Back to Journals » Cancer Management and Research » Volume 12
Clinicopathological and Prognostic Significance of EML4-ALK Rearrangement in Patients with Surgically Resected Lung Adenocarcinoma: A Propensity Score Matching Study
Authors Shi J, Gu W, Zhao Y, Zhu J, Jiang G, Bao M, Shi J
Received 30 August 2019
Accepted for publication 3 January 2020
Published 24 January 2020 Volume 2020:12 Pages 589—598
DOI https://doi.org/10.2147/CMAR.S229217
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Jinghan Shi, 1,* Weiqing Gu, 2,* Yanfeng Zhao, 1 Junjie Zhu, 1 Gening Jiang, 1 Minwei Bao, 1 Jingyun Shi 3
1Department of Thoracic Surgery, Shanghai Pulmonary Hospital Affiliated to Tongji University, Shanghai, People’s Republic of China; 2Department of Oncology, Shanghai Pulmonary Hospital Affiliated to Tongji University, Shanghai, People’s Republic of China; 3Department of Radiology, Shanghai Pulmonary Hospital Affiliated to Tongji University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Minwei Bao
Department of Thoracic Surgery, Shanghai Pulmonary Hospital Affiliated to Tongji University, 507 Zhengmin Road, Shanghai 200433, People’s Republic of China
Tel +86 138 1808 9090
Email [email protected]
Jingyun Shi
Department of Radiology, Shanghai Pulmonary Hospital Affiliated to Tongji University, 507 Zhengmin Road, Shanghai 200433, People’s Republic of China
Tel +86 136 7170 1139
Email [email protected]
Objective: The echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) fusion gene is a key oncogenic driver in non-small cell lung cancer (NSCLC). This study analyzed the clinicopathological characteristics and prognostic significance of EML4-ALK fusion gene in patients with surgically resected adenocarcinoma.
Methods: The clinicopathological characteristics of 1056 consecutive patients with surgically resected stage I–IIIA adenocarcinoma were collected from February 2014 to October 2014, and EML4-ALK rearrangement was detected using real-time polymerase chain reaction (RT-PCR) technology. To compare the imaging and pathological features, a propensity score matching (PSM) method was performed. The follow-up information was collected to evaluate the long-term outcomes of patients with EML4-ALK rearrangement.
Results: The prevalence of EML4-ALK rearrangement was 6.6% in 1056 consecutive patients. A total of 70 EML4-ALK-positive and 210 EML4-ALK-negative patients were identified after PSM. Imaging and pathological analyses showed that EML4-ALK rearrangement was significantly associated with less ground-glass opacity (GGO) (adjusted OR=1.38, 95% CI=1.03– 1.85, Ptrend=0.029) and higher prevalence of non-invasive mucinous adenocarcinoma mucin-laden adenocarcinomas (non-IMA MLA, adjusted OR=6.79, 95% CI=2.69– 17.17, P< 0.001). EML4-ALK rearrangement was found to be an unfavorable prognostic factor for disease-free survival (DFS) in female patients (HR=2.26, 95% CI=1.13– 4.53, P=0.021).
Conclusion: Our results suggest that adenocarcinomas harboring EML4-ALK fusion gene exhibit specific radiological and pathological characteristics compared with EML4-ALK-negative adenocarcinomas. In female patients, EML4-ALK rearrangement was associated with shorter DFS.
Keywords: adenocarcinoma, non-small cell lung cancer, NSCLC, anaplastic lymphoma kinase, survival
Introduction
Tyrosine kinase inhibitors (TKIs) are a class of pharmaceutical drugs that show beneficial effects in non-small cell lung cancer (NSCLC) patients harboring specific molecular alterations with less toxicity compared to chemotherapy and radiotherapy. They function as anticancer agents by targeting oncogene-driven signaling pathways and thus improving patients’ survival and quality of life.1,2 The investigations on TKIs contribute to the development of therapeutic interventions for NSCLC patients.
The translocation and inversion of anaplastic lymphoma kinase (ALK) have been detected in a subset of NSCLC patients, especially the ones diagnosed with adenocarcinoma. Echinoderm microtubule-associated protein-like 4 (EML4) is the most common fusion partner for ALK in NSCLC.3,7 As the first TKI approved for ALK-positive NSCLC, Crizotinib treatment improved the survival of patients at advanced stages compared to standard chemotherapy.4 Therefore, ALK rearrangement detection is recommended for lung adenocarcinoma patients according to the guideline published by the International Association for the Study of Lung Cancer (IASLC).5
Patients harboring ALK rearrangement share certain clinicopathological features.6 ALK rearrangement is associated with younger age, never or light smoker, solid nodules, adenocarcinoma and excessive mucin production.8–10 However, the prognosis of patients with ALK-positive surgically resected adenocarcinoma in the absence of TKIs treatment remains controversial. Previous data suggested that ALK positivity was an unfavorable prognostic factor for disease-free survival (DFS).11–13 However, other studies failed to prove that ALK rearrangement was associated with the prognosis of patients with surgically resected NSCLC.14–17 The conflicting data on the prognostic significance of ALK positivity may be due to baseline difference. Some clinical characteristics, such as gender, smoking status and age might be the confounders in evaluating the prognostic significance of ALK rearrangement in surgically resected NSCLC.
In the current study, we assessed the prognostic significance of EML4-ALK rearrangement in patients underwent curative resection for lung adenocarcinoma. To minimize selection bias and reduce baseline differences, a propensity score matching (PSM) analysis was performed to compare the computed tomography (CT) features, pathological characteristics and DFS between EML4-ALK-positive and EML4-ALK-negative patients.
Materials and Methods
Patients and Follow-Up
A total of 1056 patients who were diagnosed with stage I-IIIA lung adenocarcinoma and underwent curative resection were included from February 2014 to October 2014 in Shanghai Pulmonary Hospital. They were screened for EML4-ALK fusion gene using real-time polymerase chain reaction (RT-PCR) (Figure 1). This retrospective study was approved by the Review Board of Shanghai Pulmonary Hospital and performed in accordance with the Declaration of Helsinki. The written informed consent was obtained from all patients before surgery. Patients who received preoperative chemotherapy, radiation therapy or TKI targeted therapy were excluded. None of the 1056 patients received ALK-targeted TKIs before disease recurrence. The medical records including age, gender, smoking history, tumor size and location, pathological Tumor-Node-Metastasis (pTNM) stage, histological type, preoperative carcino-embryonic antigen (CEA) level and postoperative therapy were collected from all patients. Pathologic staging was determined according to the 8th edition of TNM classification.
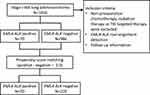 |
Figure 1 Flow diagram for this study. Abbreviations: EML4, echinoderm microtubule-associated protein-like 4; ALK, anaplastic lymphoma kinase; TKI, tyrosine kinase inhibitors. |
All patients received regular follow-up, mainly at our outpatient clinic or by telephone. The examination results were recorded in the health system. The radiological images of patients who visited our outpatient department for postoperative follow-up were reviewed by two radiologists and one physician. The disease-free survival (DFS) was defined as the time from the date of operation to the day of tumor recurrence or metastasis.
CT Examinations, Pathologic Diagnosis and EML4-ALK Fusion Gene Detection
All patients underwent preoperative chest CT in our hospital one month before surgery using SIEMENS Somatom Definition AS (Siemens Medical Systems, Erlangen, Germany). CT images were acquired at tube voltage of 120kVp, tube current of 300mA, section width of 2.0mm, reconstruction interval of 1.0mm, slice acquisition of 128×0.6 mm and rotation time of 0.5 s. Two radiologists reviewed the images and the following characteristics were recorded: estimated greatest diameter, percentage of ground-glass opacity (GGO) component, border (smooth or lobulated), speculation, cavity, bubble-like lucency, and air bronchogram. The percentage of GGO component was calculated as: (the greatest diameter of the tumor-the greatest diameter of the solid component)/(the greatest diameter for the tumor) ×100%.
Formalin-fixed and paraffin-embedded tissue sections (FFPE) were examined by two pathologists. The subtypes of adenocarcinoma were determined according to the International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) International Multidisciplinary Classification of Lung Adenocarcinoma.18
To detect EML4-ALK rearrangement, total RNA was extracted from fresh tissues using QIAamp RNeasy Kit (Qiagen, Germany). EML4-ALK rearrangement was detected using a commercially available kit (ACCB Diagnostics, Beijing, China) according to the manufacturers’ protocol. There are nine known EML4-ALK fusion transcript variants could be detected, including: E13;A20, E20;A20, E6a;A20, E6b;A20, E14ins11;del49A20, E2;A20, E2;ins117A20, E13;ins69A20, E14;del12A20 (“E”: EML4 and “A”: ALK).
Statistical Analysis and Propensity Score Matching (PSM)
Pearson’s Chi-square test (χ2 test) or the Fisher exact test was used to compare the differences in categorical variables. The differences between the means of continuous variables were compared using independent t-test. Univariate logistic regression was performed to investigate the association between the imaging and pathological features and EML4-ALK rearrangement. Multivariate logistic regression was used to determine the independent variables that could distinguish EML4-ALK-positive adenocarcinomas from the negative ones.
Kaplan–Meier curves were generated to compare the survival outcomes, and log-rank test was used to compare the survival differences between EML4-ALK -positive and EML4-ALK-negative groups. Cox regression model was performed to determine the independent prognostic factors after complete tumor resection. A P-value of less than 0.05 was considered statistically significant. All statistical analyses were processed by SPSS version 17.0 (SPSS Inc., Chicago, IL).
To minimize the selection bias, PSM method was used to balance the confounding factors with a 1:3 matching ratio between EML4-ALK-positive and negative groups, using a logistic regression model.
Results
Clinical Characteristics of Patients Harboring EML4-ALK Fusion Gene
The baseline characteristics of patients with surgically resected lung adenocarcinoma (n = 1056) are retrospectively analyzed in Table 1. Results showed that 6.6% (70/1056) of these patients harbored EML4-ALK fusion gene. The mean age of EML4-ALK-positive patients was younger than that of EML4-ALK-negative patients (mean±SD, 59.94±9.30 vs 55.96±11.35, P=0.005). Significant differences were observed in the distribution of pathological T-stage and the level of preoperative CEA between the two groups (P=0.047 and P=0.006, respectively), whereas no significant differences were found in gender, smoking history, tumor size, pathological N-stage, tumor location, and postoperative chemotherapy (P>0.05). PSM was used to reduce the selection bias in this study. It balanced the potential confounders between the two groups, including age, gender, pTNM stage, smoking history, preoperative CEA level and postoperative chemotherapy. After matching, no significant differences were observed in age, pathological T-stage and preoperative CEA level. Baseline characteristics were well balanced between EML4-ALK-negative (N=210) and EML4-ALK-positive (N=70) patients.
 |
Table 1 Clinicopathologic Characteristics of 1056 Consecutive Patients and 280 Propensity Score-Matched Patients Screened for EML4-ALK Rearrangement |
Comparison of Radiological and Pathological Characteristics
The mutation status analysis is critical for the diagnosis and treatment of NSCLC. However, the detection of molecular alteration in tumor tissue using biopsy has some limitations considering tumor location or tumor size. CT images are widely used in the prediction of pathological characteristics in many diseases, and the correlation between the image patterns and gene mutations has been widely investigated. To identify the variables that might differentiate EML4-ALK-positive lung cancer from the negative ones, the imaging and pathological features of 280 propensity score-matched patients were collected and summarized in Table 2. The CT patterns such as nodule type, lobulated border, speculated margin, bubble-like lucency and mucin-laden adenocarcinomas (MLA) in EML4-ALK-positive group were significantly different from those in EML4-ALK-negative patient (P<0.05). The results of univariate logistic regression analysis demonstrated that all positive variables in chi-squared test were significantly associated with an increased risk of EML4-ALK rearrangement (OR>1.00, P<0.05), except bubble-like lucency. The nodule type (adjusted OR=1.38, 95% CI=1.03–1.85, Ptrend=0.029) and non-invasive mucinous adenocarcinoma MLA(non-IMA MLA, adjusted OR =6.79, 95% CI =2.69–17.17, P<0.001 compared with non-MLA) were found to be important predictors of EML4-ALK-positive lung adenocarcinoma.
 |
Table 2 Association Analysis Between Imaging and Pathological Features of 280 Propensity Score-Matched Patients and EML4-ALK Rearrangement |
Prognostic Significance of EML4-ALK Fusion Gene in Surgically Resected Adenocarcinoma
To investigate the association between EML4-ALK rearrangement and survival outcomes in 280 matched patients, the analysis of DFS was performed. The median follow-up time was 49 (3–57) months. The survival curve analysis showed that there was no significant association between EML4-ALK rearrangement and DFS in the general population (log rank P=0.161) (Figure 2A). Subgroup analysis by gender showed that female patients harboring EML4-ALK fusion gene had a shorter DFS (log rank P=0.022) (Figure 2B). No significant association was found between EML4-ALK rearrangement and DFS in male patients (log rank p=0.63) (Figure 2C).
The cox regression analysis showed that EML4-ALK rearrangement was a significant prognostic factor for decreased DFS in female patients with adenocarcinoma (HR=2.10, 95% CI=1.10–4.04, P=0.026). Other variables including tumor size, pathological T-stage, pathological N-stage, preoperative CEA level, and postoperative chemotherapy were shown to be associated with a worse DFS in all groups (HR>1.00, P<0.05; Table 3). Multivariate analysis suggested that lymphatic metastasis was an unfavorable prognostic factor for DFS in all groups after adjusted for confounders (adjusted HR>1.00, Ptrend<0.05). EML4-ALK rearrangement remained to be an independent prognostic factor for a shortened DFS in the female population (adjusted HR=2.26, 95% CI=1.13–4.53, P=0.021; Table 4).
 |
Table 3 Univariate Analysis of Prognostic Factors for Disease-Free Survival in Propensity Score-Matched Patients Screened for EML4-ALK Rearrangement |
 |
Table 4 Multivariate Analysis of Prognostic Factors for Disease-Free Survival in Propensity Score-Matched Patients Screened for EML4-ALK Rearrangement |
Discussion
The identification of EML4-ALK rearrangement, an important biomarker in NSCLC, is highly recommended for patients with lung adenocarcinoma.5 The imaging characteristics of tumors are shown to have significant clinical values in pathological and survival analyses. In this retrospective study, we reported that surgically resected EML4-ALK-positive adenocarcinomas had a significantly higher proportion of solid component on CT images and were more likely to produce excessive mucin. Our data also showed that EML4-ALK rearrangement was significantly associated with decreased DFS in female patients.
The prevalence of EML4-ALK fusion genes in adenocarcinomas ranges from 3.4% to 10.2% when using the RT-PCR method.19,20 Here, we showed that 70 (6.6%) of 1056 patients with completely resected adenocarcinoma harbored EML4-ALK fusion gene, which was consistent with previously reported mutation rate (6.1%) in Asian ethnicity groups.31 Patients harboring ALK rearrangement appear to be younger and have a light or never smoking history.21 Consistently, our patients with EML4-ALK fusion gene were relatively younger compared to EML4-ALK-negative patients before PSM. In non-smokers, the incidence of EML4-ALK rearrangement was higher than EML4-ALK-negative adenocarcinomas (22.9% vs 30.3%), but not statistically significant. The negative association might be resulted from the study population and adenocarcinoma subtypes.
The use of CT image features to predicate the epidermal growth factor receptor (EGFR) mutations in NSCLC has been previously studied. Most studies showed that nodules with higher GGO proportion were more likely to harbor EGFR mutations.22,23 However, other studies reported a negative or contrary correlation between EGFR mutations and CT features,24,25 which might be explained by the difference in demographic characteristics.26,27 A recent PSM study of a large Asian population showed that patients with lung adenocarcinomas harboring EGFR exon 21 missense had the most inner GGO component, followed by the ones harboring EGFR exon 19 deletion, when compared to patients with EGFR wild-type tumors.28 However, the imaging features of EML4-ALK-positive NSCLC were rarely reported – due to its low incidence in lung cancer patients.10 We further used PSM method to minimize the potential selection bias caused by demographic confounders. Tumors harboring EML4-ALK rearrangement were more likely to appear as a pure solid nodule on CT scans in comparison to EML4-ALK-negative tumors (81.4% vs 60.0%). Also, less GGO proportion was significantly associated with EML4-ALK rearrangement in multivariate logistic regression analysis (adjusted OR=1.38, 95% CI=1.03–1.85, Ptrend=0.029). It has been revealed that solid component may be the result of abundant mucin production,29 which is an important pathological feature in lung adenocarcinomas. Here, we also found that abundant mucin production was closely related to EML4-ALK fusion gene. Therefore, our data indicated that the radiological and pathological characteristics of tumor were closely correlated with the molecular alteration.
The detection of ALK rearrangement insurgically resected tumors is superior to biopsy considering the number of tested cancer cells received for molecular analyses.30 Some studies showed that ALK-positive adenocarcinomas were more common in patients at advanced stages and with early lymph node metastasis.16,31 Shin and colleagues32 reported the incidence of more regional lymph node recurrence was higher than that of distant metastasis in patients with ALK-positive stage IA adenocarcinoma. In the 22 EML4-ALK-positive patients who developed recurrence after PSM in this study, 13 had regional recurrence, including mediastinal lymph node and pleural metastasis (not shown). These findings indicated that ALK-positive tumors may be more aggressive compared to ALK-negative lung cancer. In contrast, some previous studies failed to demonstrate the prognostic significance of ALK rearrangement in postoperative NSCLC patients.13,15–17 To minimize the inherent selection bias from the confounding variables such as age, gender, smoking history, tumor stage, and postoperative chemotherapy, we performed this PSM study in a cohort of 70 ALK-positive patients to investigate the prognostic value of EML4-ALK fusion gene in surgically resected adenocarcinomas. Our results demonstrated that EML4-ALK rearrangement was an unfavorable prognostic factor for a decreased DFS in female patients with adenocarcinoma (HR=2.26, 95% CI=1.13–4.53, P=0.021) after adjusted for other confounders, while not in the males (HR=0.89, 95% CI=0.38–2.12, P=0.894). Some studies reported that patients with EML4-ALK-positive NSCLC had a significantly worse prognosis compared with EML4-ALK-negative ones in non-smoking population.11,14 Given that most female patients are non-smokers, our data supported the prognostic significance of EML4-ALK rearrangement in lung adenocarcinoma.
Previous evidence showed that EGFR mutations and EML4-ALK rearrangement were mutually exclusive,33 and approximately 70% of female non-smoker Asian patients with adenocarcinoma harbored EGFR mutations.34 Chaft and colleagues36 reported that the postoperative DFS in ALK-positive patients was shorter compared to EGFR-mutant patients. A study also found that most EML4-ALK–negative female patients harbored EGFR mutations and patients with EGFR-positive adenocarcinomas were more likely to have a shortened postoperative DFS.35 All these findings may explain the association between EML4-ALK rearrangement with unfavorable survival outcomes, and the high risk of disease recurrence in female patients with adenocarcinoma.
This study had some limitations. First, this was a retrospective study and all clinical data were collected from a single center. Further retrospective analysis including multiple centers will be needed. Second, although a large number of patients with EML4-ALK-positive adenocarcinomas were included, the ones with inoperable tumors harboring EML4-ALK fusion gene at advanced stages were excluded. Third, more radiological and pathological characteristics, such as the presence of signet-ring cell, need to be analyzed in the future.8 Last, the fluorescence in situ hybridization and immunohistochemistry could be used to assess ALK rearrangement, as rare or unknown ALK fusion variants might not be detected by the RT-PCR method. Considering that this study was an observational research, and the commercially available kits used had covered common variant type of ALK rearrangement, our results were more practicable to be retrospectively analyzed in multiple centers.
Conclusion
In conclusion, this study suggested that EML4-ALK -positive adenocarcinoma was significantly associated with specific radiological and pathological features, including less GGO component and higher incidence of non-IMA MLA. Also, EML4-ALK rearrangement was identified as an unfavorable prognostic factor for a decreased DFS in female patients with resected adenocarcinomas.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. New Engl J Med. 2004;350(21):2129–2139. doi:10.1056/NEJMoa040938
2. Jeon JH, Kang CH, Kim H, Seong YW, Park IK, Kim YT. Prognostic and predictive role of epidermal growth factor receptor mutation in recurrent pulmonary adenocarcinoma after curative resection. Eur J Cardiothorac. 2015;47(3):556–562. doi:10.1093/ejcts/ezu177
3. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–563. doi:10.1038/nature05945
4. Solomon BJ, Kim D, Wu Y, et al. Final overall survival analysis from a study comparing first-line crizotinib versus chemotherapy in ALK-mutation-positive non-small-cell lung cancer. J Clin Oncol. 2018;36(22):2251. doi:10.1200/JCO.2017.77.4794
5. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors. J Mol Diagn. 2018;20(2):129–159. doi:10.1016/j.jmoldx.2017.11.004
6. Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31(8):1105–1111. doi:10.1200/JCO.2012.44.5353
7. Yoshida T, Oya Y, Tanaka K, et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3383–3389. doi:10.1200/JCO.2015.65.8732
8. Yoshida A, Tsuta K, Nakamura H, et al. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol. 2011;35(8):1226–1234. doi:10.1097/PAS.0b013e3182233e06
9. Gao Q, Li P, Jiang X, et al. Worse disease-free, tumor-specific, and overall survival in surgically-resected lung adenocarcinoma patients with ALK rearrangement. Oncotarget. 2017;8(49):86066–86081. doi:10.18632/oncotarget.20973
10. Kim TJ, Lee CT, Jheon SH, Park JS, Chung JH. Radiologic characteristics of surgically resected non-small cell lung cancer with ALK rearrangement or EGFR mutations. Ann Thorac Surg. 2016;101(2):473–480. doi:10.1016/j.athoracsur.2015.07.062
11. Yang P, Kulig K, Boland JM, et al. Worse disease-free survival in never-smokers with ALK+ lung adenocarcinoma. J Thorac Oncol. 2012;7(1):90–97. doi:10.1097/JTO.0b013e31823c5c32
12. Zhou JX, Yang H, Deng Q, et al. Oncogenic driver mutations in patients with non-small-cell lung cancer at various clinical stages. Ann Oncol. 2013;24(5):1319–1325. doi:10.1093/annonc/mds626
13. Kim MH, Shim HS, Kang DR, et al. Clinical and prognostic implications of ALK and ROS1 rearrangements in never-smokers with surgically resected lung adenocarcinoma. Lung Cancer. 2014;83(3):389–395. doi:10.1016/j.lungcan.2014.01.003
14. Wang Z, Yang H, Luo S, et al. Anaplastic lymphoma kinase gene rearrangement predicts better prognosis in NSCLC patients: a meta-analysis. Lung Cancer. 2017;112:1–9. doi:10.1016/j.lungcan.2017.07.029
15. Blackhall FH, Peters S, Bubendorf L, et al. Prevalence and clinical outcomes for patients with ALK-positive resected stage I to III adenocarcinoma: results from the European Thoracic Oncology Platform Lungscape Project. J Clin Oncol. 2014;32(25):2780. doi:10.1200/JCO.2013.54.5921
16. Paik JH, Choi CM, Kim H, et al. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer. 2012;76(3):403–409. doi:10.1016/j.lungcan.2011.11.008
17. Fukui T, Yatabe Y, Kobayashi Y, et al. Clinicoradiologic characteristics of patients with lung adenocarcinoma harboring EML4-ALK fusion oncogene. Lung Cancer. 2012;77(2):319–325. doi:10.1016/j.lungcan.2012.03.013
18. Travis WD, Brambilla E, Van Schil P, et al. Paradigm shifts in lung cancer as defined in the new IASLC/ATS/ERS lung adenocarcinoma classification. Eur Respir J. 2011;38(2):239–243. doi:10.1183/09031936.00026711
19. Kometani T, Sugio K, Osoegawa A, Seto T, Ichinose Y. Clinicopathological features of younger (aged ≤ 50 years) lung adenocarcinoma patients harboring the EML4-ALK fusion gene. Thorac Cancer. 2018;9(5):563–570. doi:10.1111/1759-7714.12616
20. Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3(1):13–17. doi:10.1097/JTO.0b013e31815e8b60
21. Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17(3):889–897. doi:10.1245/s10434-009-0808-7
22. Yang Y, Yang Y, Zhou X, et al. EGFR L858R mutation is associated with lung adenocarcinoma patients with dominant ground-glass opacity. Lung Cancer. 2015;87(3):272–277. doi:10.1016/j.lungcan.2014.12.016
23. Liu Y, Kim J, Qu F, et al. CT features associated with epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. Radiology. 2016;280(1):271–280. doi:10.1148/radiol.2016151455
24. Hsu JS, Huang MS, Chen CY, et al. Correlation between EGFR mutation status and computed tomography features in patients with advanced pulmonary adenocarcinoma. J Thorac Imaging. 2014;29(6):357–363. doi:10.1097/RTI.0000000000000116
25. Sugano M, Shimizu K, Nakano T, et al. Correlation between computed tomography findings and epidermal growth factor receptor and KRAS gene mutations in patients with pulmonary adenocarcinoma. Oncol Rep. 2011;26(5):1205–1211. doi:10.3892/or.2011.1412
26. Ebright MI, Zakowski MF, Martin J, et al. Clinical pattern and pathologic stage but not histologic features predict outcome for bronchioloalveolar carcinoma. Ann Thorac Surg. 2002;74(5):
27. Park JH, Lee KS, Kim JH, et al. Malignant pure pulmonary ground-glass opacity nodules: prognostic implications. Korean J Radiol. 2009;10(1):12–20. doi:10.3348/kjr.2009.10.1.12
28. Suh YJ, Lee H, Kim YJ, et al. Computed tomography characteristics of lung adenocarcinomas with epidermal growth factor receptor mutation: a propensity score matching study. Lung Cancer. 2018;123:52–59. doi:10.1016/j.lungcan.2018.06.030
29. Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15(16):5216–5223. doi:10.1158/1078-0432.CCR-09-0802
30. Liu Y, Shi Y, Hao X, et al. Analysis of clinicopathological features of the echinoderm microtubule-associated protein-like-4-anaplastic lymphoma kinase fusion gene in Chinese patients with advanced non-small-cell lung cancer. Thorac Cancer. 2014;5(3):255–260. doi:10.1111/1759-7714.12101
31. Zhao F, Xu M, Lei H, et al. Clinicopathological characteristics of patients with non-small-cell lung cancer who harbor EML4-ALK fusion gene: a meta-analysis. PLoS One. 2015;10:e01173332.
32. Shin SH, Lee H, Jeong B, et al. Anaplastic lymphoma kinase rearrangement in surgically resected stage IA lung adenocarcinoma. J Thorac Dis. 2018;10(6):3460–3467. doi:10.21037/jtd
33. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi:10.1038/nature13385
34. Ren S, Kuang P, Zheng L, et al. Analysis of driver mutations in female non-smoker Asian patients with pulmonary adenocarcinoma. Cell Biochem Biophys. 2012;64(2):155–160. doi:10.1007/s12013-012-9384-8
35. D’Angelo SP, Janjigian YY, Ahye N, et al. Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol. 2012;7(12):1815–1822. doi:10.1097/JTO.0b013e31826bb7b2
36. Chaft JE, Dagogo-Jack I, Santini FC, et al. Clinical outcomes of patients with resected, early-stage ALK-positive lung cancer. Lung Cancer. 2018;122:67–71. doi:10.1016/j.lungcan.2018.05.020
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

