUSC, respectively. But it was not correlated with tumor size and smoking history. The diagnostic value of plasma PCAT6 was also evaluated, and the results showed that PCAT6 had an AUC value of 0.9213 (95% CI 0.8663–0.9763; Figure 4B) for LUAD and 0.9583 (95% CI 0.9109–1; Figure 4C) for LUSC. The sensitivity and specificity of PCAT6 for distinguishing LUAD and LUSC from healthy controls was 87.67%/97.44% and 94.12%/100%, respectively (Table 3). Together, these results indicate that plasma PCAT6 may be exploited as a promising noninvasive biomarker of NSCLC.
Discussion
So far, numerous studies have shown that lncRNAs play an important role in tumor occurrence, invasion, and metastasis. With the implementation of next-generation sequencing platforms for molecular diagnostics, an increasing number of lncRNAs, such as MALAT1, colon cancer-associated transcript 2 (CCAT2), LINC01133, and GAS5, have been identified to be correlated with lung cancer, which were reported with the potential to be used as biomarkers for the diagnosis, prognosis, and personalized treatment indicators for NSCLC.5,14,16,23–25 However, none of the currently identified biomarkers are sensitive or specific enough for reliable NSCLC screening in clinical settings.26 Thus, identification of novel lncRNA-based biomarkers for the early diagnosis of NSCLC will have significant clinical benefits.
In a previous study, we have determined the biological role of lncRNA PCAT6 in the progression of LUAD and found that PCAT6 was significantly elevated in lung cancer tissues and positively correlated with tumor size, TNM stage, and lymph node metastasis.17 Therefore, PCAT6 may have the diagnostic potential in NSCLC. In this study, the expression and diagnostic value of PCAT6 in cancer tissues was first examined in 349 NSCLC patients from five GEO datasets (GSE19804,18 GSE18842,19 GSE30219,20 GSE19188,21 and GSE2726222). The expression of tissue PCAT6 was consistently increased and showed a great diagnostic value (AUC >0.9; sensitivity, 86.67%–100%; specificity, 78.57%–96%) in NSCLC patients included in the five GEO datasets. The result was further reconfirmed in samples from our local hospital, including 73 LUAD patients and 51 LUSC patients. Therefore, these results suggest that PCAT6 may have the potential to be used as a diagnostic marker of NSCLC patients.
Determination of blood-based circulating biomarkers is a simple, inexpensive, and noninvasive test that greatly facilitates early diagnosis of cancer, even earlier than CT imaging.27 To date, no study has quantified the expression level and the diagnostic potential of PCAT6 in plasma. Here, we first investigated the diagnostic value of PCAT6 in plasma distinguishing NSCLC patients, including LUAD and LUSC, from healthy controls. The results showed that plasma PCAT6 had an AUC value of 0.9213 (95% CI 0.8663–0.9763) for LUAD and 0.9583 (95% CI 0.9109–1) for LUSC, significantly higher than GAS5 (0.832, 95% CI 0.754–0.893) and CEA (0.700, 95% CI 0.611–0.779) according to a previous report.14 Furthermore, the sensitivity and specificity of PCAT6 for distinguishing LUAD and LUSC from healthy controls were 87.67%/97.44% and 94.12%/100%, respectively, which were significantly higher than the combination of GAS5 and CEA (86.7%/90.9%).14 Thus, these results indicated that plasma PCAT6 may be exploited as a promising noninvasive biomarker of NSCLC, even better than the previously reported biomarkers.
Conclusions
In summary, our study first systematically evaluated the diagnostic potency of PCAT6 in NSCLC patients from five GEO datasets and our local hospital, and identified that circulating PCAT6 is a potential biomarker of NSCLC diagnosis. Furthermore, PCAT6 also predicted a poor overall survival of LUAD patients in our previous study.17 Thus, PCAT6 may play a pivotal role in carcinogenesis and progression of NSCLC. Unfortunately, we only confirmed that PCAT6 regulates p53 and c-myc expressions in an indirect manner, and the exact molecular mechanisms of PCAT6 in NSCLC remain unclear and require further investigation.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant no 81000032) and the Natural Science Foundation of Hubei Province (No 02.07.16040048).
Disclosure
The authors report no conflicts of interest in this work.
References
Zhou M, Guo M, He D, et al. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. | ||
She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143(4):1117–1126. | ||
Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574–5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6(3):482–488. | ||
Heuvers ME, Hegmans JP, Stricker BH, Aerts JG. Improving lung cancer survival; time to move on. BMC Pulm Med. 2012;12(1):77. | ||
Zhu H, Zhang L, Yan S, et al. LncRNA16 is a potential biomarker for diagnosis of early-stage lung cancer that promotes cell proliferation by regulating the cell cycle. Oncotarget. 2017;8(5):7867–7877. | ||
National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. | ||
Heuvers ME, Stricker BH, Aerts JG. Generalizing lung-cancer screening results. N Engl J Med. 2012;366(2):192–193. | ||
Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA. 2012;307(22):2418–2429. | ||
Hur J, Lee HJ, Nam JE, et al. Additional diagnostic value of tumor markers in cytological fluid for diagnosis of non-small-cell lung cancer. BMC Cancer. 2012;12(1):392. | ||
Su X, Malouf GG, Chen Y, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5(20):9864–9876. | ||
Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. | ||
Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. | ||
Li Q, Shao Y, Zhang X, et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36(3):2007–2012. | ||
Han L, Ma P, Liu SM, Zhou X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumor Biol. 2016;37(5):6847–6854. | ||
Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. | ||
Yang J, Lin J, Liu T, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85(2):110–115. | ||
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Knockdown of long noncoding RNA PCAT6 inhibits proliferation and invasion in lung cancer cells. Oncol Res. 2016;24(3):161–170. | ||
Lu TP, Tsai MH, Lee JM, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2590–2597. | ||
Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129(2):355–364. | ||
Rousseaux S, Debernardi A, Jacquiau B, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Science Transl Med. 2013;5(186):186ra166. | ||
Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4):e10312. | ||
Wei TY, Juan CC, Hisa JY, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012;103(9):1640–1650. | ||
Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6(1):518. | ||
Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumor Biol. 2014;35(6):5375–5380. | ||
Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7(32):51784–51814. | ||
Hiley CT, Le Quesne J, Santis G, et al. Challenges in molecular testing in non-small-cell lung cancer patients with advanced disease. Lancet. 388(10048):1002–1011. | ||
Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature. 2017;545(7655):446–451. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






