Back to Journals » OncoTargets and Therapy » Volume 14
Construction and Analysis of a circRNA-Mediated ceRNA Network in Lung Adenocarcinoma
Authors Wang Z , Pei H, Liang H, Zhang Q, Wei L, Shi D, Chen Y, Zhang J
Received 7 February 2021
Accepted for publication 14 May 2021
Published 8 June 2021 Volume 2021:14 Pages 3659—3669
DOI https://doi.org/10.2147/OTT.S305030
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
Zhaojun Wang,1,* Hanzhong Pei,2,* Hongsen Liang,1 Qiwei Zhang,1 Li Wei,1 Donglei Shi,1 Yun Chen,2 Junhang Zhang1
1Department of Thoracic Surgery, The Seventh Affiliated Hospital, Sun Yat-sen University, Shenzhen, 518107, People’s Republic of China; 2Scientific Research Center, The Seventh Affiliated Hospital, Sun Yat-sen University, Shenzhen, 518107, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yun Chen
Scientific Research Center, The Seventh Affiliated Hospital, Sun Yat-sen University, No. 628, Zhenyuan Road, Guangming (New) Dist., Shenzhen, 518107, People’s Republic of China
Tel/Fax +86-755-81207022
Email [email protected]
Junhang Zhang
Department of Thoracic Surgery, The Seventh Affiliated Hospital, Sun Yat-sen University, No. 628, Zhenyuan Road, Guangming (New) Dist., Shenzhen, 518107, People’s Republic of China
Tel/Fax +86-755-81206874
Email [email protected]
Background: Circular RNAs (circRNAs), a new class of regulatory noncoding RNAs, are involved in gene regulation and may play a role in cancer development. The aim of this study was to identify circRNAs involved in lung adenocarcinoma (LUAD) using bioinformatics analysis.
Methods: CircRNA (GSE101684, GSE101586), miRNA (GSE135918), and mRNA (GSE130779) microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database to identify differentially expressed circRNAs (DECs), miRNAs (DEMs), and mRNAs (DEGs) in LUAD. Circinteractome and StarBase were used to predict miRNAs and mRNAs, respectively. A circRNA-miRNA-mRNA-ceRNA network was constructed. Patient survival was analyzed using UALCAN, and a sub-network was established. Real-time quantitative PCR (qRT-PCR) was used to verify the expressed of DECs between LUAD tissues and paired adjacent normal tissues.
Results: Hsa_circ_0072088 was identified as a differentially expressed (upregulated) circRNA in the two datasets. Intersection analysis identified hsa-miR-532-3p and hsa-miR-942 as the two miRNAs with the highest potential for binding to hsa_circ_0072088. Differential expression analysis and target gene prediction were performed to build a ceRNA network of hsa_circ_0072088 using Circinteractome/StarBase 3.0. Intersection analysis showed that TMEM52, IL24, POF1B, KIF1A, NHS, LBH, HIST2H2BE, ABCC3, PYCR1, CD79A, IGF2BP3, ANKRD17, GTSE1, MKI67, CLSPN, PLAU, LUC7L, MAGIX, GPATCH4, and ABAT were potential downstream mRNAs. The association between the expression level of 20 DEGs and LUAD patient survival was analyzed using UALCAN and GEPIA, which showed that IGF2BP3, MKI67, CD79A, and ABAT were related to patient survival. Hsa_circ_0072088 was verified upregulated by qRT-PCR.
Conclusion: The circRNA hsa_circ_0072088, the DEMs (hsa-miR-532-3p and hsa-miR-942-5p), and the DEGs (IGF2BP3, MKI67, CD79A, and ABAT) may serve as prognostic markers in LUAD.
Keywords: GEO, circRNA, ceRNA, lung adenocarcinoma, bioinformatics
Introduction
Lung cancer is the most common malignancy and the leading cause of cancer-related death worldwide.1 Lung adenocarcinoma (LUAD) is the most common form of lung cancer, accounting for approximately 40% of lung malignancies.2,3 Difficulties with early diagnosis and the lack of effective treatments are associated with the poor prognosis of this malignancy.4 It is therefore important to identify molecular markers associated with patient survival, which may improve early diagnosis and contribute to the development of gene-targeted therapies.
Circular RNAs (circRNAs) are a new class of noncoding RNA molecules that show a prevalent, stable, conserved, and specific expression pattern.5 CircRNAs are abundant in many eukaryotic cells and human tissues, and certain circRNAs are conserved among different species.6 Moreover, circRNAs can be detected in tissue samples, saliva, or plasma and show a cell-specific or stage-specific expression pattern.7 CircRNAs act as competitive inhibitors by binding to miRNAs (referred to as “miRNA sponges”), or they can act as target mimics to block the activity of specific microRNAs (miRNAs).8,9 CircRNAs provide binding sites for the miRNA either in the non-coding transcript or in the 3'-untranslated region of a specific gene; circRNAs are miRNA sponges and can be used as a new class of biomarker.10 CircRNAs enter the circulation and can be detected in the serum.11 Because circRNAs downregulate the expression of miRNAs, they have been investigated in many human cancers, such as colorectal cancer, lung, and stomach cancers.12 Wan et al showed that the expression level of the circRNA circ-ITCH is lower in lung cancer tissues than in adjacent normal tissues, and in the lung cancer cells lines A549 and NCI-H460, circ-ITCH inhibits miR-214 and miR-7 by acting as a sponge.13 CircRNA_100876 (circ-CER) is significantly upregulated in non-small cell lung cancer (NSCLC) tissues compared with paired adjacent non-tumorous tissues.
In this study, we analyzed the circRNA-miRNA-mRNA-competing endogenous RNA (ceRNA) regulatory network using GEO datasets. Differentially expressed circRNAs (DECs), miRNAs (DEMs), and mRNAs (DEGs) were identified and compared between LUAD samples and controls. The interaction between DEMs and predicted miRNAs, and the association between the expression of DEGs and LUAD patient survival were analyzed using The Cancer Genome Atlas (TCGA) samples. The present bioinformatics analysis may help identify novel biomarkers for LUAD.
Materials and Methods
Source of Microarray Data
The public GEO database is a functional genomic database found at http://www.ncbi.nlm.nih.gov/geo. Four datasets (GSE101684, GSE101586, GSE135918, and GSE130779) were downloaded. The circRNA dataset GSE101684 included four LUAD tissues and four adjacent normal lung tissues, and the samples were tested by the GPL21825 074301 Arraystar Human CircRNA microarray V2 platform. The circRNA dataset GSE101586 included four LUAD tissues and four lung tissues, and the samples were tested by the GPL19978 Agilent-069978 Arraystar Human CircRNA microarray V1 platform. The miRNA dataset GSE135918 included five LUAD tissues and five adjacent normal lung tissues, and the samples were tested by the GPL18058 Exiqon miRCURY LNA microRNA array, 7th generation [miRBase v18, condensed Probe_ID version] platform. The mRNA dataset GSE130779 included eight LUAD tissues and eight adjacent non-tumor tissues; the platform used for the microarray was GPL20115 Agilent-067406 Human CBC lncRNA + mRNA microarray V4.0 (Probe name version).
Identification of DECs, DEMs, and DEGs
Differential analyses of GSE101684, GSE101586, GSE135918, and GSE130779 were performed to identify DECs, DEMs, and DEGs between LUAD tissues and control normal tissues using the online tool GEO2R.14 The criterion for DECs was P <0.05 and |logFC| ≥2. The criterion for DEMs was P <0.05 and logFC ≤-1, and the criterion for DEGs was P <0.05 and logFC ≥1. A volcano plot of the DECs was constructed using the R software gplots package.
Construction of the ceRNA Network
Circinteractome is a web tool for exploring circRNAs and their interacting proteins and miRNAs.15 Circinteractome was used to predict the potential miRNAs (https://circinteractome.nia.nih.gov/). The predicted potential miRNAs intersecting with DEMs from GEO in the online website Venny (http://bioinformatics.psb.ugent.be/webtools/Venn/) were regarded as potential miRNAs of circRNAs in LUAD. StarBase 3.0 (http://starbase.sysu.edu.cn/index.php), an open-source platform for studying RNA interactomes,16 was used to predict mRNAs. The intersecting genes that commonly appear in predicted mRNAs and DEGs were regarded as potential mRNAs in LUAD. Finally, a circRNA-miRNA-mRNA network was constructed, and the data visualized using Cytoscape software 3.7.1.
Survival Analysis
The association between the mRNAs selected by the ceRNA network and survival was analyzed using UALCAN (http://ualcan.path.uab.edu/index.html) and GEPIA (http://gepia.cancer-pku.cn/). First, we performed the relative level of mRNAs in LUAD. The association between DEGs and LUAD patient survival and the expression of DEGs based on TCGA samples were analyzed using UALCAN. Pathological stage analysis and the prognostic value of mRNAs patients in the disease overall survival curve in LUAD were performed for further verification using GEPIA. P < 0.05 was considered statistically significant.
Tissue Specimens
A total of 5 pairs of LUAD tissues and adjacent normal tissues were collected from LUAD patients who had surgery in the Seventh Affiliated Hospital, Sun Yat-sen University. None of these patients received preoperative chemotherapy, radiotherapy or targeted therapy. The tissue specimens frozen and stored at liquid nitrogen until further analysis.
RNA Extraction and qRT-PCR
RNA from tissues was extracted by using TRIzol reagent (Abbion) under the manufacturer’s protocol. RNA was quantified using a NanoDrop-ND1000 (thermo) system, qPCR used Biorad CFX96 touch system. SYBR master mix (hamQ) and cDNA Synthesis Kit (TAKARA) were used in this study. The specific primers were shown as below: hsa_circ_0072088 (Forward, 5'- GGATTCACCCACAGCGG-3'. Reverse, 5'-CTACACCCGAAGCTGGCT-3'). 2−ΔΔCt was used for quantitative analysis.
Statistics
Data are displayed as mean ± SD, t-test was used to assess the differences. P < 0.05 was considered as statistically significant.
Results
Identification of DECs
The flowchart of the validation and construction of the ceRNA network is shown below (Figure 1). To identify DECs in LUAD, GSE101684 of four LUAD patients and GSE101586 of five LUAD patients were selected (Table 1). Using |logFC| ≥2 and P < 0.05 as the cut-off criteria, 34 DECs were identified in paired lung adenocarcinoma tissues and adjacent non-tumor tissues (5 in GSE101586 and 29 in GSE101684; Tables S1 and S2). A volcano plot of the DECs was plotted using the R software gplots package (Figure 2A and B). The intersection of the two datasets was determined using the online website Venny, and hsa_circRNA_103809 (hsa_circ_0072088 using the circBase database transformation) was upregulated in the results.
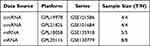 |
Table 1 Basic Information of the Four Microarray Datasets from GEO |
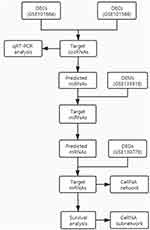 |
Figure 1 Flow chart of the approach utilized in this study. |
Identification of Potential DEMs
To identify DEMs in LUAD, GSE135918 of five LUAD patients was selected (Table 1). Using logFC≤-1 and P < 0.05 as the cut-off criteria, 431 downregulated DEMs were identified in lung cancer tissues and non-tumor lung tissues (Table S3). MiRNAs that might bind to hsa_circ_0072088 were predicted by Circinteractome; 24 miRNAs were identified (Table S4). The intersection of DEMs and predicted miRNAs was determined, hsa-miR-532-3p and hsa-miR-942 were downregulated (Figure 2C).
Identification of mRNAs of DEMs
MiRNAs negatively regulate the expression of mRNAs. To identify mRNAs regulated by hsa-miR-532-3p and hsa-miR-942, mRNAs were predicted using the StarBase database. The StarBase database can only predict the target genes of hsa-mir-942-5p, and 6674 mRNAs (3299 mRNAs in hsa-miR-532-3p and 3375 mRNAs in hsa-miR-942-5p) were predicted (Table S5). After removal of duplicate values, 5343 mRNAs were selected for further analysis. To identify DEGs in LUAD, GSE130779 of eight LUAD patients was selected (Table 1). Using logFC ≥1 and P < 0.05 as the cut-off criteria, 107 upregulated DEGs were identified in lung cancer tissues and non-tumor lung tissues (Table S6). The intersection of DEGs was used to predict mRNAs, and 20 mRNAs were upregulated (Figure 2D).
Construction of the ceRNA Network
The findings were used to build a circRNA-miRNA-mRNA network using Cytoscape (Figure 3).
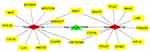 |
Figure 3 CeRNA networks of circRNA-miRNA-mRNA interactions in LUAD. Green indicate circRNA, red indicates miRNAs, and yellow indicates mRNAs. |
MRNA Analysis and Establishment of a Sub-Network
To analyze the mRNAs selected by the ceRNA network, survival analysis was performed using UALCAN and GEPIA. The analysis identified 19 mRNAs (TMEM52, IL24, POF1B, KIF1A, NHS, HIST2H2BE, ABCC3, PYCR1, CD79A, IGF2BP3 ANKRD17, GTSE1, MKI67, CLSPN, PLAU, LUC7L, MAGIX, GPATCH4, ABAT) that were upregulated in LUAD based on TCGA samples. The expression of LBH was lower in tumor tissues than in normal tissues (Figure 4). We also compared the relative level of mRNAs in LUAD tissues, and we found that the relative level of PYCR1 was the highest (Figure 5). The expression of IGF2BP3 (P = 0.001), MKI67 (P = 0.0046), CD79A (P = 0.012) and ABAT (P = 0.035) had prognostic value in LUAD patients using UALCAN (Figure 6). We then assessed the correlation between the pathological stage of LUAD patients and the four mRNAs (IGF2BP3, MKI67, CD79A and ABAT), and we found a significant correlation between pathological stage and the expression of IGF2BP3 (P=0.00901), MKI67 (P=0.000305), CD79A (P=2.5e-5) and ABAT (P=0.000807) (Figure 7). The prognostic value of the four mRNAs in LUAD patients was evaluated by using GEPIA for further verification, we found that LUAD patients with low levels of IGF2BP3 (P = 0.0017) and MKI67 (P =0.027) were associated with longer overall survival, but high levels of CD79A (P = 0.024) and ABAT (P = 0.00074) were associated with longer overall survival (Figure 8). Finally, a potential circRNA-miRNA-mRNA sub-network was successively established in LUAD (Figure 9).
 |
Figure 4 The expression of 20 DEGs in LUAD using UALCAN. “p<0.05” represents significant difference. |
 |
Figure 5 The relative level of 20 mRNAs in LUAD (GEPIA). |
 |
Figure 6 The expression level of the four DEGs in LUAD patient survival using UALCAN. “p<0.05” represents significant difference. |
 |
Figure 7 Correlation between the four mRNAs and the pathological stage of LUAD patients. (A) IGF2BP3, (B) MKI67, (C) CD79A, and (D) ABAT, p<0.05. |
 |
Figure 8 The prognostic value of 4 mRNAs in LUAD patients in the disease overall survival curve (GEPIA). (A) IGF2BP3, (B) MKI67, (C) CD79A, and (D) ABAT, p<0.05. |
 |
Figure 9 CeRNA sub-network of circRNA-miRNA-mRNA interactions in LUAD. Green indicate circRNA, red indicates miRNAs, and yellow indicates mRNAs. |
qRT-PCR Analysis
Hsa_circ_0072088 was found upregulated between LUAD tissues with paired adjacent normal tissues by qRT-PCR (Figure 10).
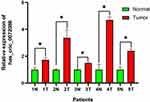 |
Figure 10 Expression of hsa_circ_0072088 in tissues. *P<0.05. |
Discussion
CircRNA is highly stable because of its covalent closed-loop structure. Many studies have suggested that circRNAs may be involved in tumorigenesis. The main way for circRNAs regulating tumors progression is to act as sponges for miRNAs and then further regulating the expression of downstream mRNAs. Although studies have reported the construction of circRNA-miRNA-mRNA regulatory networks in LUAD,17,18 the current understanding of circRNA-related ceRNA networks in LUAD is limited, and further study is needed. In this study, we identified a potential circRNA-miRNA-mRNA ceRNA regulatory network in LUAD.
Differential expression analysis led to the selection of one circRNA for further investigation. Hsa_circ_0072088, which is associated with cancer cell proliferation and progression, regulates the expression of miR-532-3P and FOXO4 in colorectal cancer.19 Liu et al demonstrated that hsa_circ_0072088 contributes to lung cancer cell proliferation and invasion by sponging miR-4302 and promoting ZNF121-dependent MYC expression.20
CircRNAs act as miRNA sponges and regulate the expression of miRNA target transcripts.9,21 For instance, the proliferation and invasion of HCC are dependent on circMAST1 binding to miR-1299 and inhibiting CTNND1.22 Circ_100565 upregulates HMGA2 expression by sponging miR-506-3p to promote migration, invasion, and proliferation.23 Hsa_circ_0043265 inhibits NSCLC progression by sponging miR-25-3p to promote FOXP2 expression.24 Here, we used Circinteractome and the intersection of DEMs to identify hsa-miR-532-3p and hsa-miR-942-5p.
MiRNAs regulate gene function by negatively downregulating gene expression.25,26 Therefore, we explored the molecular mechanisms underlying the roles of the hsa_circ_0072088-hsa-miR-532-3p and hsa_circ_0072088-hsa-miR-942-5p axis in LUAD. Firstly, target genes of hsa-miR-532-3p and hsa-miR-942-5p were predicted using the StarBase database. Then, the DEGs between LUAD tissues and normal tissues were screened. Twenty upregulated mRNAs (TMEM52, IL24, POF1B, KIF1A, NHS, LBH, HIST2H2BE, ABCC3, PYCR1, CD79A, IGF2BP3 ANKRD17, GTSE1, MKI67, CLSPN, PLAU, LUC7L, MAGIX, GPATCH4, ABAT) were selected according to the intersection of the DEG set and predicted gene set.
It is important to develop genomic detection methods for LUAD to predict the survival rate of patients. In this study, survival analysis was performed using UALCAN. The results identified 19 mRNAs (TMEM52, IL24, POF1B, KIF1A, NHS, HIST2H2BE, ABCC3, PYCR1, CD79A, IGF2BP3, ANKRD17, GTSE1, MKI67, CLSPN, PLAU, LUC7L, MAGIX, GPATCH4, ABAT) showing differential expression in LUAD. The differential expression levels of IGF2BP3, MKI67, CD79A, and ABAT indicated that they may have prognostic value for predicting survival in LUAD patients. Moreover, we found that IGF2BP3, MKI67, CD79A and ABAT increased with the tumors progressed. LUAD patients with low levels of IGF2BP3 and MKI67 were associated with longer overall survival, but high levels of CD79A and ABAT were associated with longer overall survival. A potential circRNA-miRNA-mRNA sub-network was established. According to the ceRNA mechanism, the relationship between circRNAs and mRNAs should be positive.27 Hsa_circ_0072088 expression was positively correlated with the expression of IGF2BP3, MKI67, CD79A and ABAT in LUAD. In this study, the hsa_circ_0072088 was identified to be overexpressed through experimental verification in the tissue of LUAD. In fact, IGF2BP3 and mKI67 have been experimentally confirmed, but CD79A and ABAT still need further experimental verification. Guo et al demonstrated that IGF2BP3 is abnormally highly expressed in LUAD tissue, and can lead to worse overall survival.28 Li et al found that MKI67 was significantly upregulated in human LUAD tissues was higher than that in the control tissues.29
The present study had several limitations. First, the findings need to be confirmed in additional cohorts and further experimental verification is needed. Second, the sample size was too small, as there were few samples in each microarray dataset. Third, the DAVID (https://davidd.ncifcrf.gov/) was used to the function enrichment analysis with gene count, but there were not clustered.
Conclusions
The present study revealed a potential hsa_circ_0072088-mediated circRNA-miRNA-mRNA ceRNA network in LUAD, and intersection analysis, differential expression analysis, and survival analysis were performed. But the sample size is too small, hsa_circ_0072088, the miRNAs (hsa-miR-532-3p and hsa-miR-942-5p), and the mRNAs (IGF2BP3, MKI67, CD79A, and ABAT) need further broader investigation to explore its potential to serve as prognostic biomarkers in LUAD.
Ethics Statement
This study was approved by the Ethics Committee of the Seventh Affiliated Hospital, Sun Yat-sen University, and conducted in accordance with the Declaration of Helsinki. The patients provided their written informed consent to participate in this study.
Acknowledgments
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol. 2007;25(5):561–570. doi:10.1200/JCO.2006.06.8015
3. Myers DJ, Wallen JM. Cancer, lung adenocarcinoma. 2020.
4. Zhang B, Chen M, Jiang N, Shi K, Qian R. A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the proliferation of lung adenocarcinoma. Cancer Biol Ther. 2019;20(8):1127–1135. doi:10.1080/15384047.2019.1598762
5. Pang W, Huang F, Zhang X, et al. Circular RNA hsa_circ_0072309 inhibits non-small cell lung cancer progression by sponging miR-580-3p. Biosci Rep. 2020;40(5). doi:10.1042/BSR20194237
6. Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi:10.1186/s12864-017-4029-3
7. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. doi:10.1016/j.cell.2018.12.021
8. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
9. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
10. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
11. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
12. Cortes-Lopez M, Miura P. Emerging functions of circular RNAs. Yale J Biol Med. 2016;89(4):527–537.
13. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi:10.1155/2016/1579490
14. Qin Y, Li L, Wang F, et al. Knockdown of Mir-135b sensitizes colorectal cancer cells to oxaliplatin-induced apoptosis throughincrease of FOXO1. Cell Physiol Biochem. 2018;48(4):1628–1637. doi:10.1159/000492284
15. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. Rna Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
16. Wang W, Lou W, Ding B, et al. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY). 2019;11(9):2610–2627. doi:10.18632/aging.101933
17. Chen F, Huang C, Wu Q, Jiang L, Chen S, Chen L. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J Cell Biochem. 2020;121(3):2525–2533. doi:10.1002/jcb.29475
18. Liang L, Zhang L, Zhang J, Bai S, Fu H. Identification of circRNA-miRNA-mRNA networks for exploring the fundamental mechanism in lung adenocarcinoma. Onco Targets Ther. 2020;13:2945–2955. doi:10.2147/OTT.S235664
19. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352. doi:10.1016/j.bbrc.2018.09.073
20. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
21. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928
22. Yu X, Sheng P, Sun J, et al. The circular RNA circMAST1 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-1299 and regulating CTNND1 expression. Cell Death Dis. 2020;11(5):340. doi:10.1038/s41419-020-2532-y
23. Li L, Wei H, Zhang H, Xu F, Che G. Circ_100565 promotes proliferation, migration and invasion in non-small cell lung cancer through upregulating HMGA2 via sponging miR-506-3p. Cancer Cell Int. 2020;20(1):160. doi:10.1186/s12935-020-01241-8
24. Ren T, Liu C, Hou J, Shan F. Hsa_circ_0043265 suppresses proliferation, metastasis, EMT and promotes apoptosis in non-small cell lung cancer through miR-25-3p/FOXP2 pathway. Onco Targets Ther. 2020;13:3867–3880. doi:10.2147/OTT.S235231
25. Liu Z, Pan HM, Xin L, et al. Circ-ZNF609 promotes carcinogenesis of gastric cancer cells by inhibiting miRNA-145-5p expression. Eur Rev Med Pharmacol Sci. 2019;23(21):9411–9417. doi:10.26355/eurrev_201911_19433
26. Shen X, Jiang H, Chen Z, et al. MicroRNA-145 inhibits cell migration and invasion in colorectal cancer by targeting TWIST. Onco Targets Ther. 2019;12:10799–10809. doi:10.2147/OTT.S216147
27. Ding B, Yao M, Fan W, Lou W. Whole-transcriptome analysis reveals a potential hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory sub-network in colorectal cancer. Aging (Albany NY). 2020;12(6):5259–5279. doi:10.18632/aging.102945
28. Guo W, Huai QL, Wan H, et al. Prognostic impact of IGF2BP3 expression in patients with surgically resected lung adenocarcinoma. DNA Cell Biol. 2021;40(2):316–331. doi:10.1089/dna.2020.6136
29. Li JY, Liu XY, Cui Z, Han GY. Comprehensive analysis of candidate diagnostic and prognostic biomarkers associated with lung adenocarcinoma. Med Sci Monit. 2020;26:e922070. doi:10.12659/MSM.922070
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

