Back to Journals » Infection and Drug Resistance » Volume 15
Comprehensive Surveillance and Sampling Reveal Carbapenem-Resistant Organism Spreading in Tertiary Hospitals in China
Authors Zhang Y, Yu S, Chen C, Sun F, Zhou L, Yao H, Hu J, Li S, Ai J, Jiang N, Wang J, Liu Q, Jin J, Zhang W
Received 22 March 2022
Accepted for publication 23 July 2022
Published 17 August 2022 Volume 2022:15 Pages 4563—4573
DOI https://doi.org/10.2147/IDR.S367398
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yi Zhang,1,* Shenglei Yu,1,* Chen Chen,1,* Feng Sun,1,* Lei Zhou,1 Haijun Yao,2 Jin Hu,2 Shirong Li,3 Jingwen Ai,1 Ning Jiang,1 Jing Wang,1 Qihui Liu,1 Jialin Jin,1 Wenhong Zhang1,4,5
1Department of Infectious Diseases, National Medical Center for Infectious Diseases, Shanghai Key Laboratory of Infectious Diseases and Biosafety Emergency Response, Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai, China; 2Department of Critical Care Medicine, Huashan Hospital, Fudan University, Shanghai, China; 3Department of Clinical Laboratory, Huashan Hospital, Fudan University, Shanghai, China; 4National Clinical Research Center for Aging and Medicine, Huashan Hospital, Fudan University, Shanghai, China; 5Key Laboratory of Medical Molecular Virology (MOE/MOH), Shanghai Medical College, Fudan University, Shanghai, China
*These authors contributed equally to this work
Correspondence: Wenhong Zhang; Jialin Jin, Email [email protected]; [email protected]
Purpose: Carbapenem-resistant organisms (CROs) have posed a great threat to antibiotic use and induce multi-drug resistance. Contamination of the hospital environment and infection of healthcare workers (HCWs) are reported as sources of nosocomial infections. Here, we performed a comprehensive environment sampling and timely epidemiological investigation during outbreaks to investigate the role of the environment and HCWs in CRO transmission.
Patients and Methods: We enrolled carbapenem-resistant organism outbreaks in ICU-1 of Huashan Hospital from January 2019 to March 2019, and ICU-2 located at west branch of Huashan Hospital from October 2019 to November 2019. Carbapenem-resistant Klebsiella pneumoniae (CRKP) and carbapenem-resistant Acinetobacter baumannii (CRAB) isolates were collected from the patients. We performed a real-time comprehensive environmental and HCW sampling in the two ICUs. Isolated strains from patients and the positive colonies from the screening were sent for whole-genome sequencing. Finally, phylogenetic trees were constructed.
Results: CRAB and CRKP outbreaks simultaneously occurred in ICU-1; the outbreak involved 13 patients. Meanwhile, the CRKP outbreak in ICU-2 included 11 patients. Twelve out of 146 environment and HCWs samples in ICU-1 were carbapenem-resistant bacteria, including six CRKP and six CRAB strains. For ICU-2, hospital surfaces and HCWs were negative for CRKP. Phylogenetic analyses showed that CRKP strains in ICU-1 were classified into two clades: Clade 1 and Clade 2, sharing a high similarity of isolates from the environment and HCWs. The same phenomenon was observed in CRAB.
Conclusion: A timely comprehensive sampling combined with genome-based investigation may aid in tracking the transmission route of and controlling the infections. The environment and HCWs could be contaminated during CRO transmission, which calls for strengthened prevention and control measures.
Keywords: carbapenem-resistant, environment, whole-genome sequencing, prevention, control
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) and Acinetobacter baumannii (CRAB) belong to “The ESKAPE pathogens”, which poses a great threat to antibiotic use and resistance. The increased attention to ESKAPE pathogens originates from high occurrence of outbreaks and hospital-associated infections harboring antimicrobial resistance (AMR) genes. Mutations and plasmid-mediated horizontal transfer of resistance and virulence genes accelerate the spread and transmission of infections, leading to difficulties in prevention and control. Whole-genome sequencing has been widely used in pathogen identification, genotyping, and assessment of transmission route.1,2
Contact transmission is the most common mechanism of diseases caused by bacteria and virus. In nosocomial infections, the source of contact transmissions are infected patients or contaminated surfaces or equipment near the patients.3 High-touch surfaces in wards represent a critically important multi-drug resistant organism (MDRO) reservoir,4 as well as the hospital environment including wastewater.5 In addition, infected healthcare workers play a key role in the transmission of multidrug-resistant bacteria.6
Previously, we have reported a CRKP outbreak tracing in intensive care units (ICUs) using short and long read sequencing. However, the source or media of transmission remains unclear. In the present study, we performed a comprehensive environment sampling and epidemiological investigation during the outbreak to investigate whether the environment plays an essential role in the transmission of infection and potential driving force for carbapenem-resistant organisms (CROs) transmission.
Patients and Methods
Patients and Isolates
Patients from outbreaks in ICU-1 of Huashan Hospital from January 2019 to March 2019 and ICU-2 located at west branch of the hospital from October 2019 to November 2019 were enrolled in the study. Clinical information and epidemiological data were collected. CRKP and CRAB isolates were collected from patients who were admitted into ICU-1 during outbreak period, while only CRKP strains were obtained from patients in ICU-2. Carbapenem-resistance was identified using minimum inhibitory concentration (MIC) of imipenem or meropenem ≥4 ug/mL. Isolates were identified from 13 and 11 patients admitted to Huashan Hospital and the west branch of the hospital during the outbreak period, respectively. Isolates collected from the same study site with accordant antibiotic susceptibility tests were excluded from the study. Results from the antibiotic susceptibility tests (ASTs) were collected and analyzed using the Clinical and Laboratory Standards Institute version 2019 guidelines. The study protocol and informed consent forms were approved by the Huashan Hospital Ethical committee (KY19-429).
Environment and HCW Sampling and Isolation
A comprehensive environment and HCW sampling in ICU-1 during the outbreak was conducted for eight times until June 7, 2019. Meanwhile, environment and HCW sampling was conducted in ICU-2 as well. A total of 146 samples were collected from high-touch surfaces in the patients’ rooms and wards, including the bed railing, bedside tabletop, and ventilator surface, and in the doctors’ office, including phones, keyboards, and tables. Hand sampling was conducted on available nurses, nursing workers, and doctors. All samples were cultured in lysogeny broth (LB) medium for 48 h and screened using meropenem (4 μg/mL). Subsequently, the isolates were identified using 16S rRNA sequencing. CRAB and CRKP strains were then selected for storage and sequencing.
Whole-Genome Sequencing and Plasmid Identification
The total DNA from a single colony of clinical CRKP or CRAB isolate was extracted using the TIANAmp Micro DNA Kit (TIANAmp, China), according to the manufacturer’s recommendation. DNA libraries were sequenced on the Illumina Nova-seq Platform (Illumina, USA) using a pair-end 150-base pair strategy. De novo assembly of short-read sequencing data was performed using SPAdes (v3.11.1) with default parameters. Multilocus sequences were identified using the BIGSdb-Pasteur (https://bigsdb.pasteur.fr/).
Among the strains analyzed, CRKP strain kp8 was subjected for GridlON Nanopore long-read sequencing. The average depth for the Nanopore sequencing is approximately 400X, and error rate is approximately 15%. Unicycler tool was used to assemble short and long reads, and then the assembled plasmids of kp8 were mapped to the plasmid sequences in the NCBI database.
Single Nucleotide Polymorphism Analysis and Phylogenetic Tree Construction
Klebsiella pneumoniae strain SWU01 (GeneBank CP018454.1) and Acinetobacter baumannii strain MDR-ZJ06 (GeneBank NC_017171.2) were used as the reference genomes for read mapping. Bowtie2 (v 2.3.3.1)7 was used for read mapping, and candidate SNPs were identified using SAMtools (v 1.9).8 Next, we constructed a phylogenetic tree based on the SNPs of CRKP isolated from ICU-1 and ICU-2. To compare the CRKPs in the two ICUs, we performed an evolutionary analysis based on 54,338 SNPs found in the 30 clinical CRKP isolates. Meanwhile, the phylogenetic tree of CRAB was established using 7007 SNPs among 19 strains. All phylogenetic trees were generated using Molecular Evolutionary Genetics Analysis (MEGA) X,9 with maximum-likelihood estimation and the general time reversible nucleotide substitution model. The phylogenetic trees were generated in iTOL v6.
Results
Overview of CRAB and CRKP Outbreaks in ICU-1
ICU-1 experienced CRAB and CRKP outbreaks simultaneously, which involved 13 patients (Table 1). The median age of patients was 56 years old (range: 34–78 years old). Underlying diseases included central nervous system infection (CNS infection), severe pneumonia, liver failure, brain abscesses, and disseminated nocardiosis. Five patients died, of which four died of underlying disease, while one patient died due to CRAB and severe Pneumocystis jiroveci co-infection.
 |
Table 1 Characteristics of Patients in ICU-1 |
CRAB and CRKP strains were isolated from 11 and 7 patients, respectively. Five patients had simultaneous CRKP and CRAB infections before discharge. On December 25, 2018, the index patient (Pt 1) was admitted into ICU-1 of A Hospital due to CNS infection. Eight days later, CRAB was isolated from his sputum, and the other 11 strains were found in the next two months in this ward. Most of the patients were admitted in Room 3. Admission of Pt 3 overlapped with Pt 1 and 2 from January 23 to February 6, 2019. Furthermore, Pt 6, 7, 8, and 9 shared Room 3 with Pt 3 from February 15 to March 7, 2019. Pt 4 had no evident epidemiological link with other patients. In total, rectal swab screen was performed on seven patients (Table 1), among which Pt 6, 9, 10, and 12 were positive for ESBL. Three patients had positive CRAB CSF (Pt 1–6) or sputum (Pt 10 and 12) on admission.
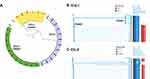
For CRKP, the first identified infection during this outbreak was Patient 6, who was admitted in ICU-1 on 15th February and diagnosed with brain abscesses and ventriculitis. The patient was implanted with Ommaya reservoir on February 18; three days later, CRKP was isolated from the cerebrospinal fluid. In the following month, a total of six patients in ICU-1 acquired CRKP infections. As shown in Figure 1A, Patients 1, 3, and 6 had an epidemiological link of shared ward stays. In addition, Patients 1, 7, 9, and 10 overlapped in Room 3. Patient 9 had a positive ESBL screen result, and two CRKP strains isolated from sputum and blood were collected.
Overview of the CRKP Outbreak in ICU-2
The CRKP outbreak in ICU-2 occurred in October 2019 until late December 2019 (Table 2). The first identified patient infected with CRKP (Pt A) in ICU-2 was hospitalized in B Hospital on October 28, 2019. Pt A was identified as an imported CRKP case. He received neurosurgery in other hospital in June 2019 and acquired CRKP infection in the CSF afterwards. Pt C, D, and E had overlapped stays of a room, and a similar epidemiological link was observed in Pt H and I (Room 5) (Figure 1B). Pt E and G shared Room 1 and Room 4, while Pt 2–10 and 2–11 had no evident epidemiological relationship with other patients. The patients were transferred in different wards in ICU-2, especially Pt D, E, and F.
 |
Table 2 Characteristics of Patients in ICU-2 |
Effects of Comprehensive Environmental Sampling and Control Measures
We performed a comprehensive environmental and medical staff sampling in ICU-1 for a total of eight times since the outbreak. Sampling on March 12, March 29, and April 4 showed positive results, while the next five sampling days reported negative results. Twelve out of 146 isolates (8.22%) were carbapenem-resistant bacteria. The CRKP strain H7 was isolated on March 12; strains E1, E13, E14, and E19 were isolated on March 29, and strain ES5 was isolated on April 4, which came from bed railing, bedside tabletop, and ventilator surfaces. CRAB strains E4, E16, and E25 were isolated on March 29, while strains ES1, ES3, and ES9 were isolated on April 14; the strains were isolated from bed railing, bedside tabletop, computer keyboards, and hands of intern doctors. Surfaces of and HCWs in ICU-2 were negative for CRKP.
Meanwhile, control measures against nosocomial infection were implemented in both ICU-1 and ICU-2 to control the outbreaks. Control measures included strengthening of the active rectal swab screening, isolating CRO-infected patients, contact precautions and hand hygiene, and environment cleaning. The outbreaks in ICU-1 and ICU-2 were attenuated in April 2019 and December 2021, respectively.
Phylogenetic Analysis and Genome Comparisons of CRKP and CRAB
A total of 54,338 SNPs detected in all CRKP strains isolated from ICU-1 and ICU-2 were used to construct the phylogenetic tree (Figure 2A). Strains W873 and W937 were considered as the outgroup strains.
Interestingly, CRKP strains from ICU-1 were classified into two clades: Clade 1 and Clade 2. As shown in Figure 2B, the ICU-1 Clade 1 (ST17) was distributed in the upper branch of the tree, while ICU-1 Clade 2 (ST11) was located at the lower group. The ICU-1 Clade 1 belonged to ST11 CRKP, which was resistant to aminoglycoside and beta-lactams. These observations were consistent with those from the AST (Supplementary Table 1). Although the fluoroquinolone-resistance gene Qnr-S1 was detected, the AST showed that the resistance of ST17 clinical strain to ciprofloxacin and levofloxacin was intermediate. Meanwhile, 11 strains belonged to ICU-1 Clade 2, and these strains shared four SNPs. Aside from multidrug-resistance genes, all strains harbored the following virulence genes: iutA, rmpA, and rmpA2.
Among the CRKP strains isolated from ICU-2, strains W873 (ST2250) and W937 (ST15) were considered as the outgroup strains (Figure 2C). Nine strains in the lower clade shared 11 SNPs in total. The remaining strains in the phylogenetic tree belonged to ST11 and harbored the virulence genes iutA and rmpA. Although no carbapenem-resistant gene was detected in W937, the AST showed that the strain was resistant to both imipenem and meropenem, the AST results are listed in Supplementary Table 2. W909 was identified as wzi209 and KL47, while the other ST11 strains were wzi64 and KL64.
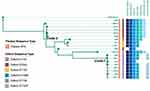
As for the CRAB outbreak, 7007 SNPs were detected in 13 clinical and 6 environmental and HCW samples (Figure 3). All strains belonged to ST2 via the Pasteur sequence type, while several Oxford sequence types were identified, including ST191 (n = 8), ST195 (n = 2), ST540 (n = 6), ST1806 (n = 1), ST136 (n = 1), and ST1837 (n = 1). All the strains carried the carbapenem-resistance gene OXA-23. The lower Clade-1 including ab2 and ab3 (Pt 8), ab5 (Pt1), ab8 (Pt12), ab11 (Pt 2), ES1, ES-3, and ES-9 shared only 20 SNPs. Similarly, in the upper clade, ab1 and E4 shared one SNP. The Clade-2 ST540 strains ab6, ab10, ab12, and E16 were closely related as well, with 43 SNPs detected among these four strains.
Plasmid Identification Through Transmission
Since the CRKP strains of ICU-1 Clade 2 were found to possess the virulence genes rmpA, rmpA2, and iuc1, we performed Nanopore sequencing to characterize the plasmids. A total of five plasmids of strain kp8 were confirmed through short- and long-read sequencing. Plasmid 1 carried the virulence genes iutA, rmpA, and rmpA2, with a mapping coverage of 93.65% to plasmid MG053312.1, which was first confirmed as a hypervirulent plasmid in Henan, China, in 2018. Additionally, Plasmid 2 is a KPC-harboring plasmid, carrying the resistance genes RmtB, SHV12, and TEM30, and had a 97.48% mapping to MF168404.1. The multidrug-resistant Plasmid 3 (MF133496, 100.00%) harbored Qnr-S1, SuIII, tetA, DfrA14, and LAP-2. The two remaining plasmids were identified as CP023932.1 and CP023936.1 (both 100.00% mapped).
Putative Source of and Driving Force for CRO Transmission
Combined with the epidemiological and genomic evidence, we could infer the potential transmission route and source of the CRO outbreaks. Among the ICU-1 infections, kp5 (Pt 1), kp8 (Pt 6), and W901 (Pt I) were considered as index cases of CRKP ICU-1 Clade 1, ICU-1 Clade 2, and ICU-2, respectively. The index cases of CRAB transmission were ab5 (pt1) and ab10 (Pt6). The environmental and HCW samples were contaminated during both CRKP and CRAB transmission in ICU-1.
The CRKP ICU-1 Clade 1 originated from kp5, while the strain isolated from Pt 1 was transmitted to kp6 (Pt 7), kp7 (Pt 3), and kp10 (Pt 5). Pt 6 contributed the index KP strain of ICU-2 CRKP Clade 2. The strain was transmitted to clinical strains kp1, kp2, and kp9. Additionally, environmental strains including H7, E1, E14, and E19 were contaminated during the CRO transmission. Subsequently, the kp4 was infected by these strains with another three SNP site mutations. In addition, the strain ES5 had two SNPs different from kp8. The outbreak of CRKP in ICU-2 might have come from strain W901 (Pt I, ICU-2), then the other seven patients got infected, which was most likely driven by frequent transferring among distinguished rooms; thus, the CRO in the environment or other media were contaminated. Furthermore, CRAB is inferred to be transmitted from ab5 (Pt 1) to ab2 and ab3 (Pt 8), and ab8 (Pt 12) and ab11 (Pt 2). The ST540 strain of ab6 (Pt 7) and ab12 (Pt 4) might have originated from ab10 (Pt 6), which was an imported case carrying CRAB upon admission.
Discussion
Here, we performed a real-time investigation of outbreaks using comprehensive sampling and whole-genome sequencing to identify the sources and driving force of CRO spreading in tertiary hospitals. Two findings were illustrated: (1) environmental and HCW contamination were common during CRO transmission, and (2) real-time comprehensive sampling combined with genome-based investigation might help track the transmission.
In this study, we performed real-time investigation using a comprehensive environmental and HCW sampling. The origin of CRKP transmission might have stemmed from dominant CRKP clones throughout the hospital, including surfaces, which infected the patients after admission. Persistent environmental contamination and HCW infection may be possible sources of carbapenem-resistance in both Klebsiella pneumoniae and Acinetobacter baumannii.10,11 Several studies have shown the genetically similarity between strains isolated from the patients, HCWs, and the environment.5,11 However, the origin of transmission between patient, HCW, and environment remains unknown, and thus should be elucidated in the future.12 The index patient of ICU-1 CRKP transmission, Pt 6, underwent a surgery before testing positive, which may be due to bacterial translocation from the gut microbiota caused by procedures.13 After detection of CRO contaminants in the environment and HCWs, we strengthened the environmental disinfections, and subsequently no more CROs were detected from the third sampling. No significant evidence was detected in the transmission of methicillin-resistant Staphylococcus aureus and Vancomycin-resistant Enterococci when the contact precaution measure was implemented.14 However, OXA-23 was still transmitted in a new environment, even after the implementation of the control measures in the ICUs. Thus, apart from the contact precaution measures and environment disinfection, additional measures should be conducted.
Studies have shown that gram-negative bacteria mainly colonize the intestinal tract, and approximately 40% of CRKP infections are caused by the patients’ own unique strain.13 Active rectal swab screen could identify CRO carriers and provide evidences in tracking and controlling multidrug-resistant bacteria infection in subsequent hospital outbreaks. However, the positive rectal swab screen could not rule out hospital-acquired infection. In the present study, we isolated the strain 932 from Pt J (rectal swab), while strain W937 was isolated from sputum. Interestingly, the two strains were genetically different. The gut microbiota is complex and might harbor different genotypes of CRO; therefore, a timely surveillance and follow-up of rectal screening might help identify nosocomial transmission index and route.
Environment disinfection, contact precaution, and rectal swab are “bundle” measures suggested to be conducted in the ICUs. Since the introduction of Israel national intervention of CRE in 2007,15 the incidence of CRE acquisition declined by a 79%.15,16 Subsequently, these bundle measures combined with real-time surveillance should be performed to observe the impact of hospital-acquired CRO infection.
In ICU-2, no pathogen indicating environmental or staff contamination was found. Although no direct evidence was observed for the transmission route, we found that this outbreak might come from an imported index patient from referring hospitals, resulting in its transmission.
In our study, whole-genome sequencing combined with real-time comprehensive sampling was performed. These techniques have been widely used in outbreak tracing and genetic analysis.17–19 The WGS-based surveillance was applied in Candida auris infection control in ICU.20 Furthermore, we utilized a combination of short and long-read sequencing for plasmids analysis and reported plasmids carrying virulence genes, indicating the spread of hypervirulent-CRKP, which is of concern recently.21–24
This study has several limitations. First, the direction of the transmission route between the environment and HCWs was unclear, although close genomic distances were identified. Thus, timely surveillance should be conducted in a new ward in the future. Second, follow-up sample collections were not performed since the first sampling of ICU-2 was negative. Third, we did not perform the previous surveillance culture in Pt 6; thus, the source of Pt 6 infection was unclear.
Conclusion
In conclusion, a timely and comprehensive sampling combined with genome-based investigation may help in tracking the transmission route of and controlling the infections. HCWs and the environment could be contaminated during CRO transmission, which calls for strengthened prevention and control of infections.
Acknowledgments
This work was supported by Research grants from the Shanghai Science and Technology Committee (20dz2260100, 20Y11900400), Key Discipline Construction Plan from Shanghai Municipal Health Commission (GWV-10.1-XK01), Sumitomo Pharmaceutical (Suzhou) Co. Ltd. (2019QD040) and Shanghai Hospital Development Center (SHDC22020214).
Disclosure
The authors declare that they have no competing interests.
References
1. Chen M, Conlan S, Lau AF, et al.;Program NCS. Whole-genome sequencing overrules a suspected case of carbapenem-resistant Enterobacter cloacae transmission. J Clin Microbiol. 2017;55(9):2868–2870. doi:10.1128/JCM.00915-17
2. Mork RL, Hogan PG, Muenks CE, et al. Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S aureus skin and soft tissue infection: a prospective cohort study. Lancet Infect Dis. 2020;20(2):188–198. doi:10.1016/S1473-3099(19)30570-5
3. Gorrie CL, Mirceta M, Wick RR, et al. Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis. 2018;67(2):161–170. doi:10.1093/cid/ciy027
4. Shenoy ES, Pierce VM, Walters MS, et al. Transmission of Mobile Colistin Resistance (mcr-1) by Duodenoscope. Clin Infect Dis. 2019;68(8):1327–1334. doi:10.1093/cid/ciy683
5. Weingarten RA, Johnson RC, Conlan S, et al. Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring carbapenem resistance. mBio. 2018;9(1). doi:10.1128/mBio.02011-17
6. Popovich KJ, Green SJ, Okamoto K, et al. MRSA transmission in intensive care units: genomic analysis of patients, their environments, and healthcare workers. Clin Infect Dis. 2021;72(11):1879–1887. doi:10.1093/cid/ciaa731
7. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923
8. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi:10.1093/bioinformatics/btp352
9. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Battistuzzi FU. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi:10.1093/molbev/msy096
10. Brondani Moreira RR, Viana GF, de Moraes ACC, et al. Dissemination of Acinetobacter baumannii OXA-23 in old and new intensive care units without transfer of colonized patients. Infect Control Hosp Epidemiol. 2018;39(9):1135–1137. doi:10.1017/ice.2018.168
11. Wei L, Wu L, Wen H, et al. Spread of carbapenem-resistant Klebsiella pneumoniae in an intensive care unit: a whole-genome sequence-based prospective observational study. Microbiol Spectr;2021. e0005821. doi:10.1128/Spectrum.00058-21
12. Qiao F, Wei L, Feng Y, et al. Handwashing sink contamination and carbapenem-resistant Klebsiella infection in the intensive care unit: a prospective multicenter study. Clin Infect Dis. 2020;71(Suppl 4):S379–S85. doi:10.1093/cid/ciaa1515
13. Gorrie CL, Mirceta M, Wick RR, et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin Infect Dis. 2017;65(2):208–215. doi:10.1093/cid/cix270
14. Khader K, Thomas A, Huskins WC, et al. Effectiveness of contact precautions to prevent transmission of methicillin-resistant staphylococcus aureus and vancomycin-resistant enterococci in intensive care units. Clin Infect Dis. 2021;72(Suppl 1):S42–S9. doi:10.1093/cid/ciaa1603
15. Schwaber MJ, Lev B, Israeli A, et al. Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin Infect Dis. 2011;52(7):848–855. doi:10.1093/cid/cir025
16. Ben-David D, Masarwa S, Fallach N, et al. Success of a national intervention in controlling carbapenem-resistant Enterobacteriaceae in Israel’s long-term care facilities. Clin Infect Dis. 2019;68(6):964–971. doi:10.1093/cid/ciy572
17. Gouliouris T, Coll F, Ludden C, et al. Quantifying acquisition and transmission of Enterococcus faecium using genomic surveillance. Nat Microbiol. 2021;6(1):103–111. doi:10.1038/s41564-020-00806-7
18. Espedido BA, Dimitrijovski B, van Hal SJ, Jensen SO. The use of whole-genome sequencing for molecular epidemiology and antimicrobial surveillance: identifying the role of IncX3 plasmids and the spread of blaNDM-4-like genes in the Enterobacteriaceae. J Clin Pathol. 2015;68(10):835–838. doi:10.1136/jclinpath-2015-203044
19. Onori R, Gaiarsa S, Comandatore F, et al. Tracking nosocomial Klebsiella pneumoniae infections and outbreaks by whole-genome analysis: small-scale Italian scenario within a Single Hospital. J Clin Microbiol. 2015;53(9):2861–2868. doi:10.1128/JCM.00545-15
20. Lesho EP, Bronstein MZ, McGann P, et al. Importation, mitigation, and genomic epidemiology of candida auris at a large teaching hospital. Infect Control Hosp Epidemiol. 2018;39(1):53–57. doi:10.1017/ice.2017.231
21. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3). doi:10.1128/CMR.00001-19
22. Turton JF, Payne Z, Coward A, et al. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and ‘non-hypervirulent’ types ST147, ST15 and ST383. J Med Microbiol. 2018;67(1):118–128. doi:10.1099/jmm.0.000653
23. Heiden SE, Hubner NO, Bohnert JA, et al. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 2020;12(1):113. doi:10.1186/s13073-020-00814-6
24. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.