Back to Journals » Infection and Drug Resistance » Volume 15
Diagnosis of Mycoplasma hominis Meningitis with Metagenomic Next-Generation Sequencing: A Case Report
Authors Dong Y , He Y, Zhou X, Lv X, Huang J, Li Y, Qian X, Hu F, Zhu J
Received 18 May 2022
Accepted for publication 1 August 2022
Published 12 August 2022 Volume 2022:15 Pages 4479—4486
DOI https://doi.org/10.2147/IDR.S371771
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Yukang Dong,1,* Yingying He,1,* Xia Zhou,1 Xia Lv,1 Jia Huang,1 Yaqi Li,1 Xin Qian,2 Fangfang Hu,3 Jiaying Zhu1
1Department of Emergency, Guizhou Provincial People’s Hospital, Guiyang, People’s Republic of China; 2Department of Pharmacy, Guizhou Provincial People’s Hospital, Guiyang, People’s Republic of China; 3Department of Clinical Laboratory, Guizhou Provincial People’s Hospital, Guiyang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiaying Zhu, Email [email protected]
Background: Mycoplasma hominis meningitis is a rare postoperative complication of neurosurgery. Accurate and early diagnosis of M. hominis remains challenging because of the limitations of traditional detection methods. Metagenomic next-generation sequencing (mNGS) is an advanced technique with high sensitivity and specificity for identifying infectious pathogens; however, its application in diagnosing M. hominis meningitis has not been widely studied.
Case Presentation: We report the case of a 61-year-old man who presented with fever and headache after neurosurgical treatment for a cerebral hemorrhage. Empiric antibiotic therapy was ineffective. Traditional culture of pathogens and serological testing yielded negative results, but M. hominis was detected in the cerebrospinal fluid by mNGS. After further verification by polymerase chain reaction (PCR), the patient’s clinical treatment was adjusted accordingly. With targeted antibiotic intervention, the patient’s symptoms were effectively alleviated, and clinical indicators returned to normal levels. Furthermore, the abundance of M. hominis decreased significantly compared to the initial mNGS reading after targeted treatment, indicating that the infection caused by M. hominis was effectively controlled.
Conclusion: Using mNGS, we found that M. hominis may be a candidate causative agent of meningitis. The technique also has the advantage of timeliness and accuracy that traditional cultures cannot achieve. A combination of mNGS with PCR is recommended to identify pathogens in the early stages of infectious diseases to administer targeted clinical medication.
Keywords: Mycoplasma hominis, meningitis, metagenomic next-generation sequencing, cerebrospinal fluid, case report
Background
Mycoplasma hominis is a commensal of the genital tract associated with various genitourinary tract infections and complications of pregnancy.1 The organism has also been reported to be involved in other postoperative infections in addition to affecting the urinary system, including infections after lung transplantation,2 total hip replacement,3 kidney transplantation,4 and heart transplantation,5 raising the possibility that M. hominis participates in unknown causes of infections in the body. Furthermore, case reports have indicated that M. hominis can cause spontaneous meningitis or post-neurosurgical infection, although these complications are uncommon.6,7 Clinically, M. hominis infection is difficult to detect and diagnose using traditional culture strategies. One of the reasons for this is the lack of a cell wall, making it difficult to observe using a light microscope.8 The invalidation of conventional gram staining is also the main reason for the failure of clinical specimen detection.9 Owing to limitations in testing methods and their lack of timeliness, the diagnosis of intracranial infection caused by this organism poses a great challenge, which may also lead to underestimation of the true incidence of M. hominis infection and a delay in definitive therapy. Therefore, the use of an effective means of detection to rapidly and accurately determine pathogens is important for optimizing treatment strategies.
Metagenomic next-generation sequencing (mNGS) is an advanced technique with high sensitivity and specificity for identifying infectious pathogens.10 This technique can rapidly identify and classify all pathogens causing central nervous system (CNS) infection and compensate for the defects of traditional culture methods, which are time consuming and yield low positivity results.11 Because of its comprehensiveness, mNGS is considered a potential diagnostic method for conventional pathogens and can partially replace traditional culture methodology.12 Herein, we report a case of M. hominis meningitis diagnosed by mNGS analysis using cerebrospinal fluid (CSF) from a lumbar puncture of a patient, whose condition was effectively controlled after targeted treatment. Our report provides an example for diagnosing meningitis caused by pathogens in adult patients, as well as recommendations for precise diagnosis and targeted treatment.
Case Presentation
A 61-year-old man with a 2-month history of hypertension presented with the sudden onset of left limb weakness for one day and was subsequently admitted to the local hospital on March 30, 2021. Head computed tomography (CT) scan revealed intracerebral hemorrhaging in the right basal ganglia–radiation crown and centrum semiovale. Emergency decompression craniotomy and hematoma evacuation were performed. The patient was then transferred to the intensive care unit, where he received mechanical ventilation and an indwelling catheter. On third postoperative day, the patient regained consciousness and was transferred to the general ward after withdrawal from the ventilator. On day 12 after operation, the patient developed a headache and fever with a maximum body temperature of 39.2 °C. After 10 days of anti-infection treatment with cefoperazone sulbactam (specific treatment information was not available), there was still no significant relief of symptoms, and no decrease in body temperature. Lumbar puncture results (including CSF culture results) suggested a white blood cell count of 320/μL, glucose level of 0.4 mmol/L, chloride level of 119.7 mmol/L, and protein level of 6.6 g/L, but the results of the etiological culture (blood and CSF) were negative for any pathogen.
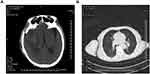
On April 29, 2021, the patient was transferred to the emergency department of Guizhou Provincial People’s Hospital for further treatment. On admission, he was drowsy but lucid, complaining of severe weakness and a mild headache. Neurological examination revealed severe paralysis of the left limb (grade 1 muscle strength), with positive meningeal irritation. Head CT after admission showed a small amount of blood and fluid accumulation in the operative area after intracerebral hemorrhage (Figure 1A), while lung CT showed no obvious signs of infection (Figure 1B). Furthermore, ceftriaxone, vancomycin, and caspofungin were used in combination to ensure coverage of the common bacterial meningitis and bloodstream infections. Lumbar puncture was performed on the second day of admission, and the intracranial pressure was elevated to 270 mmH2O. The CSF sample reported a white blood cell count of 810/μL, glucose level of 0.28 mmol/L, chloride level of 114 mmol/L, and protein level of 1.32 g/L. Subsequently, the traditional culture of peripheral blood, urine, and CSF specimens was performed, as well as the mNGS analysis of the CSF sample. Aerobic and anaerobic bacteria cultures showed negative results; however, mNGS revealed a high burden of M. hominis.
As is the case for all patients with no improvement in clinical symptoms, the anti-infection regimen was changed on May 3, 2021, to only moxifloxacin (0.4 g intravenous drops quaque die) according to the mNGS guidelines. On the sixth day of treatment for M. hominis meningitis, the patient’s headache gradually disappeared and his body temperature returned to normal. Five lumbar puncture tests performed during hospitalization also indicated a gradual recovery of routine indicators (Table 1). On the 16th day of anti-mycoplasma therapy, a CSF sample was collected for an additional mNGS assay, and the results suggested a reduced burden of M. hominis to two unique reads, which was attributed to effective treatment. After 22 days of moxifloxacin treatment, the patient was discharged on May 25, 2021, at the request of his family. Follow-up after 1 month indicated that the patient had no obvious neurological symptoms, such as fever or headache, except for left limb weakness.
 |
Table 1 CSF Indicators from Five Lumbar Punctures After Admission to Our Hospital |
mNGS Analysis
The patient’s CSF sample was delivered to Guangzhou Sagene Biotech Co., Ltd. for unbiased pathogen detection. A magnetic bead extraction kit (Sagene, Guangzhou, China) was used to extract DNA, and the Nextera XT DNA Library Prep Kit (Illumina, USA) was used to construct a metagenomic library of approximately 400 bp, as determined by electrophoresis. After fluorescence quantification, sequencing was performed using the Illumina 550 DX platform. A total of 1424 reads were identified to correspond to the M. hominis genome, with a coverage of 3.7% (Figure 2A and B). Without the human host DNA, the M. hominis reads contributed to 0.32% of the total microbial and unclassified reads.
Polymerase Chain Reaction (PCR) Validation
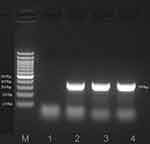
To further verify the abundance of M. hominis, the target fragment was authenticated by sequence-specific PCR using the primers of M. hominis (F-TGACCCTGAATTTGAAATCGTTG and R-CAGTTAGTAGTACATGAAGCGCCT).13 ABI PRISM 3730 (Applied Biosystems, CA, USA) was used for sanger sequencing, followed by mapping to NCBI BLAST.14,15 The results suggested a 399-bp target fragment aligned with the reference sequences of M. hominis (GenBank: CP055144.1) with 99.73% identity, thereby indicating M. hominis infection in this patient (Figure 3).
Discussion and Conclusion
M. hominis is a pathogen commonly reported in urogenital infections. Transmissible infections of this pathogen to organ systems other than the genitals are uncommon, and nearly half of the cases with invasive infections are considered related to immunosuppression. Similarly, extragenital mycoplasma infection is often accompanied by cell-mediated immune system impairment or hypogammaglobulinemia.16–18 The presence of variable adhesion-associated antigens in M. hominis may contribute to its evasion of the host immune response.19 Central nervous system infection caused by M. hominis has also been found in newborns and may be prompted by amniotic choroiditis in the womb or by exposure to infection in the birth canal.20 Infections with this pathogen have also been reported in brain abscesses, even in rare cases. Currently, the infection is thought to originate from exposure during trauma and surgery, as well as transmission of secondary bacteremia to the brain region.21,22 In brain tissues with CNS capillary damage, M. hominis can also be transmitted through the bloodstream and cause infection. In our case, the patient underwent intraocular hematoma evacuation and urethral catheterization prior to the onset of meningitis, resulting in exposure to two assumptive sources of contamination. However, it is difficult to determine the postoperative source of the infection.
In accordance with previous reports, most mycoplasma pathogens are identified using traditional culture methods, but this is inappropriate for CNS infections at an early stage.7 Owing to the barriers in detection technology, a considerable number of patients have not been able to exploit the most effective treatment opportunities. Furthermore, CSF analysis can only detect abnormal numbers of lymphocytes and the severity of the inflammatory response.23 Because of the slow growth and occult nature of pathogen isolates, the positive detection rate of plate-based culture can often be relatively low. Specifically, M. hominis can take more than a week to form an identifiable colony and is likely to evade detection by automated detection systems, such as BacT/ALERT.24 In terms of diagnosis, PCR is cheaper, faster, and more sensitive than traditional culture,25,26 but it is based on rapidly verifying predictions of suspected pathogens. Under these circumstances, we identified the microbes using mNGS, a technique becoming more widely used in identifying the causative organism of infectious diseases of the CNS. Considering the power of PCR for accurate recognition and that PCR is the gold standard for microbial identification, we further verified the candidate microorganisms using PCR and obtained a consistent result.
Metagenomic next-generation sequencing is useful for early diagnosis to provide patients with timely and accurate antimicrobial treatments. All nucleic acid information can be captured in a single operation, and the types and abundances of various pathogens can be accurately assessed through sequence analysis and comparison.27 Without speculations or assumptions of suspected pathogenic bacteria, comprehensive clinical screening is recommended. A shorter testing time allows patients to benefit from a timely diagnosis and targeted medication. The use of mNGS is believed to provide clinicians with convenient clues for diagnosis and can effectively prevent the overuse of antibiotics in viral infections.28 In addition, mNGS has advantages in pathogen typing and can accurately distinguish between different types of infection sources, including viruses, parasites, fungi, and bacteria. With the help of mNGS, Naegleria fowleri,29 Brucella spp.30 and Taenia bocina31 have been previously detected in CSF. In this study, mNGS also contributed significantly to pathogen diagnosis. Since no pathogen was found in the traditional culture, the possibility of M. hominis infection was not considered until the mNGS results were available. Therefore, mNGS is strongly recommended for diagnosis of infections with unknown etiology. Combined with the patients’ clinical characteristics and PCR verification results, this can accurately provide guidance for medication and prescription adjustments.
Because mycoplasma lacks a hard cell wall, it is not sensitive to compounds that act on the cell wall or interfere with folic acid synthesis, but it responds well to antibacterial drugs that affect cell membrane integrity.32 In addition, due to the occurrence of mycoplasma mutations and the increase in antimicrobial resistance,33 infections caused by mycoplasma require special antibiotic treatment. M. hominis is currently considered sensitive to chloramphenicol, tetracycline, lincosamide, and fluoroquinolones, but penetration of the blood–brain barrier is unfavorable for most drugs with low minimum inhibitory concentrations (MIC). Although the MIC of quinolones against M. hominis is low, reasonable CSF penetration is still present. Among the quinolones, moxifloxacin and gatifloxacin were found to have significantly higher permeability in CSF than trovafloxacin, levofloxacin, and ciprofloxacin.34 Considering that gatifloxacin is prone to cause hypoglycemia in patients, moxifloxacin may be the best treatment option. Fortunately, after 3 weeks of treatment with the moxifloxacin regimen, the patient’s disease indicators gradually returned to normal levels, and the symptoms were significantly alleviated, which further indicated the importance of mNGS detection in the clinical guidance of drug administration.
In conclusion, this case report demonstrates the clinical significance of mNGS in the diagnosis of M. hominis meningitis and illustrates the shortcomings of traditional culture methods in terms of sensitivity and specificity of pathogen diagnosis. More importantly, mNGS can accurately identify pathogens through one-time sequencing and analysis and greatly contribute to the early diagnosis of infectious diseases. PCR can be combined with the mNGS results for targeted validation to better guide clinical treatment.
Abbreviations
mNGS, metagenomic next-generation sequencing; CNS, central nervous system; CSF, cerebrospinal fluid; CT, computed tomography; PCR, polymerase chain reaction; MIC, minimum inhibitory concentrations.
Data Sharing Statement
All data generated or analysed during this study are included within the article [and its additional files]. The M.hominis DNA sequence assembled using the mNGS data have also been deposited in GenBank database with the accession number PRJNA819667 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA819667).
Ethics Approval and Consent to Participate
The study was approved by the ethics committee of Guizhou Provincial People’s Hospital (2022-27), and the patient consented for publication of his clinical data.
Consent for Publication
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Acknowledgments
The authors thank all the clinical and laboratory staffs contributed in the case.We also thank the staffs of Guangzhou Sagene Biotech Co., Ltd. for technical support.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Ahmed J, Rawre J, Dhawan N, Khanna N, Dhawan B. Mycoplasma hominis: an under recognized pathogen. Indian J Med Microbiol. 2021;39(1):88–97. doi:10.1016/j.ijmmb.2020.10.020
2. Vecchio M, Koutsokera A, Touilloux B, et al. Bronchial anastomosis dehiscence and stenosis caused by donor-transmitted Mycoplasma hominis infection in a lung transplant recipient: case report and literature review. Transpl Infect Dis. 2021;23(2):e13475. doi:10.1111/tid.13475
3. Xiang L, Lu B. Infection due to Mycoplasma hominis after left Hip replacement: case report and literature review. BMC Infect Dis. 2019;19(1):50. doi:10.1186/s12879-019-3686-z
4. Adams M, Bouzigard R, Al-Obaidi M, Zangeneh TT. Perinephric abscess in a renal transplant recipient due to Mycoplasma hominis: case report and review of the literature. Transpl Infect Dis. 2020;22(5):e13308. doi:10.1111/tid.13308
5. Givone F, Peghin M, Vendramin I, et al. Salvage heart transplantation for Mycoplasma hominis prosthetic valve endocarditis: a case report and review of the literature. Transpl Infect Dis. 2020;22(2):e13249. doi:10.1111/tid.13249
6. Zhou M, Wang P, Chen S, et al. Meningitis in a Chinese adult patient caused by Mycoplasma hominis: a rare infection and literature review. BMC Infect Dis. 2016;16(1):557. doi:10.1186/s12879-016-1885-4
7. Lee EH, Winter HL, van Dijl JM, Metzemaekers JD, Arends JP. Diagnosis and antimicrobial therapy of Mycoplasma hominis meningitis in adults. Int J Med Microbiol. 2012;302(7–8):289–292. doi:10.1016/j.ijmm.2012.09.003
8. Diab A, AlMusawi SSM, Hudhaiah D, Magzoub R, Al Rashed AS, Al Musawi TS. Iatrogenic ventriculitis due to Mycoplasma hominis: a case report and review of the literature. Am J Case Rep. 2019;20:406–411. doi:10.12659/AJCR.914284
9. Tyner HL, Virk A, Nassr A, Razonable R. Mycoplasma hominis vertebral spine infection: case report and a review of infections of bone and joints. J Infect Chemother. 2016;22(11):755–758. doi:10.1016/j.jiac.2016.04.008
10. Yang J, Xie S, Li J, Xia H, Liu X. Brain abscess caused by nocardia farcinica and diagnosed by metagenomic next-generation sequencing: a case report. Front Med. 2022;9:803554. doi:10.3389/fmed.2022.803554
11. Duan H, Li X, Mei A, et al. The diagnostic value of metagenomic next-generation sequencing in infectious diseases. BMC Infect Dis. 2021;21(1):62. doi:10.1186/s12879-020-05746-5
12. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–s240. doi:10.1093/cid/ciy693
13. Safarkar R, Mehrabadi JF, Noormohammadi Z, Mirnejad R. Development a rapid and accurate multiplex real time PCR method for the detection Chlamydia trachomatis and Mycoplasma hominis. J Clin Lab Anal. 2017;31:6. doi:10.1002/jcla.22126
14. Crossley BM, Bai J, Glaser A, et al. Guidelines for Sanger sequencing and molecular assay monitoring. J Vet Diagn Invest. 2020;32(6):767–775. doi:10.1177/1040638720905833
15. Hebert PDN, Braukmann TWA, Prosser SWJ, et al. A sequel to sanger: amplicon sequencing that scales. BMC Genomics. 2018;19(1):219. doi:10.1186/s12864-018-4611-3
16. Meyer RD, Clough W. Extragenital Mycoplasma hominis infections in adults: emphasis on immunosuppression. Clin Infect Dis. 1993;17(Suppl 1):S243–249. doi:10.1093/clinids/17.Supplement_1.S243
17. Fernández S, Nicolás D, Pericás JM, et al. A case of Mycoplasma hominis disseminated infection in a human immunodeficiency virus-1-infected pregnant woman with hypogammaglobulinemia. J Microbiol Immunol Infect. 2017;50(1):118–119. doi:10.1016/j.jmii.2014.11.006
18. Romeu Prieto JM, Lizcano Lizcano AM, Martín Consuegra IL, Largo Pau J, López Almodóvar LF, García Camacho E. Culture-negative endocarditis: mycoplasma hominis infection. Rev Esp Cardiol. 2015;68(11):1037–1038. doi:10.1016/j.rec.2015.07.018
19. Henrich B, Feldmann RC, Hadding U. Cytoadhesins of Mycoplasma hominis. Infect Immun. 1993;61(7):2945–2951. doi:10.1128/iai.61.7.2945-2951.1993
20. Hata A, Honda Y, Asada K, Sasaki Y, Kenri T, Hata D. Mycoplasma hominis meningitis in a neonate: case report and review. J Infect. 2008;57(4):338–343. doi:10.1016/j.jinf.2008.08.002
21. Kupila L, Rantakokko-Jalava K, Jalava J, et al. Brain abscess caused by Mycoplasma hominis: a clinically recognizable entity? Eur J Neurol. 2006;13(5):550–551. doi:10.1111/j.1468-1331.2006.01209.x
22. Pailhoriès H, Rabier V, Eveillard M, et al. A case report of Mycoplasma hominis brain abscess identified by MALDI-TOF mass spectrometry. Int J Infect Dis. 2014;29:166–168. doi:10.1016/j.ijid.2014.08.004
23. Waites KB, Duffy LB, Crouse DT, et al. Mycoplasmal infections of cerebrospinal fluid in newborn infants from a community hospital population. Pediatr Infect Dis J. 1990;9(4):241–245. doi:10.1097/00006454-199004000-00004
24. Waites KB, Canupp KC. Evaluation of BacT/ALERT system for detection of Mycoplasma hominis in simulated blood cultures. J Clin Microbiol. 2001;39(12):4328–4331. doi:10.1128/JCM.39.12.4328-4331.2001
25. Waites KB, Xiao L, Paralanov V, Viscardi RM, Glass JI. Molecular methods for the detection of Mycoplasma and ureaplasma infections in humans: a paper from the 2011 William Beaumont hospital symposium on molecular pathology. J Mol Diagn. 2012;14(5):437–450. doi:10.1016/j.jmoldx.2012.06.001
26. Cao X, Wang Y, Hu X, Qing H, Wang H. Real-time TaqMan polymerase chain reaction assays for quantitative detection and differentiation of Ureaplasma urealyticum and Ureaplasma parvum. Diagn Microbiol Infect Dis. 2007;57(4):373–378. doi:10.1016/j.diagmicrobio.2006.09.006
27. Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol. 2014;4:25. doi:10.3389/fcimb.2014.00025
28. Gosiewski T, Ludwig-Galezowska AH, Huminska K, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36(2):329–336. doi:10.1007/s10096-016-2805-7
29. Guo LY, Feng WY, Guo X, Liu B, Liu G, Dong J. The advantages of next-generation sequencing technology in the detection of different sources of abscess. J Infect. 2019;78(1):75–86. doi:10.1016/j.jinf.2018.08.002
30. Fan S, Ren H, Wei Y, et al. Next-generation sequencing of the cerebrospinal fluid in the diagnosis of neurobrucellosis. Int J Infect Dis. 2018;67:20–24.
31. Hu Z, Weng X, Xu C, et al. Metagenomic next-generation sequencing as a diagnostic tool for toxoplasmic encephalitis. Ann Clin Microbiol Antimicrob. 2018;17(1):45. doi:10.1186/s12941-018-0298-1
32. McCormack WM. Susceptibility of mycoplasmas to antimicrobial agents: clinical implications. Clin Infect Dis. 1993;17(Suppl 1):S200–201. doi:10.1093/clinids/17.Supplement_1.S200
33. Krausse R, Schubert S. In-vitro activities of tetracyclines, macrolides, fluoroquinolones and clindamycin against Mycoplasma hominis and Ureaplasma ssp. isolated in Germany over 20 years. Clin Microbiol Infect. 2010;16(11):1649–1655. doi:10.1111/j.1469-0691.2010.03155.x
34. McCarthy KL, Looke DF. Successful treatment of post-neurosurgical intracranial Mycoplasma hominis infection using gatifloxacin. J Infect. 2008;57(4):344–346. doi:10.1016/j.jinf.2008.06.022
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.