Back to Journals » Cancer Management and Research » Volume 13
Comparison of Different EGFR Gene Mutation Status in Patients with Metastatic Non-Small Lung Cancer After First-Line EGFR-TKIs Therapy and Analyzing Its Relationship with Efficacy and Prognosis
Authors Yuan C, Jiang H, Jiang W, Wang H, Su C, Zhou S
Received 17 July 2021
Accepted for publication 24 August 2021
Published 3 September 2021 Volume 2021:13 Pages 6901—6910
DOI https://doi.org/10.2147/CMAR.S329900
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Kattesh Katti
Chengliang Yuan, Huiqin Jiang, Wei Jiang, Huilin Wang, Cuiyun Su, Shaozhang Zhou
Department of Respiratory Oncology, Guangxi Medical University Affiliated Tumor Hospital, Nanning City, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China
Correspondence: Shaozhang Zhou; Cuiyun Su
Department of Respiratory Oncology, Guangxi Medical University Affiliated Tumor Hospital, No. 71 Heti Road, Nanning City, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China
Tel +86-0771-5320761
; +86-0771-5334955
Fax +86-0771-5300613
Email [email protected]; [email protected]
Purpose: The purpose of this study is to compare the different EGFR mutation status in patients with metastatic non-small cell lung cancer (NSCLC) after first-line EGFR-TKIs therapy and analyze its relationship with efficacy and prognosis.
Patients and Methods: This study retrospectively analyzed the data of patients with metastatic NSCLC harboring EGFR mutation in the Affiliated Tumor Hospital of Guangxi Medical University from June 2016 to December 2020. Samples were collected before treatment and at the time of disease progression after first-line EGFR-TKIs therapy. Amplification refractory mutation system (ARMS) PCR and next-generation sequencing (NGS) were used to detect EGFR mutation. ORR, DCR, and PFS of different EGFR mutation groups were compared.
Results: The EGFR mutation rate of re-biopsy was 60.23%. The inconsistency rate of EGFR mutations in the same and different simple types was 72.22% (26/36) and 92.31% (48/52), respectively. Alterations in terms of EGFR mutations were divided into four groups: Group A: EGFR-sensitive mutation negative and T790M negative (39.77%); Group B: EGFR-sensitive mutation positive and T790M negative (18.19%); Group C: EGFR-sensitive mutation negative and T790M positive (36.36%); Group D: EGFR-sensitive mutation positive and T790M positive (5.68%). The differences between the four groups in ORR and DCR were not statistically significant (P> 0.05). The median PFS of all patients was 10.65 months. PFS of Group A, B, C, and D was 12.26, 7.96, 10.55, and 13.81 months, respectively, with statistical significance (Log rank P = 0.014).
Conclusion: EGFR mutation status in metastatic NSCLC patients receiving the first- and second-generation TKIs after disease progression show diversity. Monitoring the EGFR mutation changes is of great importance for subsequent clinical decision-making and exploring the underlying mechanisms of acquired resistance.
Keywords: non-small cell lung cancer, EGFR-TKIs, re-biopsy, EGFR gene mutation status
Introduction
EGFR (epidermal growth factor receptor) gene mutation, as the most common drive mutations, occurs in Asian race patients with non-small cell lung cancer (NSCLC) with a frequency of approximately 30%–40%.1,2 Substantial previous studies support that EGFR-tyrosine kinase inhibitors (TKIs) monotherapy, including the irreversible ErbB family blocker, such as osimertinb (third-generation), afatinib (second-generation), and the reversible EGFR-TKIs (first-generation), represented by icotinib (approved in China only), gefitinib and erlotinib, has been a standard of care for patients with locally advanced or metastatic NSCLC harboring EGFR mutation. Still, almost all patients will inevitably develop acquired resistance at 9 to 14 months after treatment.3–6 The missense mutation T790M is the primary type of drug resistance mutation, substituting methionine (M) for threonine (T) at position 790 at EGFR exon 20, accounting for nearly two-thirds of patients.7,8
Therefore, dynamic detection of the EGFR gene mutation status during disease progression is of great significance for guiding patients in subsequent treatment and achieving precise therapy throughout the course. Our study enrolled the metastatic NSCLC patients with EGFR-TKIs first-line treatment and compared their EGFR gene mutations in matched specimens before treatment (baseline) and disease progression to verify the necessity of re-biopsy and to better understand dynamic changes of EGFR mutation.
Patients and Methods
Patients
We retrospectively collected data on metastatic NSCLC patients initially diagnosed with EGFR 19 del or L858R-sensitive mutation and treated with EGFR-TKIs as the first-line regiment in Guangxi Medical University Affiliated Tumor Hospital from June 2016 to December 2020. Information of age, sex, smoking history, The Eastern Cooperative Oncology Group (ECOG) score, biopsy site, sample type, EGFR mutation status, the efficacy of EGFR-TKIs, and progression-free survival were extracted from medical records. The study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the Ethics Committee of Guangxi Medical University Affiliated Tumor Hospital. The identifiable information of patients was unnamed or anonymous with the aim to protect patients’ privacy. The need for informed consent was waived owing to the retrospective nature of the study.
The inclusion criteria for enrollment were as follows: (1) histologically or cytologically diagnosed with stage IV NSCLC without receiving any anti-tumor treatment; (2) confirmed with EGFR typical (exon 19 deletion or exon 21 L858R mutation) or untypical sensitive mutations by amplification refractory mutation system (ARMS) or next-generation sequencing (NGS) before treatment; (3) patients received EGFR-TKIs as the first-line monotherapy and the EGFR mutation status were detected at the time of disease progression.
The exclusion criteria were as follows: (1) EGFR gene with non-sensitive mutation or primary T790M mutation; (2) patients used EGFR-TKIs based double- or triple-drug regimens as first-line therapy, including EGFR-TKIs combined with chemotherapy or anti-angiogenesis; (3) concurrent or secondary other primary cancer.
EGFR Gene Mutation Detection
Amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) and next-generation sequencing technology (NGS) were used to define the EGFR mutation status.
The ADx EGFR 29 Mutation Kit (Amoy Diagnostics, Xiamen, China), using AMRS-PCR to detect previously predefined point mutations, covers 29 point mutations of EGFR including exon 18 G719X (G719A, G719, G719C), exon 19 deletions, exon 20 insertions (three types of insertions), exon 20 T790M and S768I, and exon 21 L858R and L861Q mutation et al (details in Appendix Table 1). The assay was performed in accordance with the manufacturer’s protocol with the MX3000P (Stratagene, La Jolla, USA) real-time PCR system. The 25 μL RT-PCR comprised 0.4 μL template DNA, 3.6 μL deionized water, and 16 μL reaction mix (reaction buffer, dNTPs, specific oligos, and probes). PCR was carried out with an initial denaturation at 95°C for 10 min, followed by 40 cycles of amplification (at 95°C for 30 s and 61°C for 1 min). The results were analyzed according to the criteria defined by the manufacturer. Positive results were defined as Ct (sample)-Ct(control)\Ct(cut-off). The criteria are that Ct value <26 is defined as EGFR mutation-positive; Ct value ≥29 as negative. If the Ct value is in the range of 26–29, the Ct value should be determined. If the Ct value is less than the cut-off value, the EGFR mutation of the sample is viewed as weakly positive.
NSG, as a high-throughput sequencing technology, is capable of detecting mutations, indels, copy number variations, and genomic rearrangements simultaneously, was performed in the American College of Pathologists (CAP) certified labs (Shihe Gene Biotech Inc, Nanning, China, and Geneplus Technology, Beijing, China) in China. EGFR sensitizing mutations were defined according to the National Comprehensive Cancer Network (NCCN) guidelines, including typical exon 19 del and exon 21 L858R mutations and untypical exon 18 G719X, exon 20 S768I, exon 20 insertion variant A763_Y764insFQEA and exon 21 L861Q mutations.
Efficacies Evaluation and Follow-Ups
The response to EGFR-TKIs was assessed according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1, and PFS was defined as the time from diagnosis to disease progression or death from any cause. The first evaluation was conducted at 1 month, and then the follow-up interval was every 2 months. The last follow-up was on March 31, 2021.
Statistical Analysis
Continuous and categorical variables were presented as mean ±SD and frequency (%), respectively. Either Student’s t-test or Kruskal–Wallis H-test was used depending on whether the continuous variables showed normal or skewed distribution. Fisher’s exact test was applied if the theoretical frequency existing in cells of 2×2 tables was less than 5. Pearson’s chi-square test was used to assess categorical variables. The rank-sum test was used to analyze different EGFR gene mutation status effects on ORR and DCR when re-detected after first-line targeted therapy. The impact of different EGFR gene mutation status on PFS was described by Kaplan–Meier curves and compared by a Log rank test. The hazard ratios (HR) and 95% confidence intervals (CI) for the risk of disease progression were estimated using Cox proportional hazards models with adjustment for pertinent variables. The criteria for selecting variables used for adjustment was that if the change in the effect estimate was more than 10% after an adjusted variable or the P-value in the univariable analysis was less than 0.05. This variable should be adjusted in Cox proportional hazards models. A two-tailed p<0.05 was considered statistical significance. All data were analyzed using the statistical package R version 3.4.3 (R Foundation for Statistical Computing, Vienna, Austria) and Empower (X&Y Solutions, Inc. Boston, Massachusetts).
Results
Clinical Characteristics and EGFR Mutation Status
Eighty-eight patients met the criteria enrolled in the study. Clinical characteristics and EGFR mutation status before treatment and after progression are shown in Table 1. The type of pathology of all enrolled patients was adenocarcinoma. The apparent differences before treatment (biopsy) and after progression (re-biopsy) in clinical characteristics are the sample types and EGFR mutation status. Before treatment, the primary sample type was tissue, whereas, in the condition of disease progression, ctDNA-based liquid-biopsy using plasma as materials accounted for approximately two-thirds of samples (59.09%). The overall EGFR mutation rate of re-biopsy specimens was 60.23% (51/88). The primary type was the secondary T790M mutation (42.05%) and then followed by EGFR mutation clearance (39.76%), 19Del (7.96%), L858R (7.96%), and other types of mutations (2.27%). The inconsistency rate of EGFR mutations in the same and different simple types was 72.22% (26/36) and 92.31% (48/52), respectively (details in Appendix Table 2).
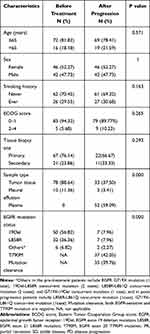 |
Table 1 Clinical Characteristics and EGFR Mutation Status of Enrolled NSCLC Patients Before Treatment and After Progression |
Treatment
Of all the 88 enrolled patients, 51 were treated with icotinib (57.95%), 32 with gefitinib (36.36%), and 5 with afatinib (5.68%). The ORR and DCR were 69.32% and 94.31%, respectively. The median PFS was 10.65 months. To clarify the different EGFR mutation status after progression on the effect of efficacies and clinical outcomes, we divided them into four groups: Group A: EGFR-sensitive mutation negative and T790M negative (39.77%); Group B: EGFR-sensitive mutation positive and T790M negative (18.19%); Group C: EGFR-sensitive mutation negative and T790M positive (36.36%); Group D: EGFR-sensitive mutation positive and T790M positive (5.68%). Group A, B, C, and D accounted for 39.77% (35/88),18.19% (16/88), 36.36% (32/88), 5.68% (5/88), respectively. The differences between the four groups in ORR and DCR were no statistical significance (P>0.05).
Comparison of the Post-Progression EGFR Mutation Status by Grouping
Clinical Characteristics and Treatment Response of Different Groups
Clinical characteristics and treatment responses of different groups are listed in Table 2. There was no statistical significance between the four groups in age, sex, smoking history, tumor biopsy site, sample type, initial EGFR mutation status, EGFR-TKIs selection (P>0.05). The difference was found in ECOG score for four groups with P <0.05. We also noticed that treatment response seems to be more negligible effect by grouping.
 |
Table 2 Clinical Characteristics and Treatment Information of Different EGFR Mutation Status Groups |
Univariate Analysis of the Relationship Between PFS and Variables
Results of univariate analysis using Cox proportional hazards regression model are listed in Table 3, demonstrating that SD comparing with PR, and Group B comparing with Group A had more hazard to develop disease progression, with HR=4.91 (95% CI=2.81–8.58, P <0.0001) and HR=2.70, (95% CI=1.44–5.07, P = 0.0021), respectively.
 |
Table 3 Univariate Analysis of the Relationship Between PFS and Variables |
Multivariate Analysis of the Relationship Between PFS and Four Groups
According to the principles of covariates selection for multivariate analysis, we conducted every two groups for comparison (shown in Table 4). Statistical significance can be found in comparing Group A and Group B with HR=2.44 (95% CI=1.32, 4.52) (P=0.0044).
 |
Table 4 Comparison PFS Between Two Groups of Different EGFR Mutation Status |
Kaplan–Meier Curve of PFS of Different Groups and Its Comparisons
Kaplan–Meier curve was used to demonstrate the PFS of 88 patients who experienced disease progression after first-line targeted therapy (shown in Figure 1). The median PFS of all patients was 10.65 months. The PFS for Group A, B, C, and D was 12.26, 7.96, 10.55, and 13.81 months, respectively (shown in Figure 2). The difference between the four groups was statistically significant (Log rank P = 0.029).
 |
Figure 1 PFS curve of patients with metastatic NSCLC with different EGFR gene mutation status after progression. |
In addition, we compared the Kaplan–Meier curve between every two groups (showed in Figure 2). The proportional COX hazard ratio model between every two groups was also conducted. The results of median PFS, Log rank values and HR with 95% confidence interval are listed in Table 4.
Discussion
Several clinical trials, such as IPASS, OPTIMAL, NEJ002, EURTAC, ARCHER1050, LUX-lung 7, and FLAURA, have established the standard role of EGFR-TKIs as the first-line setting in the treatment of patients with EGFR mutations.9–11 It is well known that mechanisms of resistance against EGFR-TKIs are complex. Heterogeneity, secondary mutation, bypass signaling pathway activation, and histology transformation are involved.10,11 To clarify the mechanisms of acquired resistance and select a corresponding therapeutical strategy, the guidelines recommend that re-biopsy should be routinely performed.12
In this study, tumor tissue was the primary source of specimens for the initial EGFR mutation detection. Nevertheless, at the time of progression, and plasma replaced tumor tissue as the primary source of specimens. Circulating tumor DNA-based liquid biopsy with non-invasive, convenient, and reproducible advantages has been widely used, especially with the emergence of highly sensitive and specific detection methods. IFUM study identified the efficacy of gefitinib in treating Caucasian NSCLC patients with EGFR mutations based on plasma-free DNA testing (ARMS testing).13 Clinical trials, such as AURA and BENEFIT, have confirmed that patients with EGFR mutations using plasma for detection can also benefit from EGFR-TKIs treatment.14,15
We also noted that the positive rate of re-biopsy was decreased by 33.77%, which may be explained by 1) the EGFR signaling pathway persistent inhibition despite progression; 2) the possibility of false negatives using ctDNA for detection;14,16,17 3) tumor cells may activate the other non-EGFR signaling pathways against TKIs.
Approximately 60% of the patients with acquired resistance to the first- and second-generation EGFR-TKIs develop a secondary T790M mutation, which has been shown to alter drug binding and enzymatic activity of the mutant EGF receptor.8,18 Osimertinib has been recommended by various authoritative guidelines for those patients as a standard of care. Meanwhile, the IMPRESS study demonstrated that continuous first-generation TKI treatment in T790M mutate patients after disease progression may impair the clinical benefit.19 In our study, the detection rate of T790M was 42.05%, which was lower than that of 60% using tumor tissue for testing.8,20 In our study, the percentage of patients who chose ctDNA of plasma for NGS detection was nearly 60%. Although the liquid biopsy can be used as an alternative in a series of clinical sceneries, including the suboptimal clinical condition or an unfavorable tumor site such as bone or central nervous system or multiple small pulmonary nodules, the sensitivity compared with tissue samples sometimes is moderately low.12 Differed from some studies, which were showed the pretty high concordance of T790M mutation detection between plasma and tumor tissue or malignant fluid specimens,19–21 Zheng et al22 explored the acquired resistance using ctDNA of 117 NSCLC patients after disease progression by digital PCR and found that 47% (55/117) of the patients developed T790M mutation. The occurrence of T790M seems to be no significant differences in age, gender, histology, smoking history, and treatment line setting, which is similar to our results.
Several reasons, including tumor size, location of primary and metastatic lesions, the timing for biopsy, and different detection methods, may be related to lower plasma T790M detection. For instance, in the AURA study, among 104 patients whose T790M were negative using plasma for testing, 47 were positive using the highly sensitive Beaming digital PCR method (Cobas) to detect tissue samples.23 Furthermore, AURA3 study also showed that 23–27% of T790M tissue-positive patients could not detect ctDNA. Further analysis revealed that ctDNA release was related to tumor size, especially in small lesions (<40 mm) before treatment.24 Besides, the positive rate of T790M blood test in patients with stage M0–M1a is significantly lower than that of patients with stage M1b.25,26 The above results suggest that the positive plasma T790M can guide clinical decision-making, but the negative result needs to be interpreted with caution.
We first divided the EGFR gene status after disease progression into four groups to the best of our knowledge. We found that the EGFR-sensitive mutation positive and T790M positive has a longer median PFS (13.81 months) than other groups, indicating secondary resistance mutation remaining on the EGFR signaling pathway has more favorable clinical outcomes. Meanwhile, we also identified that the shorter median PFS was presented in the EGFR-sensitive mutation positive and T790M negative group (7.96 months). Thus, it suggests that EGFR-TKIs may not be good at eliminating sensitive mutant cells, and other mechanisms un-relying on activation of EGFR independent pathway may be involved simultaneously.
In addition, we also identified that the PFS of patients with EGFR mutation clearance was more than 1 year with a median of 12.26 months. BENEFIT, a large-scale, prospective clinical trial, showed that the clearance of EGFR mutations after 8 weeks of gefitinib treatment significantly prolonged PFS compared with EGFR mutation persistence. Wang et al explained that these might be partly contributed by variation of multiple oncogenic driver genes and tumor suppressor genes at the time of disease progression.27 Similar phenomena have also been found in the third-generation TKIs. Zhou et al conducted an exploratory investigation based on the FLAURA study with enrolled 556 EGFR-sensitive mutate patients who used Osimertinib as the first-line setting.28 Digital PCR detected EGFR gene mutations at the third and sixth week after Osimertinib administration. The results showed that the median PFS in patients with EGFR-sensitive mutations persistence at the third week was 9.5 months, while in patients with EGFR-sensitive mutations, clearance was 13.5 months.
Of note, several limitations in our study should be acknowledged. First, the single-center property and a retrospective design are the major limitations. Second, the number of enrolled patients was not large scale, which may attenuate the power of statistics, especially after division into subgroups. For this reason, caution should be exercised in drawing the conclusions from this study. Further studies with larger sample sizes, a multi-center and prospective design should be needed. Final but not least, affected by the different detection methods and ARMS detecting abilities, we only focused on EGFR single gene and did not observe the influence of concurrent mutation beyond EGFR.
Conclusion
In summary, our study demonstrated that the subsequent EGFR gene changes show diversity in metastatic NSCLC patients receiving the first- and second-generation TKIs after disease progression. Therefore, it is essential for subsequent clinical decision-making after progression or exploration of resistance mechanisms by monitoring EGFR gene mutation status changes in the treatment process.
Acknowledgments
The authors thank all the staff of Guangxi Medical University Affiliated Tumor Hospital for their support in the work. In addition, we would like to thank Zhou S for the English language editing.
Funding
This study was supported by a “139 Talent Planning” granted by Guangxi Health Commission (grant number: 201903030) and a “Appropriate Technology Development, Promotion and Application Project” granted by Guangxi Health Commission (grant number: S2020102).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi:10.1056/NEJMoa1411817
2. Zhang X, Chang A. Somatic mutations of the epidermal growth factor receptor and non-small-cell lung cancer. J Med Genet. 2006;44(3):166–172. doi:10.1136/jmg.2006.046102
3. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR -mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi:10.1158/1078-0432.CCR-12-2246
4. Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised Phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi:10.1016/S1470-2045(11)70393-X
5. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi:10.1056/NEJMoa0810699
6. Mok TS, Wu Y, Ahn M, et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med. 2017;376(7):629–640. doi:10.1056/NEJMoa1612674
7. Yu HA, Sima CS, Huang J, et al. Local therapy with continued EGFR tyrosine kinase inhibitor therapy as a treatment strategy in EGFR-mutant advanced lung cancers that have developed acquired resistance to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2013;8(3):346–351. doi:10.1097/JTO.0b013e31827e1f83
8. Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17(6):1616–1622.
9. Martinez-Marti A, Navarro A, Felip E. Epidermal growth factor receptor first generation tyrosine-kinase inhibitors. Transl Lung Cancer Res. 2019;8:S235–46. doi:10.21037/tlcr.2019.04.20
10. Suda K, Mitsudomi T. Drug tolerance to EGFR tyrosine kinase inhibitors in lung cancers with EGFR mutations. Cells. 2021;10(7):1590. doi:10.3390/cells10071590
11. Grabe T, Lategahn J, Rauh D. C797S resistance: the undruggable EGFR mutation in non-small cell lung cancer? Acs Med Chem Lett. 2018;9(8):779–782. doi:10.1021/acsmedchemlett.8b00314
12. Wu YL, Zhang XC, Wang J, et al. Chinese experts consensus on detection of EGFR gene mutation in non-small cell lung cancer blood. Zhonghua Yi Xue Za Zhi. 2015;95(46):3721–3726.
13. Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9(9):1345–1353.
14. Ahn M, Han J, Tsai C, et al. OA 10.01 Detection of EGFR mutations from plasma ctDNA in the osimertinib Phase III trial (AURA3): comparison of three plasma assays. J Thor Oncol. 2017;12(11):S1771.
15. Wang J, Cheng Y, Wu Y, et al. MA 11.03 gefitinib as first-line treatment of plasma CtDNA EGFR mutation-positive NSCLC detected by DdPCR: BENEFIT study (CTONG1405). J Thor Oncol. 2017;12(11):S1844. doi:10.1016/j.jtho.2017.09.544
16. Jy DOUILLARD, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Brit J Cancer. 2014;110(1):55–62. doi:10.1038/bjc.2013.721
17. Mok T, Wu Y, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi:10.1158/1078-0432.CCR-14-2594
18. Mok T, Kim SW, Wu YL, et al. Gefitinib plus chemotherapy versus chemotherapy in epidermal growth factor receptor mutation-positive non-small-cell lung cancer resistant to first-line gefitinib (IMPRESS): overall survival and biomarker analyses. J Clin Oncol. 2017;35(36):4027–4034. doi:10.1200/JCO.2017.73.9250
19. Takahama T, Sakai K, Takeda M, et al. Detection of the T790M mutation of EGFR in plasma of advanced non-small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study). Oncotarget. 2016;7(36):58492–58499. doi:10.18632/oncotarget.11303
20. Su KY, Tseng JS, Liao KM, et al. Mutational monitoring of EGFR T790M in cfDNA for clinical outcome prediction in EGFR-mutant lung adenocarcinoma. PLoS One. 2018;13(11):e207001. doi:10.1371/journal.pone.0207001
21. Garcia J, Wozny AS, Geiguer F, et al. Profiling of circulating tumor DNA in plasma of non-small cell lung cancer patients, monitoring of epidermal growth factor receptor p.T790M mutated allelic fraction using beads, emulsion, amplification, and magnetics companion assay and evaluation in future application in mimicking circulating tumor cells. Cancer Med. 2019;8:3685–3697.
22. Zheng D, Ye X, Zhang MZ, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6(1):20913. doi:10.1038/srep20913
23. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non–small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. doi:10.1200/JCO.2016.66.7162
24. Ahn M, Han J, Tsai C, et al. OA 10.01 Detection of EGFR mutations from plasma ctDNA in the osimertinib Phase III trial (AURA3): comparison of three plasma assays. J Thorac Oncol. 2017;12(11):S1771.
25. Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2017;12(7):1061–1070. doi:10.1016/j.jtho.2017.04.003
26. Zhang X, Liang Z, Chen Y, et al. MA06.09 detection of EGFR T790M mutations by four testing platforms in ctDNA from Chinese patients with advanced NSCLC. J Thorac Oncol. 2017;12(11):S1823. doi:10.1016/j.jtho.2017.09.497
27. Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a Phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. 2018;6:681–690. doi:10.1016/S2213-2600(18)30264-9
28. Zhou C, Imamura F, Cheng Y, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR- TKIs in the FLAURA trial. J Clin Oncol. 2019;37(15_suppl):9020. doi:10.1200/JCO.2019.37.15_suppl.9020
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

