Back to Journals » Cancer Management and Research » Volume 11
Clinicopathological and prognostic significance of CDH1 hypermethylation in hepatocellular carcinoma: a meta-analysis
Authors Wu X, Yao X, Cao Q, Wu Z, Wang Z, Liu F, Shen L
Received 10 July 2018
Accepted for publication 31 August 2018
Published 17 January 2019 Volume 2019:11 Pages 857—864
DOI https://doi.org/10.2147/CMAR.S179710
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Xiaoyu Wu,1,* Xuequan Yao,1,* Qinhong Cao,1 Zhenfeng Wu,1 Zhaojing Wang,1 Fukun Liu,1 Lizong Shen2
1Department of Surgical Oncology, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, China; 2Department of General Surgery, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing 210029, China
*These authors contributed equally to this work
Background: The patients with hepatocellular carcinoma (HCC) have poor prognosis due to being diagnosed at late stage or recurrence following surgery. It’s critical to identify effective biomarkers that can improve overall diagnosis and treatment of HCC.
Methods: We performed a meta-analysis of all relative studies reporting the clinicopathological significance of CDH1 hypermethylation in HCC by using Review Manager 5.2. A comprehensive literature search was performed in EMBASE, PubMed, Web of Science and Google Scholar databases. Kaplan Meier Plotter online database was used for the determination of correlation between CDH1 mRNA expression and overall survival in patients with HCC. Odds Ratios (OR) with 95% corresponding confidence intervals (CIs) were calculated. A total of 12 relevant studies were included in the meta-analysis with 981 patients.
Results: The positive rate of CDH1 hypermethylation was significantly higher in HCC than in normal liver tissue; and the pooled OR was 4.34 with 95% CI 2.50–7.56, P<0.00001. CDH1 promoter in HCC was more frequently hypermethylated compared to the group of chronic liver disease (CLD); OR was 4.83 with 95% CI 2.67–8.72, P<0.00001. However, the rate of CDH1 promoter hypermethylation was not correlated with different grades as well as stages. High CDH1 mRNA expression was significantly correlated to better overall survival in all 231 HCC patients compared to 133 HCC patients with low level CDH1 mRNA expression; HR was 0.6 with 95% CI 0.42–0.85, P=0.0034.
Conclusion: In summary, CDH1 promoter hypermethylation is a risk factor and promising biomarker for HCC carcinogenesis and diagnosis, as well as a predictor of poor prognosis.
Keywords: hepatocellular carcinoma, CDH1, methylation, prognosis, diagnosis, E-cadherin
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the fourth leading cause of cancer mortality globally.1 The patients with HCC have a poor prognosis due to being diagnosed at late stage or recurrence following surgery.2 The only curative therapies are liver transplantation and resection. In order to qualify these two treatments, patients with HCC need to be diagnosed at early stage.3 On the other hand, there is no good prognostic marker to predict survival outcome apart from tumor staging classification system. Therefore, it is very critical to find effective biomarkers for early diagnosis and biomarkers to predict prognosis.
Cadherins were defined as cell surface glycoproteins responsible for the Ca2+-dependent cell–cell adhesion mechanism,4,5 including E-cadherin, neural (N-) cadherin, and the placental (P-) cadherin.5 E-cadherin functions as a mediator of cell–cell adhesion and the principal organizer of epithelial phenotype.6,7 Its adhesive function at cell surface holds cells together, facilitates other cell–cell interactions, and physically blocks the movement of cells. Additionally, E-cadherin homophilic binding leads to contact-mediated inhibition of growth via modulation of growth inhibitory signals including the Hippo, growth factor receptor tyrosine kinase (RTK), and Src family kinase signaling pathways. On the other hand, the cytoplasmic tail of E-cadherin is associated with intracellular molecules of alpha-, beta-, and gama-catenins8,9 that link to the cytoskeleton and mediate downstream growth factor signaling pathways such as Wnt, transforming growth factor β, and nuclear factor kappa-light-chain-enhancer of activated B cells.10–13 Loss of E-cadherin expression leads to induction of the epithelial to mesenchymal transition (EMT), causing cells to lose cell–cell contacts and acquire increased motility to spread into surrounding or distant tissues, also upregulates growth factor pathways and promoting proliferation. Therefore, loss of E-cadherin plays a crucial role in hepatocellular carcinogenesis and metastasis.14 Cadherin-1 (CDH1) is the gene for E-cadherin, and its downregulation with hypermethylation of CpG islands of region promoter is found in various tumors originating from epithelial cells including HCC.15,16 Aberrant DNA methylation has been demonstrated in common HCC,17,18 and recent studies showed that CpG methylation around the promoter is very important in the transcriptional inactivation of CDH1 in HCC.15 However, the positive rate of CDH1 promoter hypermethylation in HCC is inconsistent due to small size of samples in individual study.
In the present study, we conducted a meta-analysis study using the qualified studies and determined the association between CDH1 hypermethylation in HCC and its progression as well as prognosis.
Methods
Selection criteria and study search
We searched the three electronic databases EMBASE, PubMed, and Web of Science from the earliest date up to June 2018 for appropriated articles addressing the focused question. Electronic database searches were conducted by using the following terms: “liver,” “hepatocellular” and “cancer or tumor or neoplasm or carcinoma,” “methylation,” and “CDH1 or E-cadherin.” Additional studies were searched manually from the reference list of included articles. There were 37 articles from EMBASE, 73 articles found from PubMed, as well as 36 articles from Web of Science.
A total of 146 articles were reviewed and screened by article titles and abstracts. Studies were eligible for inclusion in the current analysis if they met the following criteria: 1) studies that evaluated the association between CDH1 hypermethylation and clinicopathological parameters of HCC; 2) CDH1 hypermethylation detected in the primary HCC tissues; and 3) studies published in English or Chinese. The exclusion criteria were as follows: 1) expert opinion, case reports, reviews, conference abstracts, editorials, letters; 2) all publications regarding cell lines, in vitro/ex vivo studies, and human xenografts; 3) studies in which same population was involved; and 4) studies in which CDH1 protein expression was evaluated.
After exclusion of non-relevant and/or redundant publications from different databases, the 17 remaining papers were selected and reviewed in the full-text version for inclusion and exclusion criteria. Five articles were excluded due to lack of sufficient data for the present study.
Data extraction and study assessment
Two independent authors (XW and XY) extracted the data from the included studies. Disagreement regarding selection was resolved through discussion. If they could not reach an agreement, a third reviewer (LS) was consulted. The following information was collected from each study: year of publication, the first author name, sample source, number of cases, cancer TNM (tumor-node-metastasis) stage, methylation detection method, and CDH1 methylation status. Data for study characteristics and clinical responses were collected and organized into a standard table format. Heterogeneity of the included studies was assessed to determine whether or not the data of the various studies could be analyzed for a meta-analysis.
The assessment of methodological quality of included studies was done by two assessors based on a grading system developed by the Newcastle Ottawa Quality Assessment Scale (NOQAS).19 The three reviewers graded the quality and compared them, and then they reached an agreement for each item. Those scales use a grading system to evaluate on comparability, the selection of quality, exposure, and outcomes for study participants. The NOQAS scores ranged between 0 and 9, and a study with a score of 7 or more indicated a good quality.
Statistical analysis
The meta-analysis was performed using Review Manager 5.2 (Cochrane Collaboration, Update Software, Oxford, UK). ORs with its 95% CIs were calculated. All the events represent the number of HCC with CDH1 hypermethylation. The evaluation of statistical heterogeneity was finished by using the Cochran’s Q statistic and I2 tests. When the I2 value was below 50%, fixed-effect model was generated, and when the I2 value was 50% or greater, a random-effect model was generated. Sensitivity analysis was used for exploring the reasons of statistical heterogeneity. The pooled frequency of CDH1 hypermethylation and 95% CIs were estimated. The rate of CDH1 hypermethylation was compared between different groups by tumor features. The pooled OR was estimated for the correlation between CDH1 hypermethylation and clinicopathological features. Overall survival was analyzed by using an online database Kaplan premier plotter that was established by using gene expression and the survival information of 364 liver cancer patients (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq).20 P-value less than 0.05 was regarded statistically significant. Publication bias was evaluated by using funnel plots reported by Egger et al.21
Results
Identification of relevant studies and study quality
Twelve studies were included in the meta-analysis after screening 146 studies (Figure 1). The following items were recorded from each study: first author, published year, country, tumor histology, CDH1 methylation status, differentiation and stages of tumors, as well as methylation detection methods (Table 1).
  | Figure 1 Schematic flow diagram for selection of included studies. |
The correlation of CDH1 promoter hypermethylation with clinicopathological features
The frequency of CDH1 promoter hypermethylation was significantly higher in HCC than in normal liver tissue; the pooled OR was 4.34 with 95% CI 2.50–7.56, z=5.19, P<0.00001, I2=0% (Figure 2). CDH1 promoter in HCC was more frequently hypermethylated compared to the group of chronic liver disease (CLD); OR was 4.83 with 95% CI 2.67–8.72, z=5.22, P<0.00001, I2=0% (Figure 3). However, the rate of CDH1 promoter hypermethylation was not correlated with different grades; OR was 0.84 with 95% CI 0.40–1.78, z=0.46, P=0.65, I2=0% (Figure 4). The rate of CDH1 promoter hypermethylation was not significantly different between early and advanced stages; OR was 0.79 with 95% CI 0.42–1.49, z=0.71, P=0.48, I2=0% (Figure 5). High CDH1 mRNA expression was strongly associated with better overall survival in all 231 HCC patients compared to 133 HCC patients with low-level CDH1 mRNA expression; HR was 0.6 with 95% CI 0.42–0.85, P=0.0034 (Figure 6).
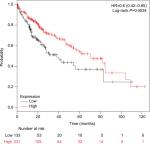  | Figure 6 Comparison of overall survival between patients with high and low expression of CDH1 mRNA. Abbreviation: HR, hazard ratio. |
Sensitivity analyses and publication bias
The quality of each study was evaluated using the NOQAS. This scale for non-randomized case-controlled studies and cohort studies was used to evaluate the quality of selection, comparability, exposure, and outcomes for study participants, with the maximum score being 9 points. Among the 12 studies, three of them scored 8 points, seven scored 7 points, and two scored 6 points (data not shown). The score for each study showed a relatively high quality. A sensitivity analysis was performed by removing one study at a time; the result remained stable, indicating the stability of the present analysis. The funnel plots were largely symmetric (Figure 7), indicating there were no publication biases in the meta-analysis of CDH1 methylation and clinicopathological features.
Discussion
E-cadherin, a Ca2+-dependent cell adhesion molecule, encoded by CDH1, regulates cell polarity and tissue morphology.8 Disturbance of cell polarity and destruction of normal tissue morphology initiate carcinogenesis. Knocking-down of CDH1 results in the activation of oncogenic signaling pathways and leads to the transition of epithelial to mesenchymal in the development of epithelial tumor.22–24 Cancer invasion is initiated by the dissociation of cells from primary cancer nests due to loosened cell adhesion. Recent studies reported that CDH1 promoter hypermethylation was observed in HCC. However, the frequency varied from 13.3% to 66.7%.25,26 We pooled nine studies including 580 patients, and evaluated the frequency of CDH1 promoter hypermethylation. The frequency of CDH1 promoter hypermethylation in HCC was 32% – 4.34 times higher than the one in normal liver tissue. Additionally, the rate of CDH1 promoter hypermethylation in HCC was 4.83 times higher than the one in CLD. Our findings suggest that CDH1 promoter hypermethylation could be a biomarker for HCC diagnosis. DNA methyltransferases (DNMTs) are pivotal regulators of the methylation of CpG islands. Previous evidence indicated that increased expression of DNMT1 was correlated with the stages, portal venous invasion, and prognosis of HCC.27,28 It has been reported that inhibitors of DNA methyltransferase inhibitors (DNMTis) such as 5-Aza-2’-deoxycytidine (5-Aza-CdR) and 5-fluoro-2-deoxycytidine (FdCyd) are applied to human breast and lung cancer cells, and currently FdCyd is in clinical trials for the treatment of breast cancer and other solid tumors.29–31 Therefore, inhibitors for DNA methylation could be a new therapy strategy for HCC patients with CDH1 promoter hypermethylation.
Overall survival was evaluated by using an online database Kaplan–Meier Plotter that was generated by using mRNA expression and the survival data of 364 liver cancer patients (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq). Our findings showed that low expression of E-cadherin mRNA was significantly correlated with poor overall survival in HCC patients. Gene methylation is a major mechanism to inactivate gene in HCC.18 Thus, CDH1 promoter hypermethylation could be a predictor of poor overall survival. Previous evidence revealed that EMT is an important process correlated with the propensity for invasion and metastases of many types of tumors including HCC.32–34 The process of EMT is initiated by suppression of the epithelial markers such as E-cadherin. Loss of E-cadherin function initiates the dissociation of cells from primary cancer nests because of loosened intercellular adhesion, thus resulting in invasion and metastasis. Therefore, CDH1 promoter hypermethylation in HCC leads to a poor overall survival in patients with HCC. However, our findings showed that CDH1 promoter hypermethylation was not significantly associated with HCC differentiation and stages. The evaluation needs to be performed when more relative studies are available in future.
Limitations
There were a few limitations of the present study. First, the selection bias was unavoidable due to the selection restricted to the articles published in English and Chinese. Second, the heterogeneity existed in the frequency of CDH1 promoter hypermethylation between HCC and normal liver tissue. This phenomenon may be caused by the moderate number of samples in the involved studies and inconsistent criteria in selection of controls. Further evaluation with a larger size of samples will be carried out in future for a more reliable conclusion.
Conclusion
In summary, CDH1 promoter hypermethylation is a promising biomarker for HCC diagnosis and a predictor of poor prognosis.
Acknowledgments
This work was supported by the National Science Foundation of China (81373990, 81402523, 81672990 to XYW), Jiangsu Province “Six talents” high peak plan (2015-WSN-052 to XYW), Project of Jiangsu Provincial Bureau of Traditional Chinese Medicine (JD201510 to XYW and XQY), and the six “1” Project of Jiangsu Province (LGY2016012 to XYW). The funding institutions did not have any roles in the study design, data collection, or analysis.
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. The corresponding author had full access to all data and the final responsibility for the decision to submit the article for publication.
Disclosure
The authors report no conflicts of interest in this work.
References
Alacacioglu A, Somali I, Simsek I, et al. Epidemiology and survival of hepatocellular carcinoma in Turkey: outcome of multicenter study. Jpn J Clin Oncol. 2008;38(10):683–688. | ||
Villanueva A, Minguez B, Forner A, Reig M, Llovet JM. Hepatocellular carcinoma: novel molecular approaches for diagnosis, prognosis, and therapy. Annu Rev Med. 2010;61:317–328. | ||
Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–458. | ||
Takeichi M. Cadherins: a molecular family essential for selective cell-cell adhesion and animal morphogenesis. Trends Genet. 1987;3(8):213–217. | ||
Takeichi M, Hatta K, Nose A, Nagafuchi A. Identification of a gene family of cadherin cell adhesion molecules. Cell Differ Dev. 1988;25(Suppl):91–94. | ||
Gumbiner B, Stevenson B, Grimaldi A. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J Cell Biol. 1988;107(4):1575–1587. | ||
Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6(8):622–634. | ||
Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251(5000):1451–1455. | ||
Mareel M, Boterberg T, Noë V, et al. E-cadherin/catenin/cytoskeleton complex: a regulator of cancer invasion. J Cell Physiol. 1997;173(2):271–274. | ||
Padua D, Massagué J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. | ||
Qian X, Karpova T, Sheppard AM, Mcnally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. Embo J. 2004;23(8):1739–1784. | ||
Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. Embo J. 1989;8(6):1711–1717. | ||
Mendonsa AM, Na TY, Gumbiner BM. E-cadherin in contact inhibition and cancer. Oncogene. 2018;37(35):4769–4780. | ||
Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. | ||
Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci U S A. 1995;92(16):7416–7419. | ||
Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55(22):5195–5199. | ||
Huang J. Current progress in epigenetic research for hepatocarcinomagenesis. Sci China C Life Sci. 2009;52(1):31–42. | ||
Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163(3):1101–1107. | ||
Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Ottawa, ON: Ottawa Hospital Research Institute; 2013. | ||
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8:e82241. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65(23):3756–3788. | ||
Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1(6):a003129. | ||
Rodriguez FJ, Lewis-Tuffin LJ, Anastasiadis PZ. E-cadherin’s dark side: possible role in tumor progression. Biochim Biophys Acta. 2012;1826(1):23–31. | ||
Kanai Y, Ushijima S, Hui AM, et al. The E-cadherin gene is silenced by CpG methylation in human hepatocellular carcinomas. Int J Cancer. 1997;71(3):355–359. | ||
Schagdarsurengin U, Wilkens L, Steinemann D, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22(12):1866–1871. | ||
Oh BK, Kim H, Park HJ, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20(1):65–73. | ||
Saito Y, Kanai Y, Nakagawa T, et al. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malignant potential and poor prognosis of human hepatocellular carcinomas. Int J Cancer. 2003;105(4):527–532. | ||
Beumer JH, Parise RA, Newman EM, et al. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2’-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU). Cancer Chemother Pharmacol. 2008;62(2):363–368. | ||
Gowher H, Jeltsch A. Mechanism of inhibition of DNA methyltransferases by cytidine analogs in cancer therapy. Cancer Biol Ther. 2004;3(11):1062–1068. | ||
Newman EM, Morgan RJ, Kummar S, et al. A phase I, pharmacokinetic, and pharmacodynamic evaluation of the DNA methyltransferase inhibitor 5-fluoro-2’-deoxycytidine, administered with tetrahydrouridine. Cancer Chemother Pharmacol. 2015;75(3):537–546. | ||
Giannelli G, Bergamini C, Fransvea E, Sgarra C, Antonaci S. Laminin-5 with transforming growth factor-beta1 induces epithelial to mesenchymal transition in hepatocellular carcinoma. Gastroenterology. 2005;129(5):1375–1383. | ||
Gao D, Vahdat LT, Wong S, Chang JC, Mittal V. Microenvironmental regulation of epithelial-mesenchymal transitions in cancer. Cancer Res. 2012;72(19):4883–4889. | ||
Lee TK, Poon RT, Yuen AP, et al. Twist overexpression correlates with hepatocellular carcinoma metastasis through induction of epithelial-mesenchymal transition. Clin Cancer Res. 2006;12(18):5369–5376. | ||
Zekri AR, Bahnasy AA, Shoeab FE, et al. Methylation of multiple genes in hepatitis C virus associated hepatocellular carcinoma. J Adv Res. 2014;5(1):27–40. | ||
Herath NI, Purdie DM, Kew MC, et al. Varying etiologies lead to different molecular changes in Australian and South African hepatocellular carcinomas. Int J Oncol. 2009;35(5):1081–1089. | ||
Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47(3):908–918. | ||
Katoh H, Shibata T, Kokubu A, et al. Epigenetic instability and chromosomal instability in hepatocellular carcinoma. Am J Pathol. 2006;168(4):1375–1384. | ||
Yuan Y, Wang J, Li J, et al. Frequent epigenetic inactivation of spleen tyrosine kinase gene in human hepatocellular carcinoma. Clin Cancer Res. 2006;12(22):6687–6695. | ||
Kwon GY, Yoo BC, Koh KC, Cho JW, Park WS, Park CK. Promoter methylation of E-cadherin in hepatocellular carcinomas and dysplastic nodules. J Korean Med Sci. 2005;20(2):242–247. | ||
Herath NI, Walsh MD, Kew MC, Young J, Leggett BA, Macdonald GA. Cadherin/catenin complex appears to be intact in hepatocellular carcinomas from Australia and South Africa. J Gastroenterol Hepatol. 2004;19(6):676–682. | ||
Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163(4):1371–1378. | ||
Matsumura T, Makino R, Mitamura K. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin Cancer Res. 2001;7(3):594–599. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






