Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 15
Detection of Cell-Dissociated Non-Typeable Haemophilus influenzae in the Airways of Patients with Chronic Obstructive Pulmonary Disease
Authors Thulborn SJ, Ceroni A, Haldar K, Mistry V, Cane JL , Brightling CE, Barer MR, Bafadhel M
Received 24 January 2020
Accepted for publication 20 April 2020
Published 12 June 2020 Volume 2020:15 Pages 1357—1365
DOI https://doi.org/10.2147/COPD.S247130
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Samantha J Thulborn,1,2 Alessandro Ceroni,3 Koirobi Haldar,4 Vijay Mistry,4 Jennifer L Cane,1,2 Christopher E Brightling,4,5 Michael R Barer,4 Mona Bafadhel1
1Respiratory Medicine Unit, Nuffield Department of Medicine, University of Oxford, Oxford, UK; 2Oxford NIHR Biomedical Research Centre, University of Oxford, Oxford, UK; 3Target Discovery Institute, Nuffield Department of Medicine, University of Oxford, Oxford, UK; 4Department of Immunity, Infection and Inflammation, University of Leicester, Leicester, UK; 5Institute for Lung Health, University of Leicester, Leicester, UK
Correspondence: Mona Bafadhel
Respiratory Medicine Unit, NDM Research Building, Old Campus Road, University of Oxford, Oxford OX3 7FZ, UK
Tel +44 1865 612898
Email [email protected]
Background: Non-typeable Haemophilus influenzae (NTHi) is the most commonly found pathogen in the lower respiratory airways of patients with COPD. NTHi is predominantly regarded as an intracellular pathogen; however, like most pathogens, it can exist and co-exist in two broad forms: cell-associated (intracellularly or adhered to cells) or cell-dissociated (biofilm dispersed or planktonic). We sought to investigate if cell-dissociated NTHi can be detected from the sputum of COPD patients and assess this relationship to disease severity and airway inflammation.
Methods: DNA was extracted from the sputum plug and cell-free supernatant to quantify absolute (cell-associated and cell-dissociated NTHi) and cell-dissociated NTHi, respectively, from 87 COPD subjects attending an observational longitudinal COPD exacerbation study. NTHi was quantified using TaqMan hydrolysis probes, targeting the OMP P6 gene using qPCR.
Results: At stable state cell-dissociated NTHi was detected 56% of subjects with a median (IQR) of 9.95x102 gene copies (1.26x102 to 1.90x104). Cell-dissociated NTHi correlated with absolute NTHi levels (r=0.34, p< 0.01) but not airway inflammation or spirometry at stable state. At exacerbation, cell-dissociated NTHi correlated with lung function (FEV1 r=0.629, p=0.005; FEV1%predicted r=0.564, p=0.015; FVC r=0.476 p=0.046) and sputum neutrophilic inflammation (% neutrophils r=0.688, p=0.002; total neutrophils r=0.518, p=0.028).
Conclusion: In patients with COPD, NTHi can exist in both cell-associated and cell-dissociated forms. Cell-dissociated NTHi is associated with neutrophilic airway inflammation during exacerbations of COPD and may be a driving factor in worsening lung function during these episodes.
Keywords: NTHi, infection, neutrophils, COPD
Introduction
The lungs are the main interface between the host and the external environment, with constant exposure to pathogens.1 Mucus production, cilia action and phagocytosis by inflammatory cells play an integral part in host-pulmonary defence. Patients with chronic obstructive pulmonary disease (COPD) have impaired pulmonary defences, as a consequence of chronic inflammation, epithelial cell damage and mucus hypersecretion.2 This is likely to play a role for increasing the susceptibility of patients with COPD to respiratory tract infections.3 Furthermore, in these patients, bacterial infection can be found in up to 30% at stable state, termed colonisation, akin to chronic infection; whilst acute bacterial infection can be found in up to 50% during an exacerbation.3 The most frequently found bacteria irrespective of the disease state is non-typeable Haemophilus influenzae (NTHi).4
Bacteria and the host can co-exist in two broad forms: un-attached from the cell, termed cell-dissociated or related to the cell, termed cell-associated.5–7 Cell-dissociated bacteria can be either planktonic or dispersed.7,8 Planktonic bacteria have been shown to infiltrate otherwise sterile sites within the host.9 Cell-associated bacteria can be attached to the host cell by bacterial adhesin and pili or can reside intracellularly.6 Bacteria that exists within a biofilm can either be cell-associated or cell-dissociated (Figure 1). Moreover, bacteria within a biofilm have been shown to evade antibiotic therapy and reside within the host for a longer duration.10,11 The ability of bacteria to adopt different forms could lead to improved bacterial survival rates leading to colonisation.12 However, there is little data examining the different existential forms of bacteria in patients with COPD. NTHi has been shown to be particularly adept at invading host tissue,13 suggesting it could be more prone to adopting a cell-associated form. Otherwise, how NTHi resides within the lungs is generally unstudied. In this study, we sought to investigate if cell-dissociated NTHi can be detected in patients with COPD and how this relates to clinical outcomes.
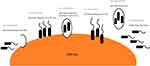 |
Figure 1 Schematic interpretation of how bacteria can reside within a host. |
Materials and Methods
Subjects and Sampling
Sputum samples from COPD subjects entering a longitudinal study looking at biomarkers in COPD were analysed. Subject inclusion and exclusion criterion, study design and measurements are as previously described.14 In brief, subjects attended a stable visit every 3 months over a 12-month period and also during an exacerbation. An exacerbation was defined according to Anthonisen criteria15 and/or health-care use.16 At each visit, pre- and post-bronchodilator spirometry, health status (chronic respiratory questionnaire, CRQ),17 symptoms (visual analogue scale, VAS)18 and blood and sputum samples were obtained. All subjects gave written informed consent and the study was approved by the Leicestershire, Northamptonshire and Rutland ethics committee (reference number: 07/H0406/157). This study was conducted in accordance with the Declaration of Helsinki.
Sputum Processing
Selected sputum plugs were dispersed with Dulbecco phosphate-buffered saline (PBS) and dithiothreitol (DTT). The PBS and DTT sputum supernatants were then stored at −80ᵒC. Cytospin preparation and quantification of cell differential count was performed. A separate sputum plug was sent to the routine microbiology laboratory for bacteria identification. An additional sputum plug was weighed and processed by SYBR green (Applied Biosystems®; Life Technologies Corp, Carlsbad, CA, USA) to determine total 16S and absolute NTHi (cell-associated and dissociated) bacterial DNA using real-time polymerase chain reaction (qPCR) as described previously.14
Cell-Dissociated NTHi Quantification
Quantification of cell-dissociated NTHi was performed by extracting DNA from confirmed acellular sputum supernatant samples using the DNeasy blood and tissue kit (QIAGEN Ltd, Hilden Germany) according to the manufacturer’s gram-negative bacteria protocol (see Supplementary Data). TaqMan qPCR was used to quantify cell-dissociated NTHi. Primers (Sigma, Poole, Dorset) previously determined to be sensitive and specific for the identification of NTHi in respiratory secretions19 were used to target the OMP P6 gene (see Supplementary Data). A plasmid was used for standards for qPCR (see Supplementary Data). All samples were processed in duplicate with standards (serial dilutions of housekeeping gene OMP P6) and negative controls. The lower limit of detection (LLD) was 126 gene copies; all samples below this were considered negative and assigned a value of 0, and included in the analysis.
Statistical Analysis
GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, CA, USA) and SPSS Statistics version 22 (SPSS Inc. Chicago, IL, USA) were used for statistical analysis. The Kolmogorov–Smirnov test for normality was applied. Parametric and non-parametric data are presented as mean (SD) and median (IQR) unless stated. The Studentʼs T test and Mann–Whitney test was performed for comparison between 2 groups in parametric and non-parametric data respectively. For paired analysis, the paired T test and Kruskal–Wallis test were used for parametric and non-parametric data, respectively. A p<0.05 was considered statistically significant.
Results
Sputum was available from 87 COPD subjects (66 males) with a mean (range) age of 69 (43–88). There were 77 sputum samples at stable state and 30 sputum samples at exacerbation available for analysis. Of the 30 exacerbation samples, 20 of these had paired stable state data. The baseline characteristics for the COPD subjects are presented in Table 1.
 |
Table 1 Baseline Characteristics of COPD Subjects Included in Analysis |
Cell-Dissociated NTHi at Stable State
Cell-dissociated NTHi was detected in 56% of samples with a median (IQR) of 9.95 x 102 gene copies (1.26 x 102–1.90 x 104) and was significantly lower than the levels of absolute NTHi detected in the sputum plug (p<0.001, Figure 2). Absolute and cell-disassociated NTHi correlated (r=0.34, p<0.01). Levels of cell-dissociated NTHi were not associated with disease severity as defined by GOLD,20 nor smoking status or inhaled corticosteroid use. Analysis of frequent exacerbators versus non-frequent exacerbators (a frequent exacerbator being more 2 or more treated exacerbations21) according to GOLD criteria showed no difference in levels of cell-dissociated NTHi, p=0.820. Univariate analysis determined that only age and percentage predicted post-bronchodilator FEV1 were statistically different between subjects that had detectable and non-detectable cell-dissociated NTHi (Table 2). Cell-dissociated NTHi levels were higher in sputum samples that were sputum culture positive compared to samples that were culture negative (median (IQR) in culture positive 1.35 x 103 (0 x 103–110.00 x 103) gene copies vs 0 (0–575.8) gene copies in culture negative, p=0.024). There were no correlations of cell-dissociated NTHi with any other parameter at stable state (Table 3).
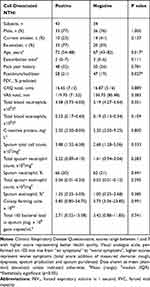 |
Table 2 Differences Observed Between Subjects That Were Positive and Subjects That Were Negative for Cell-Dissociated NTHi at Stable State |
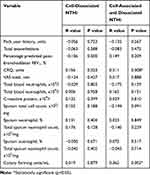 |
Table 3 Correlations Between NTHi Levels and Lung Function, Patient Symptoms and Inflammatory Mediators in Blood and Sputum at Stable State |
 |
Figure 2 Cell-dissociated NTHi levels compared to absolute NTHi levels at stable state. Note: Fifty-nine negative samples not plotted. |
Absolute NTHi at Stable State
Absolute NTHi was detected in 68% of samples with a median (IQR) of 2.59 x 105 gene copies (2.0 x 103–1.1 x 107). Similar to cell-disassociated NTHi, absolute NTHi levels were not associated with disease severity as defined by GOLD.20 Absolute NTHi levels were significantly higher in ex-smokers compared to current smokers (median (IQR) in ex-smokers 8.10 x 104 (0.41 x 104 −131.00 x 104) gene copies vs 0 (0 −24.22 x 104) gene copies in current smokers, p=0.028) and in subjects on ICS compared to those not on ICS (median (IQR) in those on ICS 2.40 x 104 (0 x 104–60.79 x 104)) gene copies vs 24.26 x 104 (8.50 x 104–6425.85 x 104) gene copies in those not on ICS, p=0.034. Bacterial load and CRQ correlated with absolute NTHi levels (Table 3).
Categorisation of absolute (a) and cell-dissociated (cd) non-detected (neg) and detected (pos) samples into 4 groups (Group 1: nega/negcd; Group 2: posa/negcd; group 3: nega/poscd; group 4: posa/poscd) no differences were apparent between these groups (Table 4).
 |
Table 4 Looking at NTHi Levels at Stable State in the Sputum Plug and Supernatant |
Cell-Dissociated NTHi at Exacerbation
At exacerbation cell-dissociated NTHi correlated with lung function (post-bronchodilator FEV1 r=0.629, p=0.005; percentage predicted post-bronchodilator FEV1 r=0.564, p=0.015; FVC r=0.476 p=0.046), blood esosinophils (r=−0.689, p=0.002) and sputum neutrophilic inflammation (sputum neutrophil percentage r=0.688, p=0.002; absolute sputum neutrophils r=0.518, p=0.028). A non-significant trend to increased levels of cell-dissociated NTHi was seen at exacerbation compared to stable state (median (IQR) at stable state 1.91 x 102 (0 x 102–1.38 x 102) gene copies vs 6.67 x 102 (0 x 102–944.75 x 102) gene copies at exacerbation, p=0.061) (Figure 3). This was also seen when looking at the paired samples (median (IQR) at stable state 62.94 (0.06–1.34 x 103) gene copies vs 4.73 x 102 (6.36–6.22 x 104) gene copies at exacerbation, p=0.081).
 |
Figure 3 Cell-dissociated NTHi levels from stable to exacerbation state. Notes: Data shown as median and interquartile range. Negative values are not displayed. |
Absolute NTHi at Exacerbation
Absolute NTHi levels correlated with sputum total cell count (r=0.498, p=0.050), sputum neutrophils (total sputum neutrophil count r=0.612, p=0.12; %sputum neutrophil r=0.612, p=0.012) and bacterial load measured by CFU (r=0.574, p=0.020). There was no significant increase in absolute NTHi at exacerbation compared to stable state (p=0.640).
Discussion
Numerous studies have examined the role of bacteria, including NTHi in patients with COPD,22–25 but these have all examined bacterial detection in sputum plugs, thereby quantifying absolute levels of bacteria which includes both cell-associated and cell-dissociated forms. To our knowledge, this is the first time cell-dissociated NTHi has been measured in sputum samples from patients with COPD. In this study, we have shown that cell-dissociated NTHi is detectable in the cell-free sputum supernatant samples from COPD subjects. Cell-dissociated NTHi was shown to be linked to lung function and airway inflammation, at exacerbation only. Associations were not seen between levels of cell-dissociated NTHi detected and disease severity. This study confirms the presence of cell-dissociated NTHi within sputum and that further investigation is warranted to investigate if this mechanism relates to exacerbations of COPD.
The microbiome in BAL and cell-free BAL supernatant have been shown to differ in their bacterial composition,7 suggesting that pathogens can reside within the host in different forms (cell-associated and dissociated). Our study has examined NTHi, the most abundant pathogen within the lungs of patients with COPD.22 NTHi is particularly adept at invading host tissue,13 suggesting it could be more prone to adopting a cell-associated form. The ability of bacteria to adopt different forms within the host appears to greatly improve the survival rate and colonisation of the pathogen. The formation of a biofilm is associated with less response to antibiotic therapy and longer survival within the host.10 Studies looking at Streptococcus pneumoniae biofilm formation have shown that within a biofilm the bacteria is non-invasive and non-toxic;7 however, once dispersed from the biofilm there is adoption of a planktonic state. This was also associated with up-regulation of genes linked to virulence and may thus be related to the transition from asymptomatic colonisation to invasive bacterial Streptococcus infection.7
Exacerbations of COPD are associated with a pronounced decline in lung function,26 an increased risk of further exacerbations27 and increased bacterial colonisation of the lower respiratory tract.28 NTHi has been shown to significantly increase at exacerbation state compared to stable state.14,28 We showed that cell-dissociated NTHi was significantly associated with lung function and airway inflammation during an exacerbation and there was a trend to increased levels during the acute event. Our findings would suggest that cell-dissociated bacteria may play a vital role during exacerbations.
Neutrophils play an important role in innate immunity24,29,30 and airway neutrophilic inflammation is dominant in COPD.14 Observations demonstrate neutrophils are drawn to NTHi by secretory immunoglobulin A (IgA) and during phagocytosis release IL-8 further stimulating neutrophil recruitment.31 The formation of neutrophil extracellular traps (NETs) aid in bacterial killing.30 NTHi biofilms have been observed throughout NET structures and show resistance to both extracellular killing within NETs and phagocytic killing by recruited neutrophils.32 Neutrophil necrosis has been observed in the process of NTHi phagocytosis, encouraging NTHi persistence and further damage to lung epithelial cells.31 In our study, we determined that the presence of cell-dissociated NTHi was associated with relative sputum neutrophil levels. Further work is needed to understand whether cell-dissociated NTHi in patients with COPD exhibits similar protective features in their interaction with neutrophils.
Studies have shown that biofilms are more resistant to immune clearance mechanisms33 and to antibiotics compared to planktonic bacteria;34,35 whilst other studies have shown bacteria biofilm dispersal has an up-regulation of genes linked to virulence.7 The presence of cell-dissociated NTHi could lead to further understanding of bacterial differentiation states in the lungs of patients with COPD.
There are several limitations in this study. Firstly, the qPCR techniques used on the sputum plug and supernatant differ, in that the plug used a SYBR Green assay and the supernatant utilised a TaqMan assay, for which the sensitivity and expression levels between the two methods are likely to differ.36 We ensured that we quantified this in a comparable way and in both instances the lower limits of detection were similar. Secondly, the detection of cell-dissociated NTHi in our study using these methods does not strictly inform whether this bacterial state has dispersed from biofilms, been released from lysed cells either prior to sampling or during processing or indeed if the bacteria have always existed in a primary planktonic state; however, this is the first study to our knowledge that has attempted to detect cell-dissociated NTHi. Due to current widely evident qPCR limitations, we cannot comment if the cell-dissociated NTHi detected is alive or dead and this is a limitation in this current study. Finally, we are unable to make comment on the importance of cell-dissociated NTHi during exacerbations, due to the small number of samples studied, but suggest further investigation is required.
Conclusion
We have shown that cell-dissociated NTHi can be detected in patients with COPD. Cell-dissociated NTHi is associated with lung function and neutrophilic airway inflammation at exacerbation state and is independent of disease severity and may play an important role during these episodes.
Abbreviations
COPD, chronic obstructive pulmonary disease; CRQ, Chronic Respiratory Disease Questionnaire; DNA, deoxyribonucleic acid; DTT, dithiothreitol; FEV1, forced expiratory volume in 1 second; GOLD, Global Initiative for Chronic Obstructive Lung Disease; ICC, interclass co-efficient variation; IQR, inter-quartile range; LLD, lower limit of detection; NETs, neutrophil extracellular traps; NTHi, non-typeable Haemophilus influenzae; OMP, outer membrane protein; PBS, phosphate-buffered saline; qPCR, quantitative polymerase chain reaction; VAS, Visual Analogue Scale.
Acknowledgments
The abstract for this paper was presented at the European Respiratory Society Conference 2015; Thulborn S, Cane J, Ceroni A, et al. The detection of free-living H. influenzae in the airways of patients with COPD; Eur Respir J. 2015;46:PA566. The authors would like to thank Mrs Beverley Hargadon, Mrs Susan McKenna, Miss Amisha Singapuri and Mrs Maria Shelley for their assistance in patient characterisation. The authors would also like to thank Mrs Carlene White, Mrs Debbie Parker and Mr Mitesh Pancholi for their assistance in sputum processing. The authors would also like to thank Mr Paul Newbold, Mr Paul Rugman, Mrs Margaret McCormick and Mr Paul Dodson for their technical assistance in the study. Finally, the authors would like to thank Dr Daniel Ebner and his team for all their help in the design of the qPCR assay.
Author Contributions
Mona Bafadhel is the guarantor of the content of the manuscript, including the data and analysis. MB, CEB, and MRB contributed to the design of the study. All authors contributed to data interpretation and analysis; took part in drafting or revising the article; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was funded by the Medical Research Council, the Wellcome Trust & Academy of Medical Sciences and 330 the Oxfordshire Health Services Research Committee. This paper presents independent research funded by the National Institute for Health Research and the views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Disclosure
CEB was funded by a Wellcome Trust Senior Fellowship. MB was funded by a National Institute for Health Research (NIHR) Fellowship. MB has received travel support from AstraZeneca, Boehringer Ingelheim and GlaxoSmithKline and consultancy honoraria outside the submitted work from AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline and Pfizer. CEB has received grant support and consultancy honoraria outside the submitted work from AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Medimmune, Novartis and Roche. IDP has received grant support and consultancy honoraria outside the submitted work from Aerocrine, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline and Novartis. The authors report no conflicts of interest in this work.
References
1. Zaas AK, Schwartz DA. Innate immunity and the lung: defense at the interface between host and environment. Trends Cardiovasc Med. 2005;15(6):195–202. doi:10.1016/j.tcm.2005.07.001
2. Bhat TA, Panzica L, Kalathil SG, Thanavala Y. Immune dysfunction in patients with chronic obstructive pulmonary disease. Ann Am Thorac Soc. 2015;12(Supplement 2):S169–S175. doi:10.1513/AnnalsATS.201503-126AW
3. Shaykhiev R, Crystal RG. Innate immunity and chronic obstructive pulmonary disease: a mini-review. Gerontology. 2013;59(6):481–489. doi:10.1159/000354173
4. Moghaddam SJ, Ochoa CE, Sethi S, Dickey BF. Nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease and lung cancer. Int J Chron Obstruct Pulmon Dis. 2011;6:113–123. doi:10.2147/COPD.S15417
5. Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA. Mechanisms of bacterial pathogenicity. Postgrad Med J. 2002;78(918):216–224. doi:10.1136/pmj.78.918.216
6. Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microb Infect. 2015;17(3):173–183. doi:10.1016/j.micinf.2015.01.004
7. Marks LR, Davidson BA, Knight PR, Hakansson AP. Interkingdom signaling induces streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio. 2013;4(4). doi:10.1128/mBio.00438-13
8. Hernández-Jiménez E, Del Campo R, Toledano V, et al. Biofilm vs. planktonic bacterial mode of growth: which do human macrophages prefer? Biochem Biophys Res Commun. 2013;441(4):947–952. doi:10.1016/j.bbrc.2013.11.012
9. Mikkelsen H, Duck Z, Lilley KS, Welch M. Interrelationships between colonies, biofilms, and planktonic cells of pseudomonas aeruginosa. J Bacteriol. 2007;189(6):2411–2416. doi:10.1128/JB.01687-06
10. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi:10.1126/science.284.5418.1318
11. Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881–890. doi:10.3201/eid0809.020063
12. Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7(3):245–257. doi:10.1586/ers.13.24
13. King P. Haemophilus influenzae and the lung (Haemophilus and the lung). Clin Transl Med. 2012;1(1):10. doi:10.1186/2001-1326-1-10
14. Bafadhel M, McKenna S, Terry S, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184(6):662–671. doi:10.1164/rccm.201104-0597OC
15. Anthonisen NR, Manfreda J, Warren CPW, Hershfield ES, Harding GKM, Nelson NA. Antibiotic therapy in exacerbations of chronic obstructive pulmonary disease. Ann Intern Med. 1987;106(2):196–204. doi:10.7326/0003-4819-106-2-196
16. Rodriguez-Roisin R. Toward a consensus definition for COPD exacerbations. CHEST. 2000;117(5):398S–401S. doi:10.1378/chest.117.5_suppl_2.398S
17. Chauvin A, Rupley L, Meyers K, Johnson K, Eason J. Outcomes in cardiopulmonary physical therapy: chronic respiratory disease questionnaire (CRQ). Cardiopulm Phys Ther J. 2008;19(2):61–67. doi:10.1097/01823246-200819020-00007
18. Brightling CE, Green RH, Parker D, Morgan MD, Wardlaw AJ, Pavord D. Induced sputum and other outcome measures in chronic obstructive pulmonary disease: safety and repeatability. Respir Med. 2001;95:999–1002. doi:10.1053/rmed.2001.1195
19. Li B, Chen J-Q. Development of a sensitive and specific qPCR assay in conjunction with propidium monoazide for enhanced detection of live Salmonella spp. in food. BMC Microbiol. 2013;13(1):273. doi:10.1186/1471-2180-13-273
20. Rabe KF, Hurd S, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;176(6):532–555. doi:10.1164/rccm.200703-456SO
21. Wedzicha JA, Brill SE, Allinson JP, Donaldson GC. Mechanisms and impact of the frequent exacerbator phenotype in chronic obstructive pulmonary disease. BMC Med. 2013;11(1):181. doi:10.1186/1741-7015-11-181
22. Bafadhel M, Haldar K, Barker B, et al. Airway bacteria measured by quantitative polymerase chain reaction and culture in patients with stable COPD: relationship with neutrophilic airway inflammation, exacerbation frequency, and lung function. Int J Chron Obstruct Pulmon Dis. 2015;10:1075–1083. doi:10.2147/COPD.S80091
23. Banerjee D, Khair OA, Honeybourne D. Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur Respir J. 2004;23(5):685–691. doi:10.1183/09031936.04.00056804
24. Barker BL, Haldar K, Patel H, et al. Association between pathogens detected using quantitative polymerase chain reaction with airway inflammation in COPD at stable state and exacerbations. Chest. 2015;147(1):46–55. doi:10.1378/chest.14-0764
25. Curran T, Coyle PV, McManus TE, Kidney J, Coulter WA. Evaluation of real-time PCR for the detection and quantification of bacteria in chronic obstructive pulmonary disease. FEMS Immunol Med Microbiol. 2007;50(1):112–118. doi:10.1111/j.1574-695X.2007.00241.x
26. Halpin DMG, Decramer M, Celli BR, Mueller A, Metzdorf N, Tashkin DP. Effect of a single exacerbation on decline in lung function in COPD. Respir Med. 2017;128:85–91. doi:10.1016/j.rmed.2017.04.013
27. Donaldson GC, Seemungal TAR, Bhowmik A, Wedzicha JA. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax. 2002;57(10):847–852. doi:10.1136/thorax.57.10.847
28. Garcha DS, Thurston SJ, Patel ARC, et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67(12):1075–1080. doi:10.1136/thoraxjnl-2012-201924
29. Stockley R. Chronic obstructive pulmonary disease, neutrophils and bacteria: from science to integrated care pathways. Clin Med (Northfield Il). 2004;4(6):567–572. doi:10.7861/clinmedicine.4-6-567
30. Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil Extracellular Traps Kill Bacteria. Science. 2004;303(5663):1532–1535. doi:10.1126/science.1092385
31. Naylor EJ, Bakstad D, Biffen M, et al. Haemophilus influenzae induces neutrophil necrosis. Am J Respir Cell Mol Biol. 2007;37(2):135–143. doi:10.1165/rcmb.2006-0375OC
32. Juneau RA, Pang B, Weimer KED, Armbruster CE, Swords WE. Nontypeable haemophilus influenzae initiates formation of neutrophil extracellular traps. Infect Immun. 2011;79(1):431–438. doi:10.1128/IAI.00660-10
33. Puig C, Domenech A, Garmendia J, et al. Increased biofilm formation by nontypeable haemophilus influenzae isolates from patients with invasive disease or otitis media versus strains recovered from cases of respiratory infections. Appl Environ Microbiol. 2014;80(22):7088–7095. doi:10.1128/AEM.02544-14
34. Murphy TF, Kirkham C. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2002;2:7. doi:10.1186/1471-2180-2-7
35. Miravitlles M, Anzueto A. Antibiotics for acute and chronic respiratory infection in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188(9):1052–1057. doi:10.1164/rccm.201302-0289PP
36. Cao H, Shockey JM. Comparison of TaqMan and SYBR green qPCR methods for quantitative gene expression in Tung tree tissues. J Agric Food Chem. 2012;60(50):12296–12303. doi:10.1021/jf304690e
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
