Back to Journals » Infection and Drug Resistance » Volume 14
Developing Machine-Learning Prediction Algorithm for Bacteremia in Admitted Patients
Authors Mahmoud E , Al Dhoayan M, Bosaeed M , Al Johani S , Arabi YM
Received 23 November 2020
Accepted for publication 14 January 2021
Published 25 February 2021 Volume 2021:14 Pages 757—765
DOI https://doi.org/10.2147/IDR.S293496
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Ebrahim Mahmoud,1 Mohammed Al Dhoayan,2,3 Mohammad Bosaeed,1,4,5 Sameera Al Johani,5,6 Yaseen M Arabi5,7
1Department of Infectious Disease, Department of Medicine, King Abdulaziz Medical City, Riyadh, Saudi Arabia; 2Department of Health Informatics, CPHHI, King Saud Bin Abdulaziz University for Health Sciences, Riyadh, Saudi Arabia; 3Data and Business Intelligence Management Department, ISID, King Abdulaziz Medical City, Riyadh, Saudi Arabia; 4King Abdullah International Medical Research Center (KAIMRC), Riyadh, Saudi Arabia; 5College of Medicine, King Saud Bin Abdulaziz University For Health Sciences, Riyadh, Saudi Arabia; 6Department of Pathology & Laboratory Medicine, King Abdulaziz Medical City, Riyadh, Saudi Arabia; 7Department of Intensive Care, King Abdulaziz Medical City, Riyadh, Saudi Arabia
Correspondence: Ebrahim Mahmoud
Division of Infectious Diseases, Department of Medicine, King Abdulaziz Medical City, Riyadh, Riyadh, Saudi Arabia
Tel +966 500081418
Email [email protected]
Purpose: Bloodstream infection among hospitalized patients is associated with serious adverse outcomes. Blood culture is routinely ordered in patients with suspected infections, although 90% of blood cultures do not show any growth of organisms. The evidence regarding the prediction of bacteremia is scarce.
Patients And Methods: A retrospective review of blood cultures requested for a cohort of admitted patients between 2017 and 2019 was undertaken. Several machine-learning models were used to identify the best prediction model. Additionally, univariate and multivariable logistic regression was used to determine the predictive factors for bacteremia.
Results: A total of 36,405 blood cultures of 7157 patients were done. There were 2413 (6.62%) positive blood cultures. The best prediction was by using NN with the high specificity of 88% but low sensitivity. There was a statistical difference in the following factors: longer admission days before the blood culture, presence of a central line, and higher lactic acid—more than 2 mmol/L.
Conclusion: Despite the low positive rate of blood culture, machine learning could predict positive blood culture with high specificity but minimum sensitivity. Yet, the SIRS score, qSOFA score, and other known factors were not good prognostic factors. Further improvement and training would possibly enhance machine-learning performance.
Keywords: bacteremia, blood culture prediction, machine learning, predictive medicine
Introduction
Bloodstream infection (BSI) is associated with adverse outcomes, including serious complications and increased mortality.1,2 Early detection of BSI is important because its absence may result in inappropriate initial therapy and lead to an increase in overall mortality.3
Though blood culture is routinely ordered for patients with suspected infections, only a small proportion of cultures yield true-positive results. Studies have demonstrated that as many as 90% of all blood cultures do not show growth of any organism.4 Published guidelines do not clearly state when blood cultures should be drawn. Usually, clinician depends on their assessment to order for blood culture if the patient has a fever or a suspected endocarditis, or defined infectious syndromes like central line-associated bloodstream infection (CLABSI).
The field of machine learning is advancing rapidly and, through occurrences and experience continuously learns and improves its skill and decision-making ability. We hypothesize that machine learning would improve the accuracy of predicting bacteremia. This study was aimed at using machine-learning algorithms with data or factors that are routinely collected while admitting patients to the hospital to calculate the probability that a requested blood culture will return a positive result.
Patients and Methods
Source of Data
The study was conducted at King Abdulaziz Medical City (KAMC), which is a tertiary care center in Riyadh, Saudi Arabia with a capacity of over 1500 beds. The hospital uses the health information system “BEST Care”. It is an electronic health record (EHR) that contains all the information about a patient, such as data on medication and physicians’ orders.
Participants
In this retrospective cohort study of blood cultures of admitted, adult patients (age more than 14 years) between July 2017 to July 2019. A flow chart is provided in Figure S1; Supplement 1. The exclusion criteria were repeated tests for the same patient on the same day and patients with “hematological malignancy and organ transplant recipients”. Also, patients who were admitted to the ICU after 48h from their initial admission to the hospital were excluded. Nevertheless, patients who were shifted to the ICU within the first 48h of their admission to the hospital were included. Data related to the patients’ blood culture results and other independent variables were acquired and analyzed to derive and validate an algorithm that could predict positive blood culture.
Study Outcome
The outcome was based on positive or negative blood cultures. Contaminant organisms according to Clinical Laboratory & Standards Institute (CLSI) Guidelines were considered as negative. Blood culture, whether positive or negative, was tracked from the time of collection to look for the vital signs and other laboratory investigations before the time of the collection by 12–24 hours.
Several descriptive and predictive analytical techniques were used to detect data patterns and establish associations between positive blood cultures and the routinely collected data.
Predictors
The list of covariates was included in the data as follows:
The following factors were considered to be predictor variables: age, admittance diagnosis, length of stay before blood sample collection, co-morbidities, presence of central line–Foley catheter and tracheostomy at the time of blood culture request, and receiving antibiotics 24h before the blood sample was taken. All of these variables were collected before blood culture collection. Vital signs: Temperature, heart rate, systolic and diastolic blood pressure, respiratory rate, and Glasgow Coma scale. Laboratory testing: WBC count, platelet count, creatinine level, lactic acid level, C-reactive protein (CRP), and procalcitonin. SIRS and qSOFA scores were also included in the analysis.
Missing Variables
After all the data pre-processing was conducted, missing values were review. To remedy for missing values, two techniques were used. First, to drop out any record with any missing data point. Second, since most of the missing values were missing completely at random (MCAR), k-Nearest Neighbors imputation was used to impute these missing values with the 3 nearest neighbors and a uniform weight function.5
All used ML algorithms were performed on data with and without missing data imputation and their performance was compared to measure the effect of missing data on the performed analyses. Furthermore, the interquartile range (IQR) rule was used to detect and remove any outliers.6
The last step in the pre-processing was to balance the distribution between negative and positive blood cultures in the outcome variable to avoid any bias in the ML models’ predictions. To do so, the training data were subjected to Synthetic Minority Over-Sampling Technique (SMOTE) using the Imbalanced-learn package in Python 3.7.7,8 Missing variables numbers are provided in Table S2; Supplement 3.
Definitions
Contamination
The following bacterial pathogens were recognized as contaminants according to Clinical Laboratory & Standards Institute (CLSI) Guidelines, namely, coagulase-negative staphylococci, Corynebacterium spp. (“diphtheroid”), Propionibacterium spp, Aerococcus spp, Micrococcus spp, or Bacillus spp. Cultures showing their presence were considered as negative blood culture result. It is to be noted that blood cultures with two organisms or more were considered as positive if an organism that is not considered as a contaminant was present.
Statistics and Machine Learning
To achieve the objective of this study, predictive analytics were used to develop classification models that could differentiate between positive and negative blood cultures using the data elements described above. Since the outcome of the blood cultures was either positive or negative, multiple binary ML algorithms were trained and validated to classify each blood culture to either positive or negative. The followed approach was to train multiple models on the same dataset and compare their performances. The first model was built using Random Forest (RF) with 100 estimators and maximum depth of 2 levels.9 The second model was built using Logistic Regression (LR, aka logit, MaxEnt) classifier, which implements regularized logistic regression using the “liblinear” solver.10
The third was built using Decision Trees (DT) with a maximum depth of 2 levels.11 The fourth model was built using Naive Bayes classifier for multivariate Bernoulli models (NB).12 The fifth was using Neural Networks (NN) with 7 hidden layers with a dropout of 20% to control for over fitting and a learning rate of 0.1.13 All layers had Elu activation function except for the last layer which had sigmoid activation function for binary outcome. The sixth model was C-Support Vector Machine Classification (SVM) with a Radial Basis Function (RBF) kernel, and 0.031 gamma.14 All these models were subjected to multiple hyperparameter tunings to select the best setup for each algorithm. All of these models (Except for NN) were developed using Scikit-learn package in Python 3.7.15
Furthermore, univariate logistic regression models were used to detect patterns and associations between the independent variables and the outcome of the blood cultures. All variables that were found to be associated with blood culture outcome at a level of significance p-value 0.05 were included in a multiple logistic regression model with a level of significance at p-value 0.05 as well.
Results
Participants
Between July 2017 and July 2019, 36,405 blood cultures were requested for 7157 admitted patients. The final analysis for the prediction model included (n=21,073 cultures).
Their mean age was 61.5 years, with almost equal numbers of males and females. The mean hospital length of stay was 18.9 days before blood culture was requested. Almost one-fifth of the patients had a central line at the time of blood culture request (18.35%) and urinary tract infections were labeled in 20.86% of the patients. Concerning comorbidities, 11.45% had heart failure and 13.63% of the patients underwent surgical procedures within 14 days of blood culture requests. The other demographics are listed in Table 1.
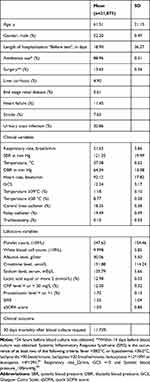 |
Table 1 Characteristics of Blood Cultures Episodes Included in Analysis (n=21,073) |
The majority of the patients were on antibiotics the day before collection (89.96%). Their mean (Quick SOFA) qSOFA score was 1, and their mean SIRS score was 1.55. Among 12% of the patients, lactic acid was more than 2 mmol/L.
Their vital signs were near-normal except for the higher mean of the respiratory rate of 21 breaths/min. Only 8.77% of the patients had a temperature of 38◦C or more, at the time of blood culture request. The presence of higher temperatures, more than 39◦C was the trigger for blood culture was in 1.18% of the patients.
Positive Blood Culture and Microbiology
The number of total true positive blood cultures was 2413 and those with contaminations were 1829. Table 2 lists the organisms found in positive blood cultures. The most common pathogen was Enterobacteriaceae (Klebsiella, Pseudomonas, and E. coli). These collectively accounted for 1525 (43%) of total cultures. Candidemia was found in 350 (14.5%) cultures and was more frequent than Staph. aureus found in 246 (10.19%) cultures. The distribution of positive blood cultures suggests that one-third of the positive blood cultures resulted from community-acquired infections (occurred in the first 72 h of the admission), while (43%) occurred after 16 days of admission.
 |
Table 2 Microbiology of True Bacteremia |
Performance of the Models and Predictor
Table 3 lists the performance of each model. The highest specificity achieved by NN was 89%, with a sensitivity of 17%. The best sensitivity was by Logistic regression (31%) with a specificity of (73%). Performance of non-imputed machine learning models is provided in Table S1; Supplement 2. Although SVM scored 100% specificity, it scored zero on sensitivity, which rendered the high specificity null. Since the SVM and NN are considered black-box algorithms, it is impossible to identify the relative importance of the independent variables to the prediction of positive blood culture. For this reason, in addition to machine learning, we undertook the examination of univariate relationships then multivariate analysis to identify the eligible covariates (see Table 4).
 |
Table 3 Machine Learning Models Performance |
 |
Table 4 Independent Univariate Predictors of Positive Blood Culture Results |
The following factors are believed to be most significantly associated:
Length of hospitalization more than 16 days (OR,1.88; 95% CI, 1.70–2.08), presence of central line catheter (OR,1.87; 95% CI, 1.67–2.09), lactic acid more than 2 (OR,1.53; 95% CI, 1.34–1.75) and Glasgow Coma Scale score (OR,1.23; 95% CI, 1.11–1.36).
Among the vital signs, only temperature was considered to be statistically significant (OR,1.23; 95% CI, 1.14–1.33): temperature at 38◦C or more (OR,1.50; 95% CI, 1.28–1.75) and temperature at 39◦C or more (OR,1.62; 95% CI, 1.11–2.38). Meanwhile, several factors such as being on antibiotics or leukocyte count did not show statistical significance. Nevertheless, the following factors were found to affect machine-learning performance:
Demographics and comorbidities: Age, antibiotics use, surgery within 14 days, Central Lines catheter, and length of hospitalization before blood culture test. Vital signs: Respiratory rate, systolic blood pressure, temperature, diastolic blood pressure, heart rate, and temperature. Laboratory test: White blood cell count, sodium level, platelet count, albumin level, and creatinine level.
The Scores of SIRS (OR,1.18; 95% CI, 1.12–1.24), and qSOFA (OR,1.22; 95% CI, 1.15–1.29), or their performance whether positive or negative (score 2 or more) (OR,1.34; 95% CI, 1.21–1.48) (OR,1.40; 95% CI, 1.26–1.55) respectively, were potential univariate predictors of bacteremia. However, they were not considered statistically significant in multivariate analysis. In the final stepwise logistic regression (Table 5), the significant predictors of bacteremia were the length of hospitalization (OR,1.30; 95% CI, 1.14–1.48), central line (OR,1.37; 95% CI, 1.21–1.55) and lactic acid more than 2 (OR,1.31; 95% CI, 1.13–1.52).
 |
Table 5 Independent Multivariate Predictors of Positive Blood Culture Results |
Discussion
In this largest analysis of the predictors of the positive blood culture using machine learning, there were nearly 34,000 negative blood cultures withdrawn during the study period, highlighting the financial waste and the unnecessary burden on the microbiology lab. However, those findings are little less than the literature rate of positivity between (8–10%)16–18 for such a serious infection that has a crude 30-days mortality rate between 13%–21%.1 This attempt at predicting positive blood culture is not a novel idea.16,18–21 However, the previous work focused mainly on community-acquired bacteremia setting (patients reporting to the emergency room), and the result was limited by low specificity. 18
In our model, very high specificity was achieved but the sensitivity was low. Thus, our model would show higher false-negative results and missing true cases of bacteremia being labeled as negative, although the ability to rule out the disease was high. We believe the limited sensitivity and performance are likely to be related to the following:
- The drawing of blood culture involves several steps and factors including the volume of the sample, which could affect the result. The volume of the blood sample has been noted in the old literature as the single most important factor for positivity and that holds in the era of highly automated blood culture machines based on the higher positive rate in the patients with higher APACHE II scores.22 Such factors are relevant in clinical practice and of value, and may explain the discordant result (positive and negative blood culture results, both collected at the same time: (n = 743 episodes)), in our analysis.
- The Heterogeneity implicated
A) The variable risk for bacteremia, which has been postulated by different studies depending on the different infectious syndromes (low risk in isolated fever, but as high as 50% in discitis, meningitis, and catheter-associated blood-stream infection).23 Thus, to cohort all of those patients in the same category assuming all patients have the same risk of bacteremia is likely to affect the machine-learning ability to predict because the risk needs to be classified based on various syndromes.
B) Variability among the patient population and the pathogens causing bacteremia. Therefore, bacteremia in the first 48 hours of admission is different from hospital-acquired infections whether in terms of the pathogen involved, risk factors, and site of infection.24 Furthermore, different etiologies of bacteremia and pathogens (gram-positive- gram-negative and candidemia) shown to make a difference to the machine-learning model’s performance. The distribution of blood culture through the admission period is provided in Figure S2; Supplement 4.
C) The variable host response to bacteremia ranges between the extremes of stable hemodynamic or shock resulting in multiorgan failure. This was strongly demonstrated by the SIRS and qSOFA score among positive blood cultures; 42% had a negative SIRS score <2 and 63% had negative qSOFA (Figure 1) in contrast with the previous works that showed good association with the SIRS score.19,25 Interestingly, since the introduction of the term SIRS in 1992,26 it was clear that bacteremia could intersect with sepsis and/or SIRS. This observation still holds. Thus, sensitivity in predicting sepsis is low and this was one of the reasons for redefining sepsis as occurring in 1 out of 8 patients (12.5%), and multiorgan failure (MOF) did not meet at least two if the SIRS criteria.27,28 Furthermore, the machine-learning model to predict sepsis by Giannini et al had a similar problem of low sensitivity.29
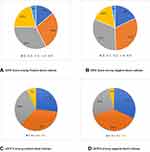 |
Figure 1 SIRS and qSOFA scores distribution among positive and negative blood culture. |
3. Important statistical factors, like a large number of missing values of some of the lab work that physicians usually do not order when suspecting bacteremia such as (Procalcitonin level – lactic acid and other inflammatory markers), which may have a role in prediction.30,31
So, What are the Best Predictors for Blood Culture and When Should We Request Blood Culture?
While fever or leukocytosis is the major clinical driver for the physician to request for blood culture, several previous studies have shown a lack of correlation between these clinical parameters and bacteremia,19,32,33 which this study confirms.
It is not surprising that the presence of central-line, which is a known risk for CLABSI, as a risk factor for bacteremia, a cumulative risk pattern (between1.1–4.8 per 1000 catheter-days).34 However, hospitalization exceeding 16 days has been shown as a strong predictor of bacteremia by various models. Therefore, though this finding is not novel,20 the majority of the previous studies, including Nielsen et al24 did not show such association as this study has shown in and reflected a community-acquired rather than nosocomial bacteremia.
What is Next?
While our study used a large sample size and an extensive analysis of the variables by different methods of machine learning, it validated the previous scores, including the majority of the variables/scores which were thought to be related based on the previous work. Not including some clinical assessment, which is compatible with the machine-learning idea and eliminating the variability and the bias in the assessment of some findings (such as suspected endocarditis, nausea or vomiting, and chills) may be a strength and a limitation of this study at the same time.
Our study has some potential limitations including missing some variables, which may lead to underestimation of some possible associations; inability to assess the risk from the blood culture and which could help to stratify the risk; and, lastly, the large percentage of the population being on antibiotics may change the hosts’ response to bacteremia.
Conclusion
Although Bacteremia is extremely complex and poorly understood, our study provides valuable insights into the predictors of bacteremia such as the duration of hospitalization as meriting attention. The machine-learning performance showed excellent specificity but still needs to improve its sensitivity.
Ethics And Consent
The study was approved by the Institutional Review Board (IRB) in King Abdullah International Medical Research Center (KAIMRC) (Protocol number RC19/325/R). Written informed consent was waived by the IRB, as the study was a retrospective chart review where research involves no more than minimal risk to the subjects. The study complied with the Declaration of Helsinki concerning maintaining the confidentiality of the patient’s data as the data were anonymized.
Disclosure
The authors report no conflicts of interest in this work.
The authors received no financial support for the research, authorship, and/or publication of this article.
References
1. Goto M, Al-Hasan MN. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect. 2013;19(6):501–509. doi:10.1111/1469-0691.12195
2. Kuderer NM, Dale DC, Crawford J, Cosler LE, Lyman GH. Mortality, morbidity, and cost associated with febrile neutropenia in adult cancer patients. Cancer. 2006;106(10):2258–2266. doi:10.1002/cncr.21847
3. Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH. Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob Agents Chemother. 2005;49(4):1306–1311. doi:10.1128/AAC.49.4.1306-1311.2005
4. Paesmans M, Klastersky J, Maertens J, et al. Predicting febrile neutropenic patients at low risk using the MASCC score: does bacteremia matter? Support Care Cancer. 2011;19(7):1001–1008. doi:10.1007/s00520-010-0925-7
5. Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17(6):520–525. doi:10.1093/bioinformatics/17.6.520
6. Analytical Methods C. Robust statistics – how not to reject outliers. Part 1. Basic concepts. Analyst. 1989;114(12):1693–1697.
7. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Int Res. 2002;16(1):321–357.
8. Lemaître G, Nogueira F, Aridas CK. Imbalanced-learn: a python toolbox to tackle the curse of imbalanced datasets in machine learning. J Mach Learn Res. 2017;18(1):559–563.
9. Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi:10.1023/A:1010933404324
10. Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: a library for large linear classification. J Mach Learn Res. 2008;9:1871–1874.
11. Quinlan JR. Induction of decision trees. Mach Learn. 1986;1(1):81–106. doi:10.1007/BF00116251
12. McCallum A. A comparison of event models for naive Bayes text classification. AAAI Workshop. 1998;1998.
13. Abadi M, Barham P, Chen J, et al. TensorFlow: a system for large-scale machine learning.
14. Wu T-F, Lin C-J, Weng RC. Probability estimates for multi-class classification by pairwise coupling. J Mach Learn Res. 2004;5:975–1005.
15. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12(null):2825–2830.
16. Bates DW, Cook EF, Goldman L, Lee TH. Predicting bacteremia in hospitalized patients. A prospectively validated model. Ann Intern Med. 1990;113(7):495–500. doi:10.7326/0003-4819-113-7-495
17. Laukemann S, Kasper N, Kulkarni P, et al. Can we reduce negative blood cultures with clinical scores and blood markers? Results from an observational cohort study. Medicine (Baltimore). 2015;94(49):e2264–e2264. doi:10.1097/MD.0000000000002264
18. Shapiro NI, Wolfe RE, Wright SB, Moore R, Bates DW. Who needs a blood culture? A prospectively derived and validated prediction rule. J Emerg Med. 2008;35(3):255–264. doi:10.1016/j.jemermed.2008.04.001
19. Coburn B, Morris AM, Tomlinson G, Detsky AS. Does this adult patient with suspected bacteremia require blood cultures? JAMA. 2012;308(5):502–511. doi:10.1001/jama.2012.8262
20. Jaimes F, Arango C, Ruiz G, et al. Predicting bacteremia at the bedside. Clin Infect Dis. 2004;38(3):357–362. doi:10.1086/380967
21. Paul M, Andreassen S, Nielsen AD, et al. Prediction of bacteremia using TREAT, a computerized decision-support system. Clin Infect Dis. 2006;42(9):1274–1282. doi:10.1086/503034
22. Bouza E, Sousa D, Rodríguez-Créixems M, Lechuz JG, Muñoz P. Is the volume of blood cultured still a significant factor in the diagnosis of bloodstream infections? J Clin Microbiol. 2007;45(9):2765–2769. doi:10.1128/JCM.00140-07
23. Fabre V, Sharara SL, Salinas AB, Carroll KC, Desai S, Cosgrove SE. Does this patient need blood cultures? A scoping review of indications for blood cultures in adult non-neutropenic inpatients. Clin Infect Dis. 2020;71(5):1339–1347. doi:10.1093/cid/ciaa039
24. Nielsen SL, Lassen AT, Kolmos HJ, Jensen TG, Gradel KO, Pedersen C. The daily risk of bacteremia during hospitalization and associated 30-day mortality evaluated in relation to the traditional classification of bacteremia. Am J Infect Control. 2016;44(2):167–172. doi:10.1016/j.ajic.2015.09.011
25. Wildi K, Tschudin-Sutter S, Dell-Kuster S, Frei R, Bucher HC, Nüesch R. Factors associated with positive blood cultures in outpatients with suspected bacteremia. Eur J Clin Microbiol Infect Dis. 2011;30(12):1615–1619. doi:10.1007/s10096-011-1268-0
26. Bone RCBR, Cerra FB, Dellinger RP, Fein AM, Knaus WA. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992;20(6):864–874.
27. Kaukonen K-M, Bailey M, Suzuki S, Pilcher D, Bellomo R. Mortality related to severe sepsis and septic shock among critically ill patients in Australia and New Zealand, 2000–2012. JAMA. 2014;311(13):1308–1316. doi:10.1001/jama.2014.2637
28. Rodríguez A, Martín-Loeches I, Yébenes JC. New definition of sepsis and septic shock: what does it give us? Med Intensiva. 2017;41(1):38–40. doi:10.1016/j.medin.2016.03.008
29. Giannini HM, Ginestra JC, Chivers C, et al. A machine learning algorithm to predict severe sepsis and septic shock: development, implementation, and impact on clinical practice. Crit Care Med. 2019;47(11):1485–1492. doi:10.1097/CCM.0000000000003891
30. Arai T, Kumasaka K, Nagata K, et al. Prediction of blood culture results by measuring procalcitonin levels and other inflammatory biomarkers. Am J Emerg Med. 2014;32(4):330–333. doi:10.1016/j.ajem.2013.12.035
31. Ratzinger F, Haslacher H, Perkmann T, et al. Machine learning for fast identification of bacteraemia in SIRS patients treated on standard care wards: a cohort study. Sci Rep. 2018;8(1):12233. doi:10.1038/s41598-018-30236-9
32. Linsenmeyer K, Gupta K, Strymish JM, Dhanani M, Brecher SM, Breu AC. Culture if spikes? Indications and yield of blood cultures in hospitalized medical patients. J Hosp Med. 2016;11(5):336–340. doi:10.1002/jhm.2541
33. Seigel TA, Cocchi MN, Salciccioli J, et al. Inadequacy of temperature and white blood cell count in predicting bacteremia in patients with suspected infection. J Emerg Med. 2012;42(3):254–259. doi:10.1016/j.jemermed.2010.05.038
34. Maki DG, Kluger DM, Crnich CJ. The risk of bloodstream infection in adults with different intravascular devices: a systematic review of 200 published prospective studies. Mayo Clin Proc. 2006;81(9):1159–1171.
35. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):801–810. doi:10.1001/jama.2016.0287
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
