Back to Journals » Infection and Drug Resistance » Volume 14
Comprehensive Genomic Survey of Antimicrobial-Resistance Bacteria in the Sewage Tank Replacement with Hospital Relocation
Authors Katagiri M , Kuroda M , Sekizuka T , Nakada N , Ito Y , Otsuka M, Watanabe M , Kusachi S
Received 6 September 2021
Accepted for publication 18 November 2021
Published 20 December 2021 Volume 2021:14 Pages 5563—5574
DOI https://doi.org/10.2147/IDR.S336418
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Miwa Katagiri,1,2 Makoto Kuroda,2 Tsuyoshi Sekizuka,2 Norihide Nakada,3 Yukitaka Ito,4 Masanobu Otsuka,4 Manabu Watanabe,1 Shinya Kusachi1
1Department of Surgery, Toho University Ohashi Medical Center, Meguro-ku, Tokyo, Japan; 2Laboratory of Bacterial Genomics, Pathogen Genomics Center, National Institute of Infectious Diseases, Shinjuku-ku, Tokyo, Japan; 3Research Center for Environmental Quality Management, Graduate School of Engineering, Kyoto University, Otsu-shi, Shiga, Japan; 4Division of Clinical Microbiology Laboratory, Toho University Ohashi Medical Center, Meguro-ku, Tokyo, Japan
Correspondence: Miwa Katagiri
Department of Surgery, Toho University Ohashi Medical Center, 2-22-36 Ohashi, Meguro-ku, Tokyo, 153-8515, Japan
Tel +81354333086
Fax +81354333091
Email [email protected]
Makoto Kuroda
Laboratory of Bacterial Genomics, Pathogen Genomics Center, National Institute of Infectious Diseases, 1-23-1 Toyama, Shinjuku-ku, Tokyo, 162-8640, Japan
Tel +81352851111
Fax +81352851150
Email [email protected]
Background: Excrement containing antimicrobial-resistant bacteria (ARB) is discharged from the hospital sewage through wastewater treatment plants (WWTP) into rivers, increasing the antimicrobial resistance (AMR) burden on the environment.
Purpose: We illustrate the contamination of hospital sewage tanks with ARB harboring antimicrobial resistance genes (ARGs) using comprehensive metagenomic sequencing. During the study period, we moved to a new hospital building constructed for renovation. Therefore, we investigated the difference in bacterial flora in the sewage tanks for each building with different departments, and the change in bacterial flora over time in new sewage tanks. Furthermore, we performed a comparative genome analysis of extended spectrum β-lactamase (ESBL)-producing organisms (EPOs) from hospital sewage and clinical samples. Residual antibiotics in the sewage tank were also measured.
Methods: Metagenomic analysis was performed on the hospital sewage samples, followed by whole genome sequencing of EPOs.
Results: The bacterial composition of new sewage tanks was comparable with that of old tanks within 1 month after relocation and was instantly affected by excrement. The bacterial composition of sewage tanks in the old and new buildings, containing rooms where seriously ill patients were treated, was similar. Selection on CHROMagar ESBL allowed detection of EPOs harboring blaCTX-M and carbapenemase genes in all sewage tanks. One of the sewage Escherichia coli strain comprising ST393 harboring blaCTX-M-27 corresponded to the clinical isolates based on core genome analysis. Moreover, the levels of levofloxacin and clarithromycin in the hospital sewage were 0.0325 and 0.0135 μg/mL, respectively.
Conclusion: Hospital sewage was contaminated with many ARB species, ARGs and residual antibiotics, which can cause a burden on WWTP sewage treatment. The bacterial flora in the sewage tank was rapidly affected, especially by the ward with seriously ill patients. AMR monitoring of hospital sewage may help detect carriers prior to nosocomial ARB-associated outbreaks and control the outbreaks.
Keywords: hospital sewage, antimicrobial resistance, antimicrobial resistance gene, metagenome analysis, whole genome sequencing, residual antimicrobial reagents
Introduction
The Global Action Plan on Antimicrobial Resistance drafted by the World Health Organization1 states that there is a need to understand the impact of human activities on the environment, particularly the spread and transfer of antimicrobial resistance genes (ARGs) and strains. The isolation rates of antimicrobial-resistant bacteria (ARB) are lower in Japan than those in other countries, but are steadily increasing.2
In several countries, extended spectrum β-lactamase (ESBL)-producing Escherichia coli or carbapenemase-producing organisms have been detected from water environments,3 wastewater treatment plants (WWTP), treated water,4,5 and hospital sewage,4,6 and Japan is no exception.7,8 In particular, hospital wastewater contaminated by ARB and residual antibiotics may generate selective pressure for the development of ARB,9,10 and is considered a hot spot for the growth and propagation of ARB.6
Similar to the general sewage system, hospital sewage is also discharged into a public WWTP and treated by filtering, microbiological treatment, and chlorination, after which the effluent is discharged into a nearby river. Although it is not yet clear whether the hospital wastewater-related ARB disseminate into the waterbodies, ARB is observed to exhibit deleterious effects on human health. In recent years, management strategies for ARB/ARGs and residual antibiotics discharged from hospitals have been studied in several countries.11,12
The aim of this study is to illustrate the contamination of hospital sewage with ARB harboring ARGs using comprehensive metagenomic sequencing.
In addition, this study was conducted in line with the relocation to a new hospital, and the formation of the bacterial flora in the new sewage tank was investigated. We also compared the whole genome sequence of extended spectrum β-lactamase (ESBL)-producing organisms (EPOs) isolated from hospital sewage and clinical samples, and analyzed the relationship between clinical and sewage isolates.
Materials and Methods
Hospital Setting
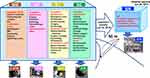
The study was conducted at the Ohashi Medical Center in Toho University, located in Jonan area, Tokyo, Japan. The Ohashi Medical Center (35.652573°N, 139.685833°E), with a capacity of 430 beds, was opened in 1973 with a single East building (BE) and expanded to Central (BC), Administration (BA), and West (BW) buildings to increase patient capacity (Figure 1). Features of each building are as follows: there were intensive care wards in BW; the number of beds was the largest among the old hospital buildings in BC; BE included the highest number of outpatient departments; and many of the rooms in BA were available only for healthcare workers.
As part of a renovation plan, a new hospital building (BN; 35.652578°N, 139.683959°E) with 319 beds was constructed approximately 50 m away from the old hospital and was inaugurated on June 20, 2018. Since May 2018, we introduced a policy of restricted hospitalization (Figure S1). We stopped providing services to outpatients from June 16 and transferred the 66 hospitalized patients to the BN on the same day (Figure S1). We started accepting patients on June 20. Both the old and new hospitals have a staff count of 2000. In BN, all outpatient departments and wards were integrated into one building (Figure 1).
In both old and new hospitals, stool and urine are stored in the underground sewage tanks without mixing with other drainage and are pumped to the sewage system several times a day (Figure 1). In the old hospital, each building had respective sewage tanks (sewage tank in BW: STW, in BA: STA, in BC: STC, and in BE: STE as shown in Figure 1); however, the new hospital had two connected storage tanks of 22.5 m3 collecting all the sewage (sewage tank in BN: STN shown in Figure 1). It was impossible to quantify daily inflow and outflow of sewage tanks as there is no system for regular measurement. The sewage discharged from the tanks is sent to the WWTP and treated using filtering, and microbiological and biochemical treatments, after which the effluent is discharged into the nearby river.
Sampling
Collection of Water Samples
In the period between May 8 and June 12, 2018, sewage samples were collected from STW, STA, STC, and STE once a week. In the period from June 6 to July 17, 2018, sewage samples were collected from the STN once a week at 9 a.m. A 20 mL of sewage samples were collected in sterile bottles from manhole of the sewage tanks and processed for analysis within 2 h.
DNA-Seq Analysis
Metagenomic Analysis for Whole Organisms in Original Hospital Sewage
First, 5 mL of sewage was centrifuged at 5000 × g for 5 min and the resultant cell pellet was vortexed with remaining 500 µL of sewage water. Next, the cell suspension was mixed with 500 µL of phenol/chloroform/isoamylalcohol (PCI) in a microcentrifuge tube with 2 mL of ZR BashingBead™ Lysis tube. Cell breaking was performed using GenoGrinder 2010 by shaking at 1500 rpm for 5 min. The PCI mixture was centrifuged at 8000 rpm for 5 min, followed by DNA purification using a Gel DNA Recovery Kit, Zymoclean-96 (ZYMO RESEARCH, Irvine, CA, USA). A metagenome DNA-seq library was prepared using the QIAseq FX DNA library prep kit (Qiagen, Venlo, Netherlands) and subsequently was performed using NextSeq 500 (Illumina) with the NextSeq 500 mid output kit v2.5 (300 cycles).
Genomic Analysis of EPOs Isolated from Hospital Sewage Using CHROMagar ESBL
To obtain EPOs from hospital sewage, 2 µL of sewage sample was mixed with 100 µL phosphate-buffered saline (PBS), plated on CHROMagar ESBL (bioMérieux, Marcy-l’Etoile, France) for selection of EPOs, and incubated at 36°C overnight. Subsequently, first appearance of color formation on each colony suggested that dark pink and metallic blue colonies were in the ratio of 1:4, respectively. Eighty colonies (20 dark pink colonies: potential ESBL-producing E. coli; 60 metallic blue colonies: potential Klebsiella, Enterobacter, Citrobacter) were selected as potential EPOs to identify a unique isolate from the first selection on CHROMagar ESBL plate. Each isolate was used for genomic analysis as described above.
In addition, all bacteria colonies on a single agar plate were harvested and mixed with 1000 µL of PBS. The cell suspension was used for metagenomic analysis as described above.
EPO Clinical Isolates
All 20 EPO clinical isolates obtained between May 8 and July 17, 2018, and reported as EPO were subjected to whole genome sequencing and comparative genomics. Among these isolates, 12 isolates were from outpatients and 11 isolates were from urology patients. They included urine, sputum, and central venous catheter samples (75%, 10%, and 10%, respectively). Some specimens were obtained from the same patient through subsequent diagnosis. The Ethics Committee of the Toho University Ohashi Medical Center waived the requirement for consent because the research was conducted without using identifiable biospecimens. Personal data related to clinical information were anonymized, and our procedure does not require a written consent from patients suffering from bacterial infections. Antimicrobial susceptibility was determined through screening (broth microdilution method) and confirmatory tests (the disk diffusion method) according to the Clinical and Laboratory Standards Institute (CLSI) recommendations (CLSI Performance Standards for antimicrobial disk susceptibility tests; Approved standard-13th edition CLSI document M02. Wayne, PA: Clinical and Laboratory Standards Institute; 2018).
Bioinformatics Analysis for Detection of Bacteria and ARG
The sequencing reads were analyzed using the MePIC2,13 Krona14 and MEGAN v6 software.15 EPO isolates were characterized using Krona,14 multi-locus sequence typing (MLST)16 and ResFinder.17
Principal Coordinate Analysis (PCoA)
The sequenced reads were assigned to a taxonomic hierarchy using MEGAN v6 software based on a megaBLAST nucleic acid homology search.
Core Genome Single-Nucleotide Variation Phylogenetic Analysis
Comparative genomics among obtained E. coli isolates (16 isolates from patients and 21 isolates from hospital sewage) were performed using BWA-MEM18 against the complete chromosome sequence of E. coli STN0717-11, which is the longest genome size among available complete genomes, followed by extraction of single nucleotide variants (SNVs) using VarScan v2.3.4.19 The prophage and repeat regions were predicted using PHASTER20 and MUMmer 3,21 respectively, and the detected SNVs in these regions were excluded. Regions of recombination in the chromosome were predicted using Gubbins v. 2.3.4,22 followed by masking of SNVs in the recombination regions. A maximum likelihood phylogenetic tree was constructed from SNV sites in the core genome region using FastTree2. De novo assembly was performed using SKESA v.2.3.023 with short reads of each strain, followed by analysis of sequence type, putative serotype, and AMR gene prediction using pubMLST (https://pubmlst.org/escherichia/), SeroTypeFinder,24 and Bacterial Antimicrobial Resistance Reference Gene Database (BioProject ID: PRJNA313047), respectively.
Measurement of Chemical Compound Concentrations
Thirty-nine drug components (Table 1) in the sewage samples were analyzed using solid-phase extraction (SPE) and ultra-performance liquid chromatography–tandem mass spectrometry (LC-MS/MS) based on a previously described method25 with minor modifications. Briefly, the sample was filtered using a polyethersulfone membrane (0.22 µm pore size, Merck) and 100 mL of the filtrate was spiked with 1 g/L ascorbic acid, 1 g/L EDTA, and a surrogate standard mixture, and then concentrated using SPE cartridge (Oasis HLB cartridges, 200 mg/6 cc, Waters, Japan). The analytes concentrated on the cartridge were extracted with 6 mL of methanol, following which they were measured using LC-MS/MS and quantified by the alternative surrogate method.25
 |
Table 1 Concentrations of Chemical Compounds in the Hospital Sewage Tank |
Results
Metagenomic Analysis of Tank Bacterial Flora in Each Building
Metagenome DNA-seq analysis of sewage samples was conducted to elucidate the differential microbial flora in the hospital sewage tank. The dominant bacteria in the sewage were classified according to the metagenomic data (Supplemental Data Set S1), wherein the proportion of genera varied depending on the building (Figure S2a and S2b).
Similarity and diversity of bacterial population among tanks were analyzed using PCoA based on bacterial genus level (Figure 2). A total of 25 sewage samples from each tank excluding STA were used for PCoA, and the results showed that the STN and STW groups were closely plotted by the presence of Aeromonas, Citrobacter, and Comamonas.
Analysis of β-Lactamase Genes Harbored by the Bacterial Flora of Tanks
Metagenome next-generation sequence (NGS) reads corresponding to β-lactamase genes were identified in original hospital sewage samples (Figure S3a) and EPOs from each tank selected on CHROMagar ESBL (Figure S3b). In the original hospital sewage samples, the blaIMP gene was detected in STC and STW samples, and the blaCTX-M gene was detected in STW and STN samples (Figure S3a). CHROMagar ESBL selection facilitated the detection of blaIMP and blaCTX-M from all sewage tanks (Figure S3b).
Genome Comparison of Sewage and Clinical Isolates
Whole genome sequencing was performed for 80 EPO isolates from STW0522 and STN0717, and 20 EPO clinical isolates (May 8 to July 17, 2018) for comparison of EPOs from sewage tanks and clinical sources. In sewage samples, E. coli, Klebsiella, Enterobacter, Citrobacter, and Achromobacter were detected (Table S1). Clinical isolates included only E. coli and Klebsiella spp.
A pairwise SNV analysis of the core genome was conducted for all E. coli strains (Figure 3). The E. coli STs included ST393, ST38, ST131, ST1011, ST12, ST73, ST9586, and ST224. E. coli ST12, ST73, ST131, and ST1011 were detected exclusively in clinical isolates (Table S2). Clinical isolates (THO-008 and −019 from the same patient) comprised ST393 harbouring blaCTX-M-27 and there was no difference in SNVs between these isolates and those obtained from sewage samples (14 isolates; STN0717-1 to −11, 14, 15, and 19) (Figure 3, Supplemental Data Set S2).
 |
Figure 3 Core genome phylogeny using single-nucleotide variations (SNVs) of ESBL-producing E. coli isolates. Core genome phylogeny was constructed using ESBL-producing E. coli isolates; 20 clinical isolates (THO-number, orange highlighted), one sewage isolate from STW0522 (brown highlighted), and 20 sewage isolates from STN0717 (blue highlighted). The complete genome sequence of STN0717-11 was used as a genome reference and 39.48% of the genome sequence was used as core genome regions among all tested strains. Few clinical isolates were obtained from same patient (§, patient No.8; ¶, patient No.9; †, patient No.7 in Supplement Data Set S2). Heatmap of pairwise differences of core genome SNVs are shown using a colour gradient with pink and red. The lower half part indicates core genome SNVs among all strains, and the upper half part shows core genome SNVs between indicated two strains. THO-008 and -019 from the same patient showed no SNVs with sewage isolates (14 isolates; STN0717-1 to −11, 14, 15, and 19). There were no identical clones between different patients. |
In three ST38 clinical isolates (THO-002, THO-007 and THO-020) harboring blaCTX-M-14, SNV analysis revealed marked 21–110 SNVs in the core genome. It was reported that molecular evolution of E. coli genome is possible with less than five SNVs within a 60-day duration.26 By contrast, ST38 sewage isolates harboring blaCTX-M-55 exhibited strict clonality with 3 SNVs, and ≥123 SNVs were detected in clinical isolates (Figure 3). Five ST131 clinical isolates harboring various CTX-M genes (blaCTX-M-15, CTX-M-27, CTX-M-44) were not identical (Figure 3). Among the 156 CHROMagar ESBL-positive strains from hospital sewage tanks (STW0522 and STN0717; Supplemental Data Set S2), carbapenemase gene (blaIMP-11) was identified only in Pseudomonas monteilii (Table 2).
 |
Table 2 ESBL-Producer in Hospital Sewage and Clinical Isolate |
Concentration of Residual Antimicrobial Agents in Hospital Sewage Tanks
The measurement of the concentrations of chemical contaminants in the tank (Table 1) showed that the most predominant antimicrobial agents were levofloxacin (32,500 ng/L) and clarithromycin (13,500 ng/L), although their concentrations were below minimal inhibitory concentration breakpoints. β-Lactam antibiotics were not measured in this study; as they are known to be almost undetectable in environmental samples.27,28
Discussion
The composition of bacterial flora in hospital sewage has been reported to comprise components of the human gut flora, including Bacteroides, Faecalibacterium, Bifidobacterium, and Blautia, in addition to Klebsiella, Aeromonas, and Enterobacter.6 The bacterial flora in each tank in the old hospital exhibited different bacterial compositions, and that of STC and STE mainly comprised the members of the human gut flora. However, the STW was mainly composed of Citrobacter and Acinetobacter, which are minimally detected in the gut of healthy individuals (Figure S2a and b).29,30 Furthermore, Comamonas and Arcobacter were detected in significant numbers in the STN. Comamonas is generally considered as environmental bacteria with less pathogenicity. Arcobacter spp. are detected in WWTP in several countries.31,32 The hospital sewage is influenced by the patients’ gut flora,29,33 but it is unclear whether the difference in the bacterial composition of each tank reflects the characteristics of the patients in each building.
Thus, the bacterial composition of STN appears similar to that of STW (Figure 3). BW and BN contain rooms where seriously ill patients are treated (Figure 1). The distribution of bacterial flora in the tanks can be strongly influenced by the severity of illness of the patients in the wards. Furthermore, bacterial flora in the tanks can be instantly affected by excrement because the bacterial composition of STN was comparable to that of STW within 1 month after the relocation. For monitoring department-specific ARB, it may be beneficial to install department-specific sewage tanks.
In Japan, the detection rate of EPO has been reported at 12.2% in healthy adult volunteers.34 Particularly, ST131 is an E. coli strain responsible for a worldwide pandemic and it carries a broad range of pathogenicity and ARGs, including a variety of β-lactamase genes on a transferable plasmid.35–37 In Japan, it has been reported that 92.9% of EPOs are blaCTX-M gene positive.38 The CTX-M genes (blaCTX-M-14, blaCTX-M-15, blaCTX-M-27 and blaCTX-M-2, listed in descending order of size)39 have been identified in Japan as well, and gene sequences obtained in this study are similar to that (Table 2).
A pairwise SNV analysis showed that the sequences of certain clinical EPO isolates had no difference compared to the SNVs of sewage isolates, suggesting that these sewage isolates may have originated from the patient. Fortunately, there was no strong evidence of a nosocomial outbreak associated with clinical EPOs (Figure 3). Monitoring of ARB/ARGs in hospital sewage may enable detection of latent carriers or nosocomial infections.
The carbapenemase gene (blaIMP-11) in the hospital sewage tanks (STW0522 and STN0717; Supplemental Data Set S2) was identified only in Pseudomonas monteilii (Table 2). P. monteilii was isolated from the environment,40 clinical samples,41 and hospital environment.40,42 P. monteilii is less pathogenic to humans, but may play a role as a metallo-β-lactamase (MBL) reservoir, and transfer of MBL genes to other species may be a cause of concern, especially in hospital sewage tanks.41,42 Many of these potential EPOs harboring ARGs in the sewage tanks were different from the clinical isolates. It is not clear whether these EPOs were excreted by healthy carriers or were transformed by acquiring the ARGs in the sewage tank.
The concentrations of ciprofloxacin and clarithromycin in hospital wastewater43,44 reported previously were similar to those in the present study. Hospital sewage tanks may promote the development of AMR by high selective pressure on bacteria,9,10,45 even at very low concentrations46 and provide optimal conditions47 for horizontal gene transfer, which is one of the mechanisms associated with the spread of AMR in the environment.48 It is known that the microbial gut flora function as a reservoir for ARGs and horizontal plasmid transfer between bacteria is common.49 This is plausible as sewage tanks consist of an accumulation of excrement and acquisition of resistance may occur frequently. We presume that selective pressure of antibiotics exists in our hospital sewage tanks; however, this will be verified in future studies.
In Japan, KPC-2-producing Klebsiella7 and NDM-5-coproducing E. coli8 were detected in the effluent of WWTP. Effective actions should be taken, including advanced wastewater treatment processes such as ozone and UV treatment11,50 and ultrafiltration51 to accelerate the removal of ARB in WWTP. However, even the above methods do not ensure a complete removal of ARB. Therefore, treatment processes may be introduced prior to the release of hospital sewage into the main sewage to reduce ARB. In Japan, there are a few reports of contamination of hospital sewage tanks with ARB/ARGs.52,53 Nevertheless, this study is the first comprehensive description of AMR in a hospital setting using metagenomic and whole genome analysis.
Conclusion
Our study reveals the presence of ARB/ARGs in the hospital sewage tanks and suggests that every hospital patient/staff/visitor can be a potential source of ARB. Monitoring of ARB/ARGs in hospital sewage is expected to identify the presence of carriers, and control nosocomial outbreaks and dissemination of ARB/ARGs in the environment.
Acknowledgments
The authors would like to thank Dr Yoshinobu Sumiyama, Chairman, Toho University and Dr Satoshi Iwabuchi, Hospital Director for giving us permission to conduct this research. We are grateful to Mr Umezu Masahiro for helping for sampling of hospital sewage. We gratefully acknowledge the staff members of the Laboratory of Bacterial Genomics, Pathogen Genomics Center, National Institute of Infectious Diseases, Tokyo, Japan.
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. Global action plan on antimicrobial resistance; 2016. Available from: https://www.who.int/publications/i/item/9789241509763.
2. Gekenidis MT, Qi W, Hummerjohann J, Zbinden R, Walsh F, Drissner D. Antibiotic-resistant indicator bacteria in irrigation water: high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS One. 2018;13(11):e0207857. doi:10.1371/journal.pone.0207857
3. Nascimento T, Cantamessa R, Melo L, et al. International high-risk clones of Klebsiella pneumoniae KPC-2/CC258 and Escherichia coli CTX-M-15/CC10 in urban lake waters. Sci Total Environ. 2017;598:910–915. doi:10.1016/j.scitotenv.2017.03.207
4. Proia L, Anzil A, Borrego C, et al. Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J Hazard Mater. 2018;358:33–43. doi:10.1016/j.jhazmat.2018.06.058
5. Makowska N, Philips A, Dabert M, et al. Metagenomic analysis of beta-lactamase and carbapenemase genes in the wastewater resistome. Water Res. 2020;170:115277. doi:10.1016/j.watres.2019.115277
6. Ng C, Tay M, Tan B, et al. Characterization of Metagenomes in Urban Aquatic Compartments Reveals High Prevalence of Clinically Relevant Antibiotic Resistance Genes in Wastewaters. Front Microbiol. 2017;8:2200. doi:10.3389/fmicb.2017.02200
7. Sekizuka T, Yatsu K, Inamine Y, et al. Complete Genome Sequence of a blaKPC-2-Positive Klebsiella pneumoniae Strain Isolated from the Effluent of an Urban Sewage Treatment Plant in Japan. mSphere. 2018;3(5). doi:10.1128/mSphere.00314-18.
8. Sekizuka T, Inamine Y, Segawa T, Kuroda M. Characterization of NDM-5- and CTX-M-55-coproducing Escherichia coli GSH8M-2 isolated from the effluent of a wastewater treatment plant in Tokyo Bay. Infect Drug Resist. 2019;12:2243–2249. doi:10.2147/IDR.S215273
9. Lien TQ, Lan PT, Chuc NTK, et al. Antibiotic Resistance and Antibiotic Resistance Genes in Escherichia coli Isolates from Hospital Wastewater in Vietnam. Int J Environ Res Public Health. 2017;14(7):699. doi:10.3390/ijerph14070699
10. Kummerer K, Henninger A. Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin Microbiol Infect. 2003;9(12):1203–1214. doi:10.1111/j.1469-0691.2003.00739.x
11. Jager T, Hembach N, Elpers C, et al. Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment, and the disseminated loads released to the environment. Front Microbiol. 2018;9:2599. doi:10.3389/fmicb.2018.02599
12. Paulus GK, Hornstra LM, Alygizakis N, Slobodnik J, Thomaidis N, Medema G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int J Hyg Environ Health. 2019;222(4):635–644. doi:10.1016/j.ijheh.2019.01.004
13. Takeuchi F, Sekizuka T, Yamashita A, Ogasawara Y, Mizuta K, Kuroda M. MePIC, metagenomic pathogen identification for clinical specimens. Jpn J Infect Dis. 2014;67(1):62–65. doi:10.7883/yoken.67.62
14. Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011;12:385. doi:10.1186/1471-2105-12-385
15. Huson DH, Beier S, Flade I, et al. MEGAN Community Edition - Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput Biol. 2016;12(6):e1004957. doi:10.1371/journal.pcbi.1004957
16. Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi:10.1128/JCM.06094-11
17. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
18. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi:10.1093/bioinformatics/btp698
19. Koboldt DC, Chen K, Wylie T, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–2285. doi:10.1093/bioinformatics/btp373
20. Arndt D, Grant JR, Marcu A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–21. doi:10.1093/nar/gkw387
21. Kurtz S, Phillippy A, Delcher AL, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi:10.1186/gb-2004-5-2-r12
22. Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi:10.1093/nar/gku1196
23. Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19(1):153. doi:10.1186/s13059-018-1540-z
24. Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J Clin Microbiol. 2015;53(8):2410–2426. doi:10.1128/JCM.00008-15
25. Narumiya M, Nakada N, Yamashita N, Tanaka H. Phase distribution and removal of pharmaceuticals and personal care products during anaerobic sludge digestion. J Hazard Mater. 2013;260:305–312. doi:10.1016/j.jhazmat.2013.05.032
26. Lee K, Izumiya H, Iyoda S, Ohnishi M. Effective surveillance using multilocus variable-number tandem-repeat analysis and whole-genome sequencing for enterohemorrhagic Escherichia coli O157. Appl Environ Microbiol. 2019;85:17. doi:10.1128/AEM.00728-19
27. Li D, Yang M, Hu J, Zhang Y, Chang H, Jin F. Determination of penicillin G and its degradation products in a penicillin production wastewater treatment plant and the receiving river. Water Res. 2008;42(1–2):307–317. doi:10.1016/j.watres.2007.07.016
28. Zhou LJ, Ying GG, Liu S, et al. Occurrence and fate of eleven classes of antibiotics in two typical wastewater treatment plants in South China. Sci Total Environ. 2013;452–453:365–376. doi:10.1016/j.scitotenv.2013.03.010
29. Tang WHW, Li DY, Hazen SL. Dietary metabolism, the gut microbiome, and heart failure. Nat Rev Cardiol. 2019;16(3):137–154. doi:10.1038/s41569-018-0108-7
30. Kurokawa K, Itoh T, Kuwahara T, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14(4):169–181. doi:10.1093/dnares/dsm018
31. Merga JY, Royden A, Pandey AK, Williams NJ. Arcobacter spp. isolated from untreated domestic effluent. Lett Appl Microbiol. 2014;59(1):122–126. doi:10.1111/lam.12256
32. Ghaju Shrestha R, Sherchan SP, Kitajima M, Tanaka Y, Gerba CP, Haramoto E. Reduction of Arcobacter at Two Conventional Wastewater Treatment Plants in Southern Arizona, USA. Pathogens. 2019;8(4). doi:10.3390/pathogens8040175
33. Usami M, Miyoshi M, Yamashita H. Gut microbiota and host metabolism in liver cirrhosis. World J Gastroenterol. 2015;21(41):11597–11608. doi:10.3748/wjg.v21.i41.11597
34. Higa S, Sarassari R, Hamamoto K, et al. Characterization of CTX-M type ESBL-producing Enterobacteriaceae isolated from asymptomatic healthy individuals who live in a community of the Okinawa Prefecture, Japan. J Infect Chemother. 2019;25(4):314–317. doi:10.1016/j.jiac.2018.09.005
35. Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–2155. doi:10.1093/jac/dkx146
36. Harris PNA, Ben Zakour NL, Roberts LW, et al. Whole genome analysis of cephalosporin-resistant Escherichia coli from bloodstream infections in Australia, New Zealand and Singapore: high prevalence of CMY-2 producers and ST131 carrying blaCTX-M-15 and blaCTX-M-27. J Antimicrob Chemother. 2018;73(3):634–642. doi:10.1093/jac/dkx466
37. Noguchi T, Matsumura Y, Kanahashi T, et al. Role of TEM-1 beta-lactamase in the predominance of ampicillin-sulbactam-nonsusceptible Escherichia coli in Japan. Antimicrob Agents Chemother. 2019;63(2). doi:10.1128/AAC.02366-18.
38. Luvsansharav UO, Hirai I, Niki M, et al. Prevalence of fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae among healthy adult people in Japan. J Infect Chemother. 2011;17(5):722–725. doi:10.1007/s10156-011-0225-2
39. Nakane K, Kawamura K, Goto K, Arakawa Y. Long-term colonization by bla(CTX-M)-harboring Escherichia coli in healthy japanese people engaged in food handling. Appl Environ Microbiol. 2016;82(6):1818–1827. doi:10.1128/AEM.02929-15
40. Remold SK, Brown CK, Farris JE, Hundley TC, Perpich JA, Purdy ME. Differential habitat use and niche partitioning by Pseudomonas species in human homes. Microb Ecol. 2011;62(3):505–517. doi:10.1007/s00248-011-9844-5
41. Ocampo-Sosa AA, Guzman-Gomez LP, Fernandez-Martinez M, et al. Isolation of VIM-2-producing Pseudomonas monteilii clinical strains disseminated in a tertiary hospital in northern Spain. Antimicrob Agents Chemother. 2015;59(2):1334–1336. doi:10.1128/AAC.04639-14
42. Scotta C, Juan C, Cabot G, et al. Environmental microbiota represents a natural reservoir for dissemination of clinically relevant metallo-beta-lactamases. Antimicrob Agents Chemother. 2011;55(11):5376–5379. doi:10.1128/AAC.00716-11
43. Wiest L, Chonova T, Berge A, et al. Two-year survey of specific hospital wastewater treatment and its impact on pharmaceutical discharges. Environ Sci Pollut Res Int. 2018;25(10):9207–9218. doi:10.1007/s11356-017-9662-5
44. Le TH, Ng C, Chen H, et al. Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country. Antimicrob Agents Chemother. 2016;60(12):7449–7456. doi:10.1128/AAC.01556-16
45. Coutu S, Rossi L, Barry DA, Rudaz S, Vernaz N. Temporal variability of antibiotics fluxes in wastewater and contribution from hospitals. PLoS One. 2013;8(1):e53592. doi:10.1371/journal.pone.0053592
46. Gullberg E, Cao S, Berg OG, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7(7):e1002158. doi:10.1371/journal.ppat.1002158
47. Acuna V, Casellas M, Font C, Romero F, Sabater S. Nutrient attenuation dynamics in effluent dominated watercourses. Water Res. 2019;160:330–338. doi:10.1016/j.watres.2019.05.093
48. Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 2018;42(1). doi:10.1093/femsre/fux053
49. Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg Infect Dis. 2010;16(6):1014–1017. doi:10.3201/eid1606.091671
50. Krzeminski P, Schwermer C, Wennberg A, Langford K, Vogelsang C. Occurrence of UV filters, fragrances and organophosphate flame retardants in municipal WWTP effluents and their removal during membrane post-treatment. J Hazard Mater. 2017;323(Pt A):166–176. doi:10.1016/j.jhazmat.2016.08.001
51. Hembach N, Alexander J, Hiller C, Wieland A, Schwartz T. Dissemination prevention of antibiotic resistant and facultative pathogenic bacteria by ultrafiltration and ozone treatment at an urban wastewater treatment plant. Sci Rep. 2019;9(1):12843. doi:10.1038/s41598-019-49263-1
52. Okubo T, Hasegawa T, Fukuda A, et al. Screening of hospital-manhole sewage using MacConkey agar with cefotaxime reveals extended-spectrum β-lactamase (ESBL)-producing Escherichia coli. Int J Antimicrob Agents. 2019;54(6):831–833. doi:10.1016/j.ijantimicag.2019.08.004
53. Azuma T, Otomo K, Kunitou M, et al. Environmental fate of pharmaceutical compounds and antimicrobial-resistant bacteria in hospital effluents, and contributions to pollutant loads in the surface waters in Japan. Sci Total Environ. 2019;657:476–484. doi:10.1016/j.scitotenv.2018.11.433
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.