Back to Journals » International Journal of General Medicine » Volume 16
Detection of Inflammatory Biomarkers Among Patients with Sepsis of Gram-Negative Bacteria: A Cross-Sectional Study
Received 8 May 2023
Accepted for publication 16 August 2023
Published 31 August 2023 Volume 2023:16 Pages 3963—3976
DOI https://doi.org/10.2147/IJGM.S415200
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Thikra Qader Khana,1 Khanda Abdulateef Anwar2
1Microbiology Department, Shar Teaching Hospital, Sulaimaniyah Directorate of Health, Sulaimaniyah, Iraq; 2Microbiology Department, College of Medicine, University of Sulaimani, Sulaimaniyah, Iraq
Correspondence: Khanda Abdulateef Anwar, Microbiology Department, College of Medicine, University of Sulaimani, Sulaimaniyah, Iraq, Email [email protected]
Background: Sepsis is a highly mixed ailment that affects patients with numerous conditions of infectious sources and can lead to multi-organ failure with dysregulated host immune response.
Objective: To determine inflammatory biomarkers in patients with sepsis caused by Gram-negative bacteria and compare their role in the early detection of sepsis.
Methods: This cross-sectional study was conducted on patients with sepsis admitted to the intensive care unit at different hospitals in Sulaimaniyah, Iraq, from May to December 2021. Patients (n=147) were enrolled in this study according to the primary diagnosis of sepsis by Sequential Organ Failure Assessment scores. Blood samples were taken from patients to investigate white blood cells, inflammatory biomarkers (pentraxin-3, procalcitonin, adrenomedullin, lipopolysaccharide binding protein, interleukin-17A, lactate dehydrogenase, and C-creative protein), blood culture, antibiotic susceptibility test, and coagulation biomarkers (Prothrombin time, activated partial thromboplastin time, and international normalized ratio). Then, isolated Gram-negative bacteria were tested for extended-spectrum β-lactamase enzymes production by screening and combined disc tests.
Results: A total of 51.7% samples were blood culture positive for different Gram-negative bacteria, and P. aeruginosa (51.95%) was a more isolated bacterium. Both males and females were affected by sepsis in a ratio of 1.23:1 with different age groups. Extended-spectrum β-lactamase was estimated to be 77.2% by antibiotic profile, and the rate decreased using two double-disc synergy tests. This was confirmed by combined disc test at a rate of 41.35%. The most prevalent biomarkers were procalcitonin (88.16%), adrenomedullin (84.21%), pentraxin-3 (22.37%), and lipopolysaccharide binding protein (11.84%).
Conclusion: Sepsis is a life-threatening condition that can be diagnosed early by several blood biomarkers such as procalcitonin, adrenomedullin, and pentraxin-3 combined with a standard blood culture technique to improve the patient outcome.
Keywords: β-Lactamase, double-disc synergy test, SOFA score, sepsis biomarkers, Gram-negative bacteria
Graphical Abstract:
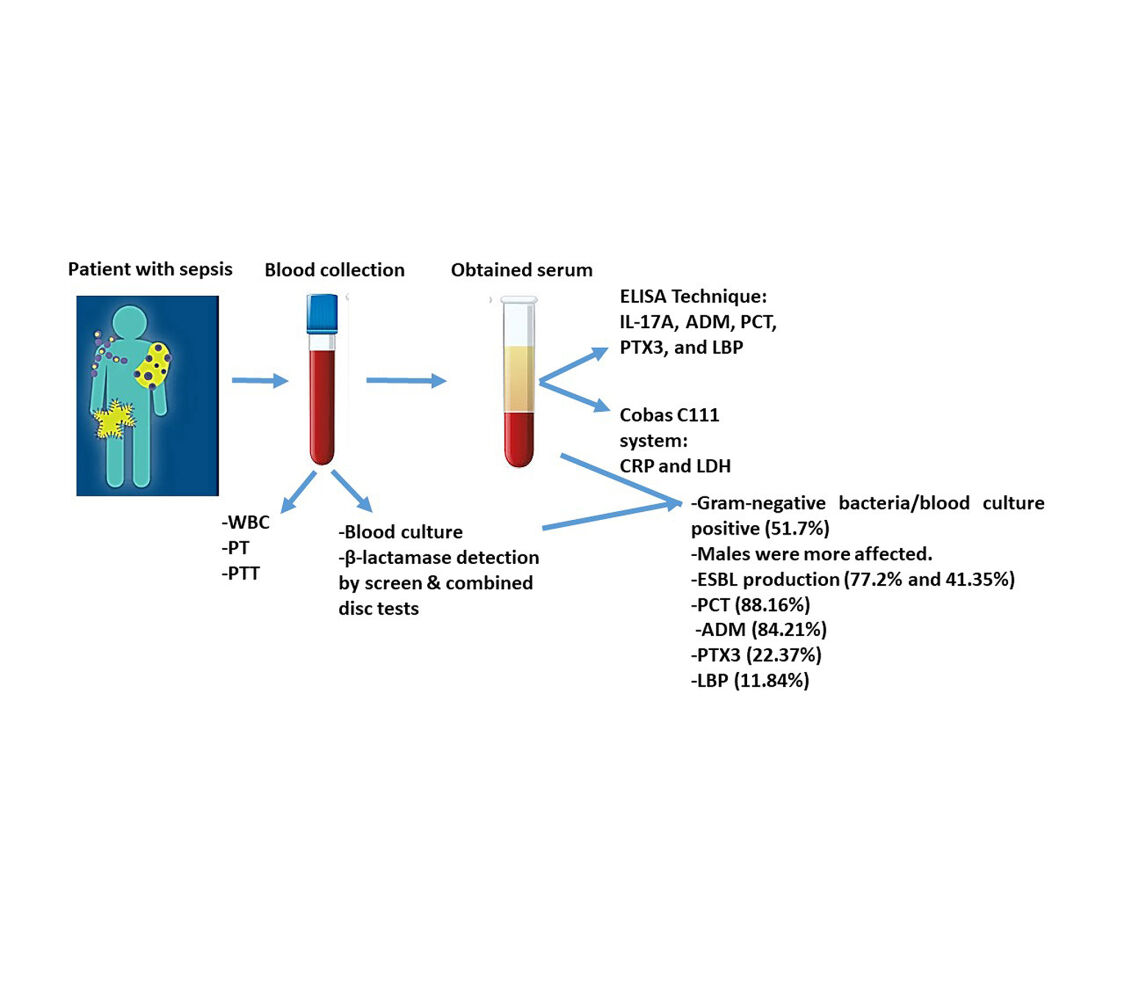
Introduction
Sepsis is a pathological syndrome initiated by a definite or suspected infection that leads to a high global mortality rate among people of all ages. Sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection and “septic shock”. The term “severe sepsis” was replaced by this new definition of sepsis.1 The 2016 consensus definitions also recommend that the Sequential (Sepsis-related) Organ Failure Assessment (SOFA) criteria and “quick” (q)SOFA criteria to be used to identify sepsis, in place of the currently used systemic inflammatory response syndrome (SIRS) criteria, which was the basis for the previous definition of sepsis.2 Sepsis results in a complex immune response characterized by pro-inflammatory and compensatory anti-inflammatory mechanisms. Therefore, most patients with sepsis rapidly exhibit signs of profound immunosuppression, with harmful consequences such as acute kidney injury and multi-organ failure caused by hospital-acquired Gram-negative bacilli or Gram-positive cocci, among immunocompromised patients and patients with chronic and debilitating diseases specifically ESBL producing Enterobacterales, multiple drug-resistant Pseudomonas aeruginosa (MDR-PA) with carbapenem-resistant Enterobacterales.3,4 A study in the Rozh Halat Emergency Hospital, Erbil, Iraq, shows that sepsis is associated with high overall mortality rate (68%) and factors associated with high mortality were female gender, older age group, positive blood culture, wrong antibiotics therapy, less fluid resuscitation, multisource of infection, multi-organ failure, high lactic acid level, and high qSOFA score.2
The inflammatory response of sepsis starts by activation of the innate immune system, which is the primary immune response to microbial infection that produces a range of pro-inflammatory cytokines that trigger cytokines storm,5 and these pro-inflammatory mediators and cytokines overproduction result in cell death in the form of apoptosis and necrosis.6
Several sepsis biomarkers had been commercialized over the past decade. They can promise the accurate detection of infection and prediction of the development of organ dysfunction and guide to antibiotic therapy.7,8 In this respect, procalcitonin (PCT) is used as an indicator for antibiotic treatment as its level is higher in bacterial infections than in viral infections. Early detection of PCT in sepsis had been recommended to be associated with an unfavorable prognosis.9,10 The pentraxin (PTX) family plays an important role in regulating inflammation, an acute phase protein, and it is a novel early diagnostic/prognostic biomarker in patients with sepsis.11
Another critical hormone involved in regulating the endothelium barrier and vascular tone is adrenomedullin (ADM), which associates with improvements in organ dysfunction scores and lowers mortality.12,13 A novel predictor of sepsis progression and an attractive therapeutic target is interleukin-17A (IL-17A) which confers powerful protective effects against various infectious agents.14 Moreover, lipopolysaccharide-binding protein (LBP) is widely reported as a biomarker to differentiate infected from non-infected patients; however, its diagnostic use for sepsis remains a matter of debate.15 Thus, this study aimed to investigate inflammatory biomarkers in patients with sepsis and compare their role in the early detection of sepsis in Sulaimaniyah, Iraq.
Patients and Methods
Sample Size and Study Setting
This cross-sectional study was conducted on 147 patients suspected to have sepsis and admitted to the ICU and Dialysis Unit in Shar Teaching Hospital, Hiwa Hospital, Burn and Plastic Surgery Hospital, and Anwar Shexa Medical City in Sulaimaniyah, Iraq, from the beginning of the May to the end of December 2021. This study was conducted in accordance with the Declaration of Helsinki and signed informed consent was obtained from all patients before enrollment. Patients with suspected sepsis were allocated and diagnosed based on SOFA. Patients with two or more SOFA scores considered to have sepsis and these scores include several variables (alteration of partial arterial oxygen pressure (PaO2)/fraction of inspired oxygen (FiO2) ratio, worsening thrombocytopenia, high total bilirubin level, hypotension with/without the use of vasopressors, increasing serum creatinine and declining urinary output and deteriorating, mean arterial pressure of ≤70 mmHg, and alteration in the level of consciousness by Glasgow Coma Scale, which recommended to help in sepsis diagnosis and prognosis.16
Inclusion Criteria
All patients confirmed to have sepsis with gram negative bacterial infection (adults and elderly) were enrolled in this study regardless of gender, ethnicity, and nationality.
Exclusion Criteria
Only pediatric age group had been excluded from this study.
Questionnaire
A well-designed questionnaire was used to collect the patient’s sociodemographic data, including age, sex, and occupation, together with hospitalization duration and antibiotic profile history.
Study Protocol
An expert phlebotomist obtained about 10 mL of blood from each patient. First, a part of the blood (3.0 mL) was used to determine white blood cell (WBC) using the Medonic Beckman Coulter system (Boule, Sweden) and coagulation biomarkers such as prothrombin time (PT) and partial thromboplastin time (PTT) using Solea (Biolab, France). Then, INR (international normalized ratio) was calculated. Another part of the blood (7.0 mL) was used for serum collection after centrifugation at 3000 rpm for 15 minutes, aliquoted, and frozen at −80°C. Consequently, serum was used to estimate inflammatory biomarkers (CRP, PCT, ILT-17A, PTX3, ADM, LBP, and LDH).
Blood Culture
Two sets of blood cultures (10 mL of blood per blood culture bottle) were obtained before the initiation of antimicrobial therapy by locating of the peripheral venous site before skin antisepsis which achieved by using 2% chlorhexidine in 70% isopropyl alcohol. For this purpose, blood samples were inoculated directly into blood culture bottles specific for BacT/Alert (3D) automated blood culture system (Marcyl’Étoile, France) and processed based on the standard guidelines of the manufacturer. Then, bottles with positive signals were subjected to routine Gram-stain and further sub-cultured on different culture media called MaCconkey agar, Blood agar, and Chocolate agar (Oxoid Ltd., Basingstoke, United Kingdom) and incubated at 37°C for 18–24 hours. After incubation, the colony was characterized by colony appearance, Gram stain, and biochemical tests.17
Screen Test 1 (Antimicrobial Susceptibility Test; AST)
Bacterial isolates were identified using VITEK 2 compact automated system (bioMérieux, France) using Gram-Negative Identity Card (GN ID card) and VITEK 2 AST card except for colistins, cefepime, ceftazidime–avibactam (CZA), ceftriaxone, aztreonam, and amoxiclav, that were processed manually on Muller Hinton agar (MHA) plates using Kirby–Bauer disc diffusion method according to CLSI 2020 guideline.18 In addition, standard bacterial strains of E. coli ATCC 25922 and P. aeruginosa ATCC 25853 were used as quality control.
Screening for Potential ESBL-Producing Isolate (Screen Test-1)
Isolated Gram-negative bacteria were tested for ESBL enzyme production by screen and confirmatory (combined disc; CD test) methods. The isolates that showed an inhibition zone of ≤22 mm with ceftazidime (30 µg), ≤27 mm with cefotaxime (30 µg), ≤25 mm with ceftriaxone (30 µg), and ≤27 mm with aztreonam (30 µg) were considered potential ESBL enzyme producers. On the other hand, combined disc of ceftazidime–clavulanate (30/10 µg) was put 20 mm apart from ceftazidime disc alone (30 µg). Positive results were considered as an increase in the inhibition halo of the combined disc (≥5 mm) compared with the ceftazidime disc alone.19 Klebsiella pneumoniae ATCC 700603 was used as ESBL-positive control strain.20
Screen Test 2 (Double-Disc Synergy Test 1; DDST-1)
The ESBL production in the selected pathogens was identified by placing amoxicillin-clavulanic acid (20/10 µg), and 30 µg disc of each third-generation cephalosporin (ceftazidime, cefotaxime, ceftriaxone, and cefixime) at a distance of 20 mm from center to center to amoxicillin–clavulanic acid on MHA plates stroked with the tested organism. Development of the inhibition (keyhole phenomenon) towards the clavulanate disc indicated a potential ESBL positive21 (Figure 1).
 |
Figure 1 Double-disc synergy test 1. Positive test: Enhancement of zone of inhibition from amoxicillin–clavulanic acid towards ceftazidime disc and ceftriaxone disc (black arrow). |
Screen Test 3 (Double-Disc Synergy Test 2; DDST-2)
DDST-2 was used for the first time to screen ESBL enzyme production using ceftazidime–avibactam (CZA) 30/20 µg at the center of 20 mm apart from 30 µg disc of each 3rd generation cephalosporins (ceftazidime, cefotaxime, ceftriaxone, and cefixime) on MHA plates stroked with the tested organism. The extension zone of inhibition (keyhole phenomenon) toward the inhibitor disc was recorded as a positive result (Figure 2).
 |
Figure 2 Double-disc synergy test 2. Positive test: Enhancement of zone inhibition from ceftazidime–avibactam towards ceftriaxone (black arrow). |
Biomarker Estimation
Several biomarkers were assessed, including CRP, PTX3, IL-17A, lactate dehydrogenase (LDH), PCT, ADM and LBP. A commercial ELISA kit (Elabscience ELISA kit, China) was used to measure serum levels of IL-17A, ADM, PCT, PTX3, and LBP. In contrast, CRP and LDH were measured using an automated multipara metric analyzer Cobas C111 (Roche Diagnostics, Mannheim, Germany).
Statistical Analysis
The collected data were entered into the Statistical Package for the Social Sciences (SPSS, version 25.0, IBM Corporation, Armonk, NY, USA). The Chi-square test was used for the correlation between variables. Descriptive statistics were presented as mean ± standard deviation (SD) and as frequency/percentages for categorical variables. A P-value was set as very highly significant (P<0.000), highly significant (P<0.001), significant (P<0.05), and non-significant (P>0.05).
Results
Out of 147 studied patients, 76 (51.7%) were positive blood cultures, while 71 (48.3%) were negative. All age groups were reported to have positive blood cultures with the highest value (47.37%) for >50 years; however, a significant correlation was found between age and blood culture results (P=0.001). The ratio of males to females affected by sepsis was 1.23:1 with no significant difference between age groups and blood culture results (P=0.3001). Regarding the patients’ occupation, workers reported having more positive blood cultures (34.21%), followed by housewives (31.58%), while employers were the least (3.95%). In this regard, a positive correlation was found between patients’ occupation and blood culture results (P=0.028). On the other hand, the hospitalization duration was 1 week for most patients (39.47%) with positive blood cultures; however, for blood culture negatives was >2-weeks in most patients (46.5%). No significant correlation was found between hospital stay duration in patients and their blood culture status (P=0.081) (Table 1).
 |
Table 1 Socio-Demographic Characteristics Among Patients with Sepsis |
Additionally, different clinical conditions were recorded as a source of infection to induce sepsis, including skin and soft tissue infection (42.18%), followed by respiratory tract infection (22.45%), then device-related infection (21.77%), bone and joint infection (9.52%), and chronic kidney disease (2.72%). At the same time, intra-abdominal infection was reported to be the least source of sepsis (1.36%). A significant relationship was found between sources of infection and sepsis rate (P=0.041) (Table 2).
 |
Table 2 Causes of Infection Among Patients with Sepsis |
Moreover, the most frequently isolated bacteria in this study were Enterobacterales, such as Escherichia coli (47.06%), followed by Klebsiella pneumoniae (41.18%), Proteus mirabilis (4.71%), Raoultella ornithinolytica (3.53%), Enterobacter cloacae (2.35%) and Salmonella (1.18%). Among the non-fermenter group, the most prevalent type was Pseudomonas aeruginosa (51.95%), followed by Acinetobacter baumannii (42.86%), Burkholderia cepacia (2.6%), then Stenotrophomonas maltophilia and Alcaligenes faecalis (1.3% each). A significant relationship was not found between Gram-negative bacterial isolates and using various screening tests (P>0.05) (Table 3).
 |
Table 3 Isolated Gram-Negative Bacteria from Patients with Sepsis |
Furthermore, we found the most resistant antibiotics to be third-generation cephalosporin at the rate of 72.5%, 70%, and 62.5% for cefotaxime, ceftazidime, and ceftriaxone, respectively, against E. coli. The resistance rate increased to 88.6% and 82.8% for ceftriaxone, cefotaxime, and ceftazidime against K. pneumoniae. The sensitivity patterns of E. coli and K. pneumoniae for colistins were 90% and 85.7%, respectively, followed by CZA (85% and 77.1%, respectively) and 4th generation cephalosporin (cefepime) (75% and 54.2%, respectively) (Supplementary Table 1).
In the non-fermenter group, P. aeruginosa showed resistance to at least four classes of antibiotics, and the most resistance was against amoxicillin–clavulanic acid (75%), followed by ceftriaxone (67.5%), ceftazidime (65%), and trimethoprim–sulfamethoxazole (52%). On the other hand, colistin was found to be the most effective drug (90%), followed by CZA (80%) and cefepime/aztreonam (72%). In this regard, the resistance pattern was observed against most of the third-generation cephalosporins, quinolones, gentamicin and amikacin. In addition, carbapenem resistance was recorded specifically against pseudomonas and Acinetobacter (42–66%) (Supplementary Table 2).
In the current study, 162 isolates were screened for ESBL enzyme using three different screening methods. About 125 isolates (77.2%) were found to be ESBL producers among isolated Gram-negative bacteria. The rate of ESBL producers in the 2nd screen test (DDST-1) was 20.4%, and 69.8% in the 3rd screen test (DDST-2). However, the detection of ESBL was decreased to 41.35% among all isolates by CD test (P>0.05) (Table 4).
 |
Table 4 Number and Percentage of Phenotypic Tests for ESBL Detection |
We tested several serum biomarkers and their correlation to the early detection of sepsis. PCT was detected among both culture positives and culture negatives at a rate of 88.16% and 74.6%, respectively (P=0.035), while LBP was 11.84% in positive blood cultures and 1.4% in negative blood cultures (P=0.012). Furthermore, ADM was raised among blood culture-positive (84.21%) and at nearly the same rate (88.7%) among culture-negative patients (P=0.424). IL-17A reported a non-significant correlation with a low rate of positivity (P=0.861), but PTX3 was found to be higher among culture-positive cases than negative cultures at a rate of 22.37% and 8.5%, respectively (P=0.02). LDH had an equal percentage among positive and negative culture groups (P=0.371).
Additionally, a higher rate (35.53%) for PT among positive blood cultures than negative blood cultures (19.7%) with a significant correlation (P=0.033), while insignificant differences for both PPT and INR in positive blood cultures compared to negative blood cultures (P>0.05) was reported. Finally, WBC showed higher account in positive blood cultures than negative blood cultures (21.05% vs 2.8%, respectively) with a significant correlation between them (P=0.001) (Table 5), also ROC curve has been calculated for sensitivity and specificity of the biomarkers (Figure 3). Moreover; both mean±SD and t-test was performed for each biomarker with their P-value (Table 6).
 |
Table 5 Sensitivity and Specificity of Inflammatory and Coagulation Biomarkers Among Patients with Sepsis |
 |
Table 6 Comparison of Mean Score of Different Biomarkers with Blood Culture Results Among Patients with Sepsis |
 |
Figure 3 ROC curve of biomarkers. |
Discussion
Early identification of infection severity and organ dysfunction is crucial in improving outcomes of patients with sepsis, as sepsis is a life-threatening and pathological syndrome mainly caused by a dysregulated host response to infection.22 Thus, we aimed to find the blood biomarkers that are directly related to the early stage of sepsis development.
In the current study, males were more affected by sepsis than females (1.23:1). These findings were agreed with that found by Liu et al1 and Xu et al23 in China with Te Marvelde et al in Australia24 who mentioned that most of the patients with sepsis were males. On the contrary, Xue et al in China found that most patients diagnosed with sepsis were females.8 Also, we found the highest value of sepsis in patients aged >50 years, which is inconsistent with that found by Te Marvelde et al in Australia,24 who found the highest sepsis rate in young patients with cancer and Lewis et al in sub-Saharan Africa who saw most patients with HIV infection aged 27–39 years.25 However, these outcomes agreed with that observed by Liu et al in China (age range, 54–82 years)1 and Luhr et al in Sweden (mean age= 62.7±4.2 years).22 Regarding the hospitalization stay for patients, most patients (39.47%) with positive blood cultures stayed for 1 week; while most patients (46.5%) with blood culture negatives was stayed for >2-weeks. In this respect, Xu et al stated the median length of hospitalization and ICU stay for male patients to be higher (19.54 and 7.54 days) than female patients (16.49 and 6.75 days) (P<0.001 and P=0.002, respectively).23
On the other hand, we found that skin and soft tissue infection was the common cause of sepsis, followed by respiratory tract infection. In contrast, intra-abdominal infection was the least source of sepsis. These findings disagreed with that of Liu et al in China, who reported that soft tissue infection is the least cause of sepsis and pneumonia, followed by kidney diseases as the leading causes of sepsis.1 Moreover, Xue et al found the most common cause of infection in sepsis to be pneumonia, followed by urinary tract infection, liver abscess, abdominal infection, and soft tissue infection.8
Additionally, we found that most (51.7%) patients’ blood samples were culture-positive (Gram-negative bacteria), which agreed with Xue et al, who reported Gram-negative bacteria (54.9%) as the main bacteria in sepsis.8 Among isolated Gram-negative bacteria, P. aeruginosa (51.95%) was the most profound bacteria. This outcome agreed with those found by Aljanaby and Aljanaby in Iraq26 and Emami et al in Iran,27 who found P. aeruginosa as the most frequent pathogen 27.6% and 49.9%, respectively in burned patients with sepsis.
In this study, the prevalence rate of ESBL enzyme producers was recorded through an antibiotic profile in which isolates of the family Enterobacterales were produced ESBL enzyme at the highest level of than non-fermenter group (42.6%), especially Klebsiella pneumonia (46.38%) and P. aeruginosa (51.79%). In this regard, P. aeruginosa was found to be the most common ESBL-producer (35.9%) among septic patients in Iraq,26 while Mansouri et al in Iran found ESBL-producer by 59.4% of P. aeruginosa isolates.28 Thus, bacterial resistance is mostly observed against amoxicillin-clavulanic acid, followed by third-generation cephalosporin (cefotaxime, ceftazidime, and ceftriaxone) and then imipenem. These outcomes might be related to excessive use of these antibiotics by populations that cause selective pressure and the emergence of plasmid-mediated mutation of antibiotic resistance gene.29,30 This is a worrying signal in low-income countries and is considered a major public health problem due to the limited laboratory services and therapeutic options despite inadequate infection control measures, especially in the healthcare setting.31
Infections caused by MDR Gram-negative organisms resulted in prolonged hospital stay with increased mortality and cost of management. Thus, new antimicrobial agents such as CZA were developed, combining the third-generation cephalosporin (ceftazidime) and non-β-lactam-β-lactamase inhibitor (avibactam). The effectiveness and safety of CZA globally have been demonstrated in managing MDR Gram-negative infections, including carbapenem-resistant Enterobacterales.32 Due to the presence of CZA in medicine, most manufacturers use this antibiotic as a disc for susceptibility testing. In this study, the disc of CZA was used in AST and as a first trial in DDST-2 for ESBL as an inhibitor disc with a high rate of positive results compared to classical DDST-1. Also, no reported data supports this result and it is not even mentioned in CLSI or EUCAST.
During the last decade, the potential effort had been directed toward the identification and usefulness of biomarkers for early detection of sepsis that can help clinicians to predict and distinguish infection from host response to inflammation.33 Accordingly, we found the highest expression level for CRP, and PCT, followed by ADM, LDH, PTX-3, LBP, and then IL-17A. In this regard, Zhang et al in China reported that IL-10, IL-17, and PCT biomarkers had a high diagnostic value for sepsis patients, particularly those admitted to the ICU.34 In contrast, Piccioni et al in Italy concluded that ADM biomarker detection directly in the emergency department could contribute to improving the prognostic assessment of patients with sepsis.35 Also, Wang et al in China reported the upregulation of the CRP and IL-17A in sepsis patients compared with those in healthy individuals,36 while Vijayan et al in India suggested PCT as a promising diagnostic marker for sepsis37 which increases in the presence IL-1, IL-6, or tumour necrosis factor α (TNF-α), probably due to the inhibition of PCT proteolysis.38,39 Furthermore, Tian et al in China suggested that PTX-3 might be an early predictor to evaluate the severity of sepsis,40 and Youness and Nahla in Egypt mentioned that LDH is a valuable biomarker in predicting sepsis in critically ill pediatric patients, especially when combined with predictive scoring systems.41
Moreover, LBP plays an essential role in innate immunity mechanisms, as it binds to the amphipathic lipid A of bacterial lipopolysaccharide (LPS), and transfers LPS to CD14 protein. By facilitating binding to the CD14 cell membrane molecules, LBP enhances the sensitivity of macrophages and other cells.6 In this study, LBP had a weak expression in septic patients, which is similar to the findings of Chen et al’s meta-analysis study in Taiwan, who reported a weak sensitivity and specificity of LBP in the detection of sepsis and they suggested that LBP was not recommended for clinical utilization as a single biomarker.15
Regarding the PT, PPT, INT, and WBC results, we observed a high level for each PT and WBC in culture-positive blood samples with no noticeable increase in the values of PPT and INR. These findings were also stated by40 in China.
Conclusion
We concluded that sepsis is common among patients with pre-existing infection with 2 weeks of hospitalization. All age groups and both genders could get sepsis; however, elderly and male gender were predominance. Consequently, as a life-threatening condition, sepsis can be caused by various sources. Since most of our patients had a higher level of several cytokines and developed endothelial cell injury in the initial phase of sepsis, thus it can be diagnosed by several biomarkers such as PCT, ADM, and PTX3 combined by standard culture techniques as these biomarkers may be helpful to evaluate severity and prognosis of sepsis in patients.
Abbreviations
ICU, intensive care unit; MDR, multidrug-resistant bacteria; SOFA, Sequential Organ Failure Assessment; WBC, white blood cell count; PT, prothrombin time; PTT, partial thromboplastin time; INR, international normalized ratio; LBP, lipopolysaccharide binding protein; CRP, C-reactive protein; PCT, procalcitonin; PTX3, pentraxin-3; ADM, aAdrenomedullin; IL-17A, interleukin-17A; LDH, lactate dehydrogenase; MHA, Muller–Hinton agar; CLSI, Clinical and Laboratory Standards Institute; ATCC, American Type Culture Collection; CD test, combined disc test; DDST, double-disc synergy test; COVID-19, coronavirus disease 2019; ESBL, extended-spectrum β-lactamase; ELISA, enzyme linked immunosorbent assay.
Data Sharing Statement
The data used to support the findings of this study are included within the article.
Ethical Approval and Consent to Participate
The protocol of this study was accepted with the approval of the Ethics Committee from the Directorate of Health in Sulaimani, Iraq, and the local medical ethics committee of the College of Medicine, University of Sulaimani (No. 78-UoS on May 18, 2021). The participants were enrolled in the study after being informed about the procedure and providing their written consent.
Acknowledgments
The authors would like to appreciate the healthcare staff of the Dialysis Unit in Shar Teaching Hospital, Hiwa Hospital, Burn and Plastic Surgery Hospital, and Anwar Shexa Medical City, Sulaimaniyah, Iraq, and special thanks to laboratory staff of microbiology department in Shar Teaching Hospital for their kind help and assistance by providing facilities and services to this study.
Funding
This work was not funded by any national/international agency, company, or organization.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu J, Bai C, Li B, et al. Mortality prediction using a novel combination of biomarkers in the first day of sepsis in intensive care units. Sci Rep. 2021;11(1):1275. doi:10.1038/s41598-020-79843-5
2. Ahmad S, Ismaeel SM, Mohammed DA, et al. Outcome and management of sepsis at Rozhhalat emergency hospital in Erbil –Kurdistan region of Iraq. J Appl Res. 2018;2018:21–30.
3. Gudiol C, Albasanz-Puig A, Cuervo G, et al. Understanding and managing sepsis in patients with cancer in the era of antimicrobial resistance. Front Med. 2021;8:636547. doi:10.3389/fmed.2021.636547
4. Venet F, Monneret G. Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat Rev Nephrol. 2018;14(2):121–137. doi:10.1038/nrneph.2017.165
5. Hung Y-L, Suzuki K. The pattern recognition receptors and lipopolysaccharides (LPS)-induced systemic inflammation. Int J Med Sci Public Health. 2017;2(7):1–7.
6. Zamyatina A, Heine H. Lipopolysaccharide recognition in the crossroads of TLR4 and caspase-4/11 mediated inflammatory pathways. Front Immunol. 2020;11:585146. doi:10.3389/fimmu.2020.585146
7. Xu C, Li S, Wang Y, et al. Biomarkers in intensive care unit infections, friend or foe? J Emerg Crit Care Med. 2019;3:27. doi:10.21037/jeccm.2019.06.02
8. Xue M, Xu F, Yang Y, et al. Diagnosis of sepsis with inflammatory biomarkers, cytokines, endothelial functional markers from SIRS patients. Medicine. 2022;101(7):e28681. doi:10.1097/MD.0000000000028681
9. Kyriazopoulou E, Liaskou-Antoniou L, Adamis G, et al. Procalcitonin to reduce long-term infection-associated adverse events in sepsis. A randomized trial. Am J Respir Crit Care Med. 2021;203(2):202–210. doi:10.1164/rccm.202004-1201OC
10. Ryoo SM, Han KS, Ahn S, et al. The usefulness of C-reactive protein and procalcitonin to predict prognosis in septic shock patients: a multicenter prospective registry-based observational study. Sci Rep. 2019;9(1):6579. doi:10.1038/s41598-019-42972-7
11. Hamed S, Behnes M, Pauly D, et al. Diagnostic value of Pentraxin-3 in patients with sepsis and septic shock in accordance with latest sepsis-3 definitions. BMC Infect Dis. 2017;17(1):554. doi:10.1186/s12879-017-2606-3
12. Geven C, Kox M, Pickkers P. Adrenomedullin and adrenomedullin-targeted therapy as treatment strategies relevant for sepsis. Front Immunol. 2018;9:292. doi:10.3389/fimmu.2018.00292
13. van Lier D, Kox M, Pickkers P. Promotion of vascular integrity in sepsis through modulation of bioactive adrenomedullin and dipeptidyl peptidase 3. J Intern Med. 2021;289(6):792–806. doi:10.1111/joim.13220
14. Ge Y, Huang M, Yao YM. Biology of interleukin-17 and its pathophysiological significance in sepsis. Front Immunol. 2020;11:1558. doi:10.3389/fimmu.2020.01558
15. Chen KF, Chaou C-H, Jiang J-Y, et al. Diagnostic accuracy of lipopolysaccharide-binding protein as biomarker for sepsis in adult patients: a systematic review and meta-analysis. PLoS One. 2016;11(4):e0153188. doi:10.1371/journal.pone.0153188
16. Fuchs A, Tufa TB, Hörner J, et al. Clinical and microbiological characterization of sepsis and evaluation of sepsis scores. PLoS One. 2021;16(3):e0247646. doi:10.1371/journal.pone.0247646
17. Murray PR, Rosenthal KS, Pfaller MA. Medical Microbiology E-Book. Elsevier Health Sciences; 2020.
18. Melvin P, James S, April M; CLSI. Performance standards for antimicrobial susceptibility testing. In: CLSI Supplement M100.
19. Rajivgandhi G, Maruthupandy M, Ramachandran G, et al. Detection of ESBL genes from ciprofloxacin resistant Gram negative bacteria isolated from urinary tract infections (UTIs). Front Lab Med. 2018;2(1):5–13. doi:10.1016/j.flm.2018.01.001
20. Teklu DS, Negeri AA, Legese MH, et al. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8(1):39. doi:10.1186/s13756-019-0488-4
21. Naseer F. Phenotypic Cofirmatory Disc Diffusion Test (PCDDT), Double Disc Synergy Test (DDST), E-Test Os Diagnostic Tool for Detection of Extended Spectrum Beta Lactamase (ESΒL) Producing Uropathogens. J Appl Biotechnol Bioeng. 2017;3(3):344–349. doi:10.15406/jabb.2017.03.00068
22. Luhr R, Cao Y, Söderquist B, et al. Trends in sepsis mortality over time in randomised sepsis trials: a systematic literature review and meta-analysis of mortality in the control arm, 2002–2016. Crit Care. 2019;23(1):1–9. doi:10.1186/s13054-019-2528-0
23. Xu J, Tong L, Yao J, et al. Association of sex with clinical outcome in critically ill sepsis patients: a retrospective analysis of the large clinical database MIMIC-III. Shock. 2019;52(2):146–151. doi:10.1097/SHK.0000000000001253
24. Te Marvelde L, Whitfield A, Shepheard J, et al. Epidemiology of sepsis in cancer patients in Victoria, Australia: a population‐based study using linked data. Aust N Z J Public Health. 2020;44(1):53–58. doi:10.1111/1753-6405.12935
25. Lewis JM, Feasey NA, Rylance J. Aetiology and outcomes of sepsis in adults in sub-Saharan Africa: a systematic review and meta-analysis. Crit Care. 2019;23(1):1–11. doi:10.1186/s13054-019-2501-y
26. Aljanaby AAJ, Aljanaby IAJ. Prevalence of aerobic pathogenic bacteria isolated from patients with burn infection and their antimicrobial susceptibility patterns in Al-Najaf City, Iraq-a three-year cross-sectional study. F1000 Res. 2018;7(1157):1157. doi:10.12688/f1000research.15088.1
27. Emami A, Pirbonyeh N, Keshavarzi A, et al. Three year study of infection profile and antimicrobial resistance pattern from burn patients in southwest Iran. Infect Drug Resist. 2020;13:1499–1506. doi:10.2147/IDR.S249160
28. Mansouri S, Razavi M, Norouzi F, et al. Prevalence of β-Lactamase production and antimicrobial susceptibility of multidrug resistant clinical isolates of non-fermenting Gram negative bacteria from hospitalized patients in Kerman/Iran. Jundishapur J Microbiol. 2012;5(2):405–410. doi:10.5812/jjm.3399
29. Kakoullis L, Papachristodoulou E, Chra P, et al. Mechanisms of antibiotic resistance in important gram-positive and gram-negative pathogens and novel antibiotic solutions. Antibiotics. 2021;10(4):415. doi:10.3390/antibiotics10040415
30. Munita JM, Arias CA, Kudva IT, Zhang Q. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2):1–15. doi:10.1128/microbiolspec.VMBF-0016-2015
31. Pana ZD, Zaoutis T. Treatment of extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBLs) infections: what have we learned until now? F1000 Res. 2018;7:1–9. doi:10.12688/f1000research.14822.1
32. Swaminathan S, Routray A, Mane A. Early and appropriate use of ceftazidime-avibactam in the management of multidrug-resistant gram-negative bacterial infections in the Indian scenario. Cureus. 2022;14(8):e28283. doi:10.7759/cureus.28283
33. Rios-Toro JJ, Márquez-Coello M, García-álvarez J-M, et al. Soluble membrane receptors, interleukin 6, procalcitonin and C reactive protein as prognostic markers in patients with severe sepsis and septic shock. PLoS One. 2017;12(4):e0175254. doi:10.1371/journal.pone.0175254
34. Zhang W, Wang W, Hou W, et al. The diagnostic utility of IL-10, IL-17, and PCT in patients with sepsis infection. Front Public Health. 2022;10:923457. doi:10.3389/fpubh.2022.923457
35. Piccioni A, Saviano A, Cicchinelli S, et al. Proadrenomedullin in sepsis and septic shock: a role in the emergency department. Medicina. 2021;57(9):920. doi:10.3390/medicina57090920
36. Wang L, Zhao H, Wang D. Inflammatory cytokine expression in patients with sepsis at an intensive care unit. Exp Ther Med. 2018;16(3):2126–2131. doi:10.3892/etm.2018.6376
37. Vijayan AL, Saikant R, Ravindran S, et al. Procalcitonin: a promising diagnostic marker for sepsis and antibiotic therapy. J Intensive Care. 2017;5(1):51. doi:10.1186/s40560-017-0246-8
38. Pierrakos C, Velissaris D, Bisdorff M, et al. Biomarkers of sepsis: time for a reappraisal. Crit Care. 2020;24(1):287. doi:10.1186/s13054-020-02993-5
39. Teggert A, Datta H, Ali Z. Biomarkers for point-of-care diagnosis of sepsis. Micromachines. 2020;11(3):286. doi:10.3390/mi11030286
40. Tian R, Wang X, Pan T, et al. Plasma PTX3, MCP1 and Ang2 are early biomarkers to evaluate the severity of sepsis and septic shock. Scand J Immunol. 2019;90(6):e12823. doi:10.1111/sji.12823
41. Youness E, Nahla A. Clinical role of serum lactic dehydrogenase assessment in critically ill pediatric patients with sepsis. Med J Cairo Univ. 2021;89(September):1917–1927. doi:10.21608/mjcu.2021.203303
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
