Back to Journals » International Journal of General Medicine » Volume 16
Clinical Application and Evaluation of Metagenomic Next-Generation Sequencing for Lower Respiratory Tract Infections and Human Tumor Screening
Authors Huang J, Weng H, Ye L, Jiang M, Li Y, Li H
Received 30 August 2023
Accepted for publication 23 November 2023
Published 7 December 2023 Volume 2023:16 Pages 5763—5777
DOI https://doi.org/10.2147/IJGM.S437800
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Jinbao Huang,1,* Heng Weng,1,* Ling Ye,1 Meiqin Jiang,1 Yangyu Li,2 Hongyan Li3
1Department of Respiratory Medicine, People’s Hospital Affiliated to Fujian University of Traditional Chinese Medicine, Fuzhou, 350004, People’s Republic of China; 2Department of Clinical Laboratory Medicine, People’s Hospital Affiliated to Fujian University of Traditional Chinese Medicine, Fuzhou, 350004, People’s Republic of China; 3Department of Critical Care Medicine, People’s Hospital Affiliated to Fujian University of Traditional Chinese Medicine, Fuzhou, 350004, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hongyan Li, Department of Critical Care Medicine, People’s Hospital Affiliated to Fujian University of Traditional Chinese Medicine, No. 602 Middle 817 Road, Taijiang District, Fuzhou, 350004, People’s Republic of China, Email [email protected]
Background: To evaluate the clinical value of metagenomic next-generation sequencing (mNGS) in screening of lower respiratory tract infections (LRTIs) and human tumors.
Methods: Human samples included bronchoalveolar lavage fluid (BALF), sputum, lung biopsy tissue, and peripheral blood from 188 patients who were admitted to our hospital between January 2020 and September 2022 were analyzed using mNGS for simultaneous pathogen and chromosome copy number variation (CNV) detection. Traditional microbial culture and comprehensive microbial test (CMT) were also conducted. The diagnostic efficiencies of the three methods (mNGS, traditional culture, and CMT groups) were compared.
Results: Among the 188 patients, 149 (79.3%) were in the LRTIs group and 39 (20.7%) were in the non-LRTIs group. The diagnostic sensitivity and accuracy of the mNGS group were higher than those of the traditional culture and CMT groups (P < 0.001; P < 0.001; P < 0.001; P < 0.001), and the specificity was higher than that of the CMT group (P = 0.039) but lower than that of the traditional culture group (P = 0.006). The positive predictive values of the mNGS and traditional culture groups were higher than that of the CMT group (P = 0.004; P = 0.011). The negative predictive value of the mNGS group was higher than that of the CMT group (P = 0.003). In addition, all samples were subjected to simultaneous chromosome CNV detection, and 8% (15/188) were positive for CNV. Of the 15 patients, 10 were initially misdiagnosed as non-neoplastic diseases, with a misdiagnosis rate of 66.7% (10/15). The BALF CNV test was performed on 13 patients diagnosed with primary or metastatic lung cancer, with a positivity rate of 38.5%.
Conclusion: The sensitivity and accuracy of pathogen diagnosis using mNGS were better than those of traditional culture and CMT. CNV detection is an important auxiliary diagnostic tool for cancer, particularly for screening occult tumors.
Keywords: metagenomics, next-generation sequencing, respiratory infectious diseases, pathogen diagnosis, genomic instability
Graphical Abstract:
Introduction
Respiratory tract infections are common and occur frequently. Rapid and accurate microbial detection is essential for timely and appropriate treatment. Traditional microbial detection methods have some limitations such as dependence on morphology, long duration, low sensitivity, and high variability.1,2
Metagenomic next-generation sequencing (mNGS) is a new detection technology characterized by the unbiased high-throughput sequencing of nucleic acids extracted from biological samples, independent of in vitro microbial culture. The nucleic acid sequence of the suspected pathogen was obtained through bioinformatics comparison and analysis.1 Since mNGS can detect from a variety of clinical samples that cover almost all pathogens, and its value in the diagnosis and differential diagnosis of multiple systemic diseases has been proven.3–7 Today, clinical applications of mNGS are increasing.8 For pathogens that are not easy to culture, are infrequent, rare, or emerging, as well as in cases of severe and intractable disease where quick aetiology identification is necessary, mNGS has a higher detection advantage.1,2,9 Currently, previous studies have shown that mNGS is highly sensitive and can be effectively used to identify pathogens associated with lower respiratory infections (LRTIs) in clinical settings. Zhang et al10 indicated that bronchoalveolar lavage fluid (BALF) testing using mNGS showed a diagnostic sensitivity of 80%, with an agreement rate of 44.68% between mNGS and culture methods. mNGS may improve sensitivity to viruses and atypical pathogens. Liang et al11 tested 140 samples (BALF, blood, and pleural effusion) using mNGS and identified pathogenic microbes in 71.4% of the patients. The results also showed that mNGS could potentially improve outcomes for patients with LRTIs. Yan et al12 showed that the use of mNGS was associated with lower rates of antibiotic escalation and may facilitate the cessation of antivirals in patients with LRTIs. Zhao et al13 demonstrated that the pathogen-positive rate of mNGS in BALF samples was higher than that of culture in patients despite prior antibiotic use, and the results also showed that mNGS of BALF was a potentially useful tool for testing mixed pathogens in severe pneumonia.
In addition, because mNGS can sequence human and microbial DNAs simultaneously, previous studies have shown that a method based on deep sequencing can detect host chromosome instability, which manifests as chromosome copy number variation (CNV) (duplication or deletion) compared with that in healthy individuals and can predict the occurrence of human malignant tumors.14–16 Generally, chromosomal CNVs can be caused by genetic diseases or tumors, whereas large CNVs (>10 M) containing multiple chromosomes are highly unlikely to be caused by genetic diseases and often indicate the presence of tumor lesions.15 In recent years, CNV detection by mNGS has been applied clinically to assist in the diagnosis of human tumors, such as lung cancer,14,17 colorectal cancer,18 lymphoma,19 and meningeal carcinomatosis (MC).20,21 Su et al22 conducted a prospective, multi-center study on the simultaneous detection of pathogens and tumors in patients with suspected infections using mNGS, and the results showed that the positive percent agreement of mNGS testing compared with the clinical diagnosis was significantly higher than that of conventional microbiological testing (CMT) (53.0% vs 20.9%). Chromosomal CNV signals were detected in 17 patients, which indicated clinically confirmed malignant lesions. Another multi-center-retrospective study of chromosome instability in pulmonary cancer using mNGS found that 30 chromosome instability-positive BALF samples were confirmed as malignant tumors based on pathological findings, and the diagnostic sensitivity, specificity, and accuracy of CNV on mNGS for lung cancer were 61.22%, 99.65%, and 83.17%, respectively.23 Ren et al20 confirmed that the CNV testing of cerebrospinal fluid by mNGS exhibited a high positivity rate (80%, 8/10) for the diagnosis of MC, indicating that this method could serve as a diagnostic marker for MC.20,21
However, to date, only a few papers have been published regarding mNGS testing to search for potential pathogens and malignant tumors simultaneously in patients with lower respiratory tract diseases (LRTDs),14,15,17,22 and additional CNV detection for human tumor screening could add value to this new method. Herein, we reviewed the data of 188 patients with LRTDs who underwent mNGS screening, analyzed the clinical application of this detection technology in respiratory medicine, and evaluated the potential value of mNGS in assisting in the screening of human malignant tumors.
Methods
Patients
The data of enrolled patients with lower respiratory tract diseases who were admitted to the respiratory department, thoracic surgery department, and comprehensive ICU of our hospital between January 2020 and September 2022 were retrospectively analyzed.
Inclusion Criteria
The following patients were included in the study: (1) hospitalized adult patients, (2) patients with the initial diagnosis of LRTIs, (3) patients with complete set of clinical data, (4) patients whose samples analyzed were peripheral blood and lower respiratory tract samples, including sputum, BALF, and lung tissue; and (5) all patients or authorized clients who agreed to undergo mNGS testing and provided signed informed consent.
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the People’s Hospital affiliated with the Fujian University of Traditional Chinese Medicine. Since this was a retrospective study, informed consent was waived, and all data were analyzed anonymously.
Exclusion Criteria
The following were excluded: (1) patients with RNA infection (mNGS for RNA virus detection was performed only in a few patients without data analysis), (2) patients with unclear final diagnosis of pulmonary lesions, (3) patients with an uncertain pathogen (it is not clear whether the detected pathogen is pathogenic or colonising bacteria), (4) patients with incomplete medical data; and (5) cases where NGS specimens submitted for inspection had failed quality control.
Diagnostic Criteria for LRTIs
(1) The etiology was determined by mNGS, traditional detection methods, and histopathology. Comprehensive clinical analysis was consistent with manifestations of lung infection, including symptoms of infection (fever, cough, sputum, and shortness of breath), laboratory indicators, abnormal changes in imaging (lung shadow and space occupation), and evidence of effective anti-infection treatment. (2) The etiology was unclear, but the clinical manifestations were consistent with pulmonary infection.14,24–26
Diagnostic Criteria for Non-LRTIs
(1) There were manifestations of lower respiratory tract lesions, but etiology was unidentified in the laboratory; comprehensive clinical analysis was not consistent with manifestations of lung infection, such as non-infectious lesions including lung tumors and interstitial pneumonia confirmed by histopathology. (2) The etiology was detected but attributed to non-pathogenic bacteria; clinical comprehensive analysis did not conform to manifestations of lung infection, such as non-respiratory infectious lesions confirmed by histopathology or improvement with treatment other than for infection.26
Routine Screening for Infectious Diseases
CMT for direct detection of pathogenic microorganisms included general bacterial and fungal smears, general bacterial and fungal cultures, acid-fast bacilli smears, polymerase chain reaction (PCR), gene chip technology,27 and GeneXpert. Serum (1,3)-β-D-glucan antigen detection (G test), serum galactomannan antigen detection (GM test), serum Cryptococcus capsular polysaccharide antigen detection, and serum pathogen IgG and IgM antibody detection indirectly detected pathogenic microorganisms.
mNGS Testing
All human samples (BALF, sputum, lung biopsy tissue, and peripheral blood) were sent to a genetic testing company (Hangzhou Jieyi Biology, Hangzhou, China) for mNGS DNA testing to identify pathogenic microorganisms and chromosome CNV. The pathogen detection scope included the whole genome sequences of 11,093 bacteria (including 66 species of mycoplasma/chlamydia), 1324 fungi, 21 Rickettsia spp., 48 spirochetes, 229 parasites, 6575 DNA viruses. Simultaneously, a depth-based approach16 was used to analyze the presence of host DNA CNV (duplicates and deletions) compared with that in healthy individuals.
Specimen Collection, Storage, and Transportation for mNGS Testing
Adequate amounts of specimens were collected according to the test requirements, including peripheral blood (≥4 mL), lung biopsy tissue (≥3×3×3 mm3), BALF (≥5 mL), and sputum (≥2 mL). Blood specimens were stored at room temperature (18–25 °C) for no more than 24 h (without freezing) and transported at room temperature; other specimens were stored at 2–8 °C for no more than 24 hours, and stored at −20±5 °C for no more than 96 hours (without repeated freeze-thaw), and were transported in cold storage.
Nucleic Acid Extraction, Library Construction, and Sequencing
Peripheral blood was centrifuged at 1600 × g for 10 min and 1.2 mL of the supernatant was centrifuged at 14,000 × g for 5 min to obtain plasma. For sputum, BALF, and lung biopsy tissue, 1.2 mL of the samples was first homogenized in a shock tube for cell-wall breaking, then centrifuged at 12,000 × g for 3 min to remove cell debris. Subsequently, the experimental procedures of DNA sequencing were performed. Using the mNGS automatic detection equipment NGS master™ (Cat. MAR002, MatriDx Biotech Corp. Hangzhou, China), which was independently developed by Jieyi Biology, all samples were subjected to DNA extraction and library construction, which were completed in 2 h using the PCR-free library construction scheme. The steps of specimen cracking, nucleic acid adsorption, nucleic acid purification and fragmentation, end repair, terminal adenylation, adaptor ligation, and library purification were integrated with one-stop automation, effective contamination prevention, and control. The following laboratory reagents were used: Nucleic Acid Extraction Kit (Cat. MD013, MatriDx Biotech Corp. Hangzhou, China), Cell-free DNA Library Preparation Kit (blood samples) (Cat. MD007, MatriDx Biotech Corp. Hangzhou, China), and Total DNA Library Preparation Kit (other samples) (Cat. MD001T, MatriDx Biotech Corp. Hangzhou, China).22 The concentration of the libraries was quantified by performing real-time PCR (KAPA) and was pooled.28 Then, the libraries were sequenced on the Illumina NextSeq 550 sequencer (San Diego, California, USA) using a 75-cycle sequencing kit. A total of 10–20 million reads were obtained for each sample for subsequent bioinformatics analysis.22 For each run, one negative control (artificial plasma mixed with fragmented human genomic DNA) and one positive control (a mixture of inactivated bacteria, fungi, and pseudoviral particles containing synthesised DNA or RNA fragments of adenovirus and influenza A virus, respectively) were used.28
Quality Control and Bioinformatics Analysis
The process of experimental quality control applies to sample receiving, experimental processes, experimental parameters, biological information processes, laboratory reagents, and environmental and microbial contaminants. Negative control, positive control, and internal reference methods in the same batch were used to monitor and evaluate contamination in real-time. Various laboratory background microbial databases have been established to monitor microorganisms in the reagents and the environment to prevent and control contamination.
Self-built analysis software (mngslibs-3.0.6) was used for bioinformatics analysis. The analysis steps included base identification, data quality control (removal of joints and low-quality bases [Q-score cut off, 20]), removal of host nucleic acids by mapping to the human reference genome (GRCh38.p13) using Burrows–Wheeler alignment (BWA), removal of low-complexity reads, and comparison of the remaining sequencing data with the reference database (NCBI nt database and GenBank29) by BWA for identification at the species level.28
For samples with a population index of more than 80% of the same species, human sequence processing was performed at the end of the experiment to avoid missing detections caused by an excessive population and decreased mNGS sensitivity.
mNGS Reporting Criteria
Microbial reads identified from a library were reported if: (1) the sequencing data passed quality control filters (library concentration > 50 pM, Q20 > 85%, and Q30 > 80%); (2) negative control (NC) in the same sequencing run did not contain the species or the RPM (sample)/RPM (NC) ≥ 5, which was determined empirically as a cut off for discriminating true positives from background contaminations.30
Chromosome CNV Detection and Analysis
The NGS-based methods for clinical samples used to detect chromosomal CNV using experimental procedures, including DNA extraction, purification, database construction, and high-throughput sequencing, were consistent with those used for pathogen detection using mNGS.22 In this study, all samples were subjected to simultaneous pathogen and chromosome CNV detection using homo-read data, and the self-built analysis software (mngslibs-3.0.6) was used for bioinformatics and CNV analyses. The raw sequencing data for chromosome CNV analysis included the following steps:22 (1) The human genome database (hg19) was referenced, and only uniquely mapped sequencing reads were selected for subsequent analysis. The reference genome was segmented into continuous windows of fixed length to calculate the read depth of each window, which was then normalized to the total reads of each sample. The copy number ratio of each window was obtained by dividing the normalized read depth by the average read depth of the reference dataset. (2) The fused lasso method (a generalization of the lasso penalty for sequential signal smoothing with sparsity) was applied to the log2-transformed copy number ratios. Smoothed adjacent windows with similar ratios were merged into segments with chromosomal positions and average annotated ratios. The copy number of each segment was calculated according to the average ratio and normal copy number of the corresponding chromosomes and then compared with predefined thresholds to validate CNVs. CNVs > 10 M were screened using a read count algorithm. Chromosomal abnormalities may be caused by tumors or genetic factors,15 and the conclusion should be confirmed using imaging, cytological, and pathological techniques.
Interpretation of Pathogenicity
The interpretation of the mNGS and traditional testing results was independently completed by three senior physicians, and a consensus was reached through negotiation when there was disagreement.
Criteria for Pathogenicity from mNGS Results
Pathogens detected by mNGS can be identified as pathogenic bacteria only when combined with clinical data to exclude symbiotic, colonizing microorganisms, or contaminating bacteria from the open parts of the human skin, mouth, respiratory tract, and intestine. Specifically, the results were divided into four types (definite, probable, possible, and unlikely) described previously by Guo et al14 and Blauwkamp et al31 as follows: (1) definite, mNGS-based pathogen was consistent with results from microbiological tests (traditional culture, nucleic acid-based detection, and pathological examination) performed within seven days of specimen collection; (2) probable, mNGS-based result was likely the cause of LRTI based on clinical, radiologic, or laboratory findings; (3) possible, mNGS-based pathogen possessed the pathogenic potential and was consonant with clinical manifestation but an alternate explanation was more likely; and (4) unlikely, mNGS-based pathogen possessed pathogenic potential but was not concordant with clinical manifestation. The definite and probable results were judged positive for clinical diagnosis, whereas the possible and unlikely results were judged negative for clinical diagnosis.
Criteria for Pathogenicity from Culture Results
Pathogens cultured using the traditional method can be identified as pathogenic bacteria only when combined with comprehensive clinical analysis to exclude contaminated bacteria and normal colonizing bacteria.
Statistical Methods
SPSS 19.0 statistical software was used for data analysis. Measurement data was expressed as median (interquartile range) and inter-group comparison was analyzed by non-parametric rank sum test. Count data was expressed as component ratio (%) and comparisons between the groups were performed using the Chi-square or Fisher’s exact probability method. The diagnostic efficiency of the different detection methods was expressed in terms of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). A P-value of <0.05 was considered statistically significant.
Results
Characteristics of Patients with LRTIs and Non-LRTIs
Between January 2020 and September 2022, 225 patients underwent mNGS testing, of which 188 met the inclusion criteria, including 149 LRTIs, 39 non-LRTIs (Figure 1). The incidence of underlying diseases in LRTIs, utilization rate of antibiotics before mNGS testing, and levels of C-reactive protein (CRP) and procalcitonin (PCT) were higher than those in non-LRTIs (P < 0.05) (Table 1).
 |
Table 1 Comparison of General Data Between LRTIs and Non-LRTIs |
Sample Types for mNGS Testing
Among the 188 samples, the proportion of BALF was the highest (83.5%, 157/188), followed by blood (8.5%, 16/188), lung biopsy tissue (6.4%, 120/188), and sputum (1.6%, 3/188).
Results of mNGS Testing
Among 149 LRTIs, pathogens were identified using mNGS in 98 cases, as shown in Tables S1 and S2. The average number of reads generated for all the samples, average host reads generated and average microbial reads generated were 17,758,047.5 (12,488,898.25, 24,851,393), 17,356,649.5 (12,103,507.75, 24,714,317.75), 192,243.5 (117,538.25, 434,154). The infection rate of single pathogens was higher than that of mixed infections (81.6% vs 18.4%), and the infection rates of two, three, and multiple pathogens (more than three pathogens, ie, microbiota) were 55.6%, 27.7%, and 16.7%, respectively. Co-infectious agents include bacteria, fungi, and viruses such as Epstein-Barr virus (EBV) and Cytomegalovirus (CMV). The infection rate with a single species of bacteria was higher than that with a single species of fungi (70.0% vs 30.0%, respectively). The former was dominated by general bacteria (32.1%), Mycobacterium tuberculosis (MTB) (30.4%), and non-tuberculous mycobacteria (NTM) (14.3%), whereas the latter was dominated by Pneumocystis jirovecii (PJ) (54.2%) and Cryptococcus neoformans (C. neoformans) (20.8%).
Comparison of Different Etiological Detection Methods
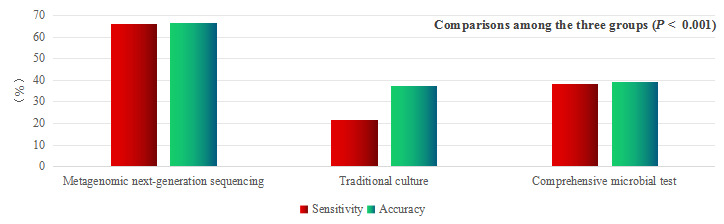
Among the 149 LRTIs, mNGS was performed in all cases (mNGS group), and pathogens were identified in 98 cases (65.8%). In total, 142 patients underwent traditional pathogen culture (traditional culture group), in whom pathogens were identified in 31 (21.8%) cases. A total of 147 patients underwent CMT (CMT group), and pathogens were identified in 56 (38.1%) cases (Table 2). Among the 149 cases of mNGS combined with CMT, pathogens were identified in 103 (69.1%) cases, and 46 patients with LRTIs were diagnosed in the absence of etiological findings.
 |
Table 2 Comparison of the Pathogen Detection Techniques |
Pairwise comparisons among the three groups: (1) sensitivity: mNGS group > CMT group > traditional culture group, with statistical significance (P < 0.001); (2) specificity: traditional culture group > mNGS group > CMT group, with statistical significance (P < 0.001); and (3) accuracy: mNGS group had higher accuracy than CMT and traditional culture groups (P < 0.001; P < 0.001). (4) PPV: both the mNGS and traditional culture groups had a higher PPV than the CMT group (P = 0.004; P = 0.011). (5) NPV: mNGS group had higher NPV than the traditional culture and CMT groups, but only the difference between the mNGS and CMT groups was statistically significant (P = 0.003).
In terms of the detection rate of different types of pathogens, including single bacteria, MTB/NTM, MTB, atypical pathogens (Chlamydia and Legionella), Chlamydia psittaci (CP), single fungi, Pneumocystis jirovecii (PJ), and mixed infections, the mNGS group showed higher detection rates than the CMT group, and the differences between the two were statistically significant (P < 0.05) (Table 3).
 |
Table 3 Comparison of Detection Rates of Different Pathogen Types Between mNGS and CMT |
Characteristics of 15 Patients with CNV Detected by mNGS
Among the 188 patients with CNV detected by mNGS, 15 were positive for CNV and histopathologically diagnosed with malignant tumors (six patients had a history of advanced tumor or tumor surgery prior to admission), including seven cases of primary lung cancer, one case of primary liver cancer, and seven cases of metastatic lung cancer, including prostate cancer, breast cancer, liver cancer, stomach cancer, urothelial cancer, and adenoid cystic cancer of the jaw. Among the 15 cases, 11 were males and 4 were females, with ages from 29 to 86 years, averaging 66 (62, 75) years. Positive samples included blood (6 cases), lung biopsy tissue (4 cases), and BALF (5 cases). Ten patients were initially diagnosed with liver abscess complicated with pneumonia (1 case), pulmonary abscess (2 cases), pneumonia (6 cases), and pneumonia secondary to pulmonary metastatic cancer (1 case), and were later confirmed to have primary liver cancer complicated with pneumonia (1 case), primary lung cancer (6 cases), and pulmonary metastatic cancer (3 cases), with a misdiagnosis rate of 66.7% (10/15) (Table 4). Serum tumor markers were detected in 12 cases, and some markers were elevated in 10 cases (Table S3).
 |
Table 4 Clinical Data of 15 Cases of Malignant Tumor with CNVs and Pathogens Detected by mNGS |
Among the 188 patients, 13 were diagnosed with primary lung cancer (9 cases) and metastatic lung cancer (4 cases) on pathology; BALF CNV test was performed, and 5 cases (38.5%) were positive. The blood CNV tests of six patients with advanced tumors were positive, the CNV tests of the lung biopsy tissue of four patients with primary lung cancer or lung metastatic cancer were positive; no false negatives were found.
Microorganisms of concern were detected in 6 of the 15 samples (cases 1, 6, 11, 13, 14, and 15) (Table 4), of which two were peripheral blood and four were BALF. According to the diagnostic criteria for LRTIs, Pneumocystis jirovecii pneumonia secondary to lung cancer was considered in case 13, and the other test results indicated non-pathogenic bacteria. These patients with primary or metastatic lung cancer were cases of non-LRTIs.
Typical Case (Case 11)
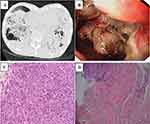
A 75-year old male complaining of cough, phlegm, and shortness of breath for one month was admitted to our hospital. Blood examination showed a white blood cell count of 14.3×109/L, neutrophil count of 13.1×109/L, and CRP level of 191.25 mg/L. Serum G, GM, and Cryptococcus capsular polysaccharide antigen test results were negative. The tumor marker of serum carcinoembryonic antigen (CEA) was 11.94 μg/L. Chest computed tomography (CT) (Figure 2A) revealed multiple pulmonary bullae, multiple patches, and large flake consolidations in both lungs, with predominance in the lower lobes. Tracheoscopic examination revealed no significant abnormalities. No pathogens or tumor cells were detected in the BALF. A lung biopsy was not performed because of pulmonary bullae. Initial diagnosis was a pulmonary bulla complicated by pneumonia. The cause of elevated CEA levels was undetermined. Gastroscopy was recommended; however, the patient refused. After 3 weeks of empirical anti-infective treatment, the patient was discharged with improvement in cough and sputum production. Three months later, a repeat chest CT revealed partial absorption of both lung lesions. However, the shortness of breath worsened. Considering the possibility of organizing pneumonia, methylprednisolone was administered; however, the patient’s condition failed to improve. Owing to the elevated serum CEA level, an abdominal enhanced CT examination was performed, and the results showed irregular thickening of the gastric wall from the cardia area to the bottom of the stomach, suggesting a suspected malignant tumor. Electronic gastroscopy revealed an irregular mass in the fundus involving the upper body of the stomach (Figure 2B). Stomach biopsy revealed a poorly differentiated adenocarcinoma. (Figure 2C). Because the chest imaging could not be explained by typical gastric cancer metastasis, tracheoscopy was performed again. Pathogens or tumor cells were not detected in BALF. mNGS was also performed, and five pathogens were detected (Table 4), which were considered as non-pathogenic bacteria based on a comprehensive clinical analysis. However, chromosomal CNV was detected (Figure 3), indicating a highly suspected tumor. A biopsy of the left lower lobe of the lung pathologically confirmed an invasive adenocarcinoma (intestinal type) originating from the stomach (Figure 2D). The diagnosis was confirmed as gastric adenocarcinoma with bilateral lung metastases, and the metastases showed pneumonia-like changes on imaging.
 |
Figure 3 Chromosome detection revealing genome instability due to CNV, indicating a highly suspected tumor. |
Discussion
This retrospective study showed that compared with non-LRTIs, LRTIs were more common in patients with underlying disease and had higher levels of conventional indicators of infection (CRP and PCT) and initial antibiotic use, but there was no difference in white blood cell and neutrophil counts. Many previous similar studies have also found no significant difference in inflammatory indicators between infected and non-infected groups.14,26,32 In clinical practice, infections and non-infectious diseases are difficult to distinguish,26 and the traditional pathogen detection cycle is generally long or the detection rate is not ideal. This poses great challenges for precise clinical diagnosis and treatment. To distinguish LRTIs from non-LRTIs, identifying the pathogen of LRTIs at the earliest, and starting accurate anti-infection treatment quickly is critical. In this context, mNGS as a new detection technology, has been applied in LRTIs and other infectious diseases in recent years, showing good prospects.
In this study, pathogens were detected in respiratory tract specimens (BALF, lung tissue, and sputum) and blood samples of patients with lower respiratory tract lesions, and the diagnostic efficiencies of mNGS, traditional culture methods, and CMT were comprehensively compared. mNGS had the highest diagnostic sensitivity (65.8%) and accuracy (66.5%), and better specificity (69.2%) which was significantly better than that of CMT (43.6%). Although the traditional culture method had the highest specificity (94.7%) and lowest sensitivity (21.8%), the combination of mNGS and CMT further improved the detection rate (69.1% vs 65.8%). Second, the PPV and NPV of mNGS were significantly higher than those of CMT (89.1% vs 71.8% and 34.6% vs 15.7%, respectively). Xie et al33 showed that the sensitivity, specificity, PPV, and NPV of mNGS were 67.53%, 68.75%, 91.23%, and 30.56%, respectively, when testing various human specimens (mainly blood and BALF) from patients with systemic infectious diseases. The sensitivity was significantly higher than that of traditional culture (67.53% vs 27.23%), similar to the results of our study. Miao et al34 also found that the sensitivity of mNGS was much higher than that of the culture method (50.7% vs 35.2%), supporting the conclusions of this study. Huang et al32 tested 240 lower respiratory tract specimens (lung tissue, BALF, and brush specimens of protective hair), and the results showed that the diagnostic sensitivity, specificity, PPV, and NPV of mNGS for lower respiratory tract diseases were 88.3%, 81.16%, 92.07%, and 73.68%, respectively. All the indices were higher than those in our study, which may be due to the inclusion of blood samples (8.5%) in our study, which affected the detection accuracy, suggesting that mNGS detection in lower respiratory tract specimens can maximize the detection accuracy of pathogens in lower respiratory tract diseases. This study also showed that the diagnostic accuracy and sensitivity of mNGS were significantly higher than those of traditional pathogen detection (86.25% vs 43.75% and 88.30% vs 25.73%, respectively),32 which is consistent with the results of our study.
Among the patients with LRTIs, 65.8% had pathogens identified by mNGS, mainly single pathogens and single bacteria, mainly MTB/NTM (44.7%), common bacteria (32.1%), and atypical pathogens (17.9%, of which CP accounted for 80%). The main fungi were PJ (54.2%) and C. neoformans (20.8%), and special pathogens (Mycobacterium, CP, PJ, C. neoformans) accounted for a high proportion, which was considered to be related to patient selection bias. When some patients are suspected of having MTB/NTM, PJ, Cryptococcus, or PS infection based on clinical manifestations and chest imaging, or when LRTIs are considered but empirical anti-infection treatment is ineffective, and when laboratory tests often have defects such as low positive rate, insufficient laboratory detection capacity, or long reporting cycle, we prefer to conduct mNGS detection simultaneously when conducting CMT. In fact, since most medical units are non-tuberculosis-specialized hospitals, routine tuberculosis testing items are mainly ordinary acid-fast bacillus smears, which not only have a low positive rate but also cannot distinguish between MTB and NTM. The routine detection methods for PJ include sputum and BALF smears, and pathological examination of lung biopsy is performed in some patients. However, the overall diagnostic rate is not ideal because of the high technical and diagnostic requirements of the inspectors. Moreover, biopsy is an invasive procedure that is not routinely performed. The culture rate of Cryptococcus in respiratory tract specimens is low and mainly relies on the pathological examination of lung biopsy and detection of serum Cryptococcus capsular antigen. Owing to the diverse morphology of pulmonary cryptococcosis lesions, they are easily confused with common diseases, especially early lesions, which are often misdiagnosed or missed altogether. Common laboratories for PS, Legionella and other atypical pathogens mainly rely on antibody detection, which has defects, such as a low detection rate and an inability to identify pathogen strains. However, the non-deviation detection function of mNGS testing can solve many of these problems.
The data in this study shows that mNGS showed good diagnostic sensitivity in the detection of various types of pathogen infections and had the advantage of reporting immediacy (13 h), which greatly improved the diagnostic efficiency. Compared with CMT, mNGS is superior in detecting single bacteria, single fungi, and atypical pathogens, as well as in identifying special pathogen infections (such as MTB/NTM, PJ, and PS), which is consistent with previous studies.34,35 Especially in the detection rate of PJ, mNGS has obvious advantages36 and can identify Cryptococcus more effectively than traditional culture methods. In addition, mNGS had a higher detection rate for mixed infections than CMT (90.0% vs 25.0%). These results are similar to those of previous studies.36,37 This study showed that mixed infections mainly occurred in severely immunosuppressed populations; such patients often received multiple antibiotics and the positive rate of traditional in vitro culture was low. However, antibiotic use had a small impact on mNGS33,34,38 which greatly improved the positive rate of pathogen detection. This would therefore be helpful to adjust treatment plans in a timely manner. Some of the pathogens in mixed infections are microbiota,2 all being mixed infections with multiple anerobic bacteria and are found in aspiration pneumonia and lung abscesses. Owing to the low detection rate of anerobic bacteria by traditional methods, the above pathogens were detected by mNGS, which demonstrates the unique advantages of mNGS for detection of anerobic bacteria.34,35
In this study, in addition to the application of mNGS technology for detection of pathogens, chromosomal CNV detection was simultaneously performed on human specimens to assist in tumor screening. Blood, lung tissue, and BALF samples from 15 (8.0%) of the 188 patients tested positive for CNV. All of the above cases were confirmed by histopathology as malignant tumors, including primary lung cancer, primary liver cancer, and various lung metastases, with a misdiagnosis rate of 66.7% at the first diagnosis. It is often confused with infectious diseases (bacterial pneumonia, lung abscess, pulmonary fungal infection, and liver abscess). The positive rate of BALF CNVs in primary and metastatic lung cancer was 38.5%, in the lung tissue of primary lung cancer it was 100%, and in the blood of middle and advanced malignant tumors it was 100%. This study suggests that mNGS technology can detect chromosomal CNVs in a variety of human specimens and plays a role in early screening and auxiliary tumor diagnosis, which is similar to previous studies.14,15,17,19,39 Guo et al showed that with pathological results as the gold standard, the sensitivity, specificity, and accuracy of mNGS detection of CNV in lung biopsy tissue for lung cancer diagnosis reached 83.7%, 97.6%, and 92.9%, respectively.14 Leary et al also showed that whole-genome sequencing to detect total circulating free DNA in the plasma can identify chromosomal alterations in all patients with advanced cancer and is an effective noninvasive method for detecting human tumors.39 Wei et al17 analyzed BALF in patients with clinically suspected pulmonary abscesses, and no pathogenic bacteria were found; however, CNV was detected. By adding lung biopsy and pathological examination, it was confirmed that the patient had cavitary lung squamous cell carcinoma, which was similar to the typical case 11 in our study.
It is worth noting that the chest imaging of the five patients with positive BALF CNVs in this study showed pneumonia-like lesions, manifested as pneumonia-type primary lung adenocarcinoma and pneumonia-type gastric adenocarcinoma metastasis, all of which were misdiagnosed as pulmonary infections. Two cases were confirmed as lung cancer by synchronous lung biopsy, but no tumor was detected by BALF cytology in the other three cases. Lung tissue biopsy was performed again after the CNVs were found to be positive, and all cases were confirmed as malignant lung tumors. In this study, the positive rate of BALF CNVs was low, which may be related to the influence of many factors, such as lesion location, lesion size, selection of lavage site, lavage volume, and recovery volume. However, the detection accuracy of BALF may be higher for in advanced-stage malignant lung lesions.23 Meanwhile, the above cases also show that for patients who have not undergone lung biopsy for the first time and BALF tumor cytology is negative, especially those who also have a negative mNGS etiology test, the BALF CNVs test provides further clinical clues for tumor diagnosis, and this application value is also applicable to other specimens, such as blood.15,19 Gu et al15 used mNGS to detect CNV in various bodily fluid specimens. In malignant body fluid specimens with and without tumor cells, detected by routine cytology and flow cytometry, the diagnostic sensitivity of mNGS was 87% and 68%, respectively, and the specificity reached 100%. Consistent with the findings of this study, mNGS-based CNV testing has the potential to assist in the early detection of malignancies or reduce invasive procedures in cases without a definitive diagnosis.15 In particular, when a tumor is not the primary concern, a positive CNV test can alert clinicians to conduct more targeted diagnostic tests to avoid delays in diagnosis and treatment.
This study has some limitations. First, we examined a variety of human specimens, including blood, which may have reduced the ability of mNGS to detect pathogens in LRTIs. Second, since our laboratory did not perform PCR, gene chip, GeneXpert, or specialized culture technologies for MTB/NTM, although some samples were sent to third-party laboratories or tuberculosis-specialized hospitals for the above detection, the detection ability of mNGS in MTB/NTM may have been overestimated. Third, previous studies have found that EBV and CMV can exist in normal human blood,40 but this study shows that, up to 23.4% (44/188) of the samples (blood, BALF, and sputum) detected various human herpes viruses (types I, IV/EBV, V/CMV, VI, VII) and human polyomavirus (types I and III). Combined with comprehensive clinical analysis, the human herpes virus is considered a non-pathogenic pathogen in most cases; however, it still causes significant interference in clinical diagnosis. Fourth, mixed infections are more likely to occur in individuals with severe infections and immunosuppression. Although mNGS is conducive to improving the diagnostic rate of mixed infections, the variety of bacteria detected is not necessarily real pathogens. Thus, clinicians should interpret reports closely along with clinical findings to avoid antibiotic abuse based solely on NGS reports. Fifth, the number of DNA virus cases detected in this study was small and RNA virus detection was performed only in a few patients without data analysis in our study; therefore, the ability of mNGS to detect virus infections could not be evaluated comprehensively and accurately. Finally, mNGS CNV detection can help improve the sensitivity of tumor diagnosis; however, it cannot determine the primary lesion site, tumor type, or stage. In particular, positive CNV in blood sample screening can pose challenges in follow-up diagnosis, and whole-body positron emission tomography-CT is conducive to collaborative tumor screening. Additionally, this is a single-center study with a short period, and the number of patients with tumors included is small; thus, a larger sample size will be used in our future studies to better reveal the value of CNV detection using mNGS.
Conclusions
In summary, the sensitivity and accuracy of pathogen diagnosis of mNGS are superior to those of traditional culture and CMT. The specificity is higher than that of CMT, and the detection rate of various types of pathogens is higher than that of CMT, especially for special pathogens such as MTB/NTM, PS, PJ, and anerobic microflora. However, despite several advantages, mNGS has some limitations and should complement traditional detection methods to maximize the ability of laboratory etiology detection. In addition, mNGS CNV detection has important auxiliary diagnostic value for tumors, especially for the screening of occult tumors. More large-scale case studies are needed to further evaluate its clinical value.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Wang YY, Liu J. Application of metagenomic next generation sequencing for pathogen detection in respiratory infectious diseases. Chin J Clin Infect Dis. 2020;13(6):475–480.
2. Chinese Society of Laboratory Medicine. Expert consensus on clinical standardized application of metagenomics next‐generation sequencing for detection of pathogenic microorganisms. Chin J Lab Med. 2020;43(12):1181–1195.
3. Liu BB, Tian Q, Wang P, et al. Evaluating the diagnostic value of using metagenomic next-generation sequencing on bronchoalveolar lavage fluid and tissue in infectious pathogens located in the peripheral lung field. Ann Palliat Med. 2022;11(5):1725–1735. doi:10.21037/apm-21-3474
4. Kanaujia R, Biswal M, Angrup A, Ray P. Diagnostic accuracy of the metagenomic next-generation sequencing (mNGS) for detection of bacterial meningoencephalitis: a systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis. 2022;41(6):881–891. doi:10.1007/s10096-022-04445-0
5. Ramchandar N, Burns J, Coufal NG, et al. Use of metagenomic next-generation sequencing to identify pathogens in pediatric osteoarticular infections. Open Forum Infect Dis. 2021;8(7):ofab346. doi:10.1093/ofid/ofab346
6. Grundy BS, Parikh H, Jacob S, et al. Pathogen detection using metagenomic next-generation sequencing of plasma samples from patients with sepsis in Uganda. Microbiol Spectr. 2023;11(1):e0431222. doi:10.1128/spectrum.04312-22
7. Leitl CJ, Stoll SE, Wetsch WA, et al. Next-generation sequencing in critically ill COVID-19 patients with suspected bloodstream infections: a retrospective cohort study. J Clin Med. 2023;12(4):1466. doi:10.3390/jcm12041466
8. Bi MY, Wang CF, Lian JQ, Sun YT. Application of metagenomic next generation sequencing in infectious diseases. Chin J Clin Infect Dis. 2019;12(5):379–784.
9. Li Y, Ma JM. Expert consensus for the application of metagenomic next generation sequencing in the pathogen diagnosis in clinical moderate and severe infections (first edition). Chin Crit Care Med. 2020;32(5):531–536.
10. Zhang D, Yang X, Wang J, Xu J, Wang M. Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in patients with lower respiratory tract infections. J Int Med Res. 2022;50(4):3000605221089795. doi:10.1177/03000605221089795
11. Liang M, Fan Y, Zhang D, et al. Metagenomic next-generation sequencing for accurate diagnosis and management of lower respiratory tract infections. Int J Infect Dis. 2022;122:921–929. doi:10.1016/j.ijid.2022.07.060
12. Yan M ZX, Wang Y, Wang Y, et al. Metagenomic next-generation sequencing (mNGS) of bronchoalveolar lavage fluid on antimicrobial stewardship in patients with lower respiratory tract infections (LRTIs): a retrospective cohort study. J Infect Dis. 2023.;jiad296. doi:10.1093/infdis/jiad296
13. Zhao H, Zhao Y, Yan N, et al. Metagenomic next-generation sequencing of bronchoalveolar lavage fluid in non-severe and severe pneumonia patients. J Microbiol Methods. 2023;215:106848. doi:10.1016/j.mimet.2023.106848
14. Guo Y, Li H, Chen H, et al. Metagenomic next-generation sequencing to identify pathogens and cancer in lung biopsy tissue. EBioMedicine. 2021;73:103639. doi:10.1016/j.ebiom.2021.103639
15. Gu W, Talevich E, Hsu E, et al. Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids. Genome Med. 2021;13(1):98. doi:10.1186/s13073-021-00912-z
16. Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19(9):1586–1592. doi:10.1101/gr.092981.109
17. Wei P, Gao Y, Zhang J, et al. Diagnosis of lung squamous cell carcinoma based on metagenomic next-generation sequencing. BMC Pulm Med. 2022;22(1):108. doi:10.1186/s12890-022-01894-3
18. Xu JF, Kang Q, Ma XY, et al. A novel method to detect early colorectal cancer based on chromosome copy number variation in plasma. Cell Physiol Biochem. 2018;45(4):1444–1454. doi:10.1159/000487571
19. Liu K, Gao Y, Han J, et al. Diffuse large B-cell lymphoma of the mandible diagnosed by metagenomic sequencing: a case report. Front Med Lausanne. 2021;8:752523. doi:10.3389/fmed.2021.752523
20. Ren HT, Liu S, Fang KC, et al. Detection of meningeal carcinomatosis by metagenomic next-generation sequencing and copy number variation analysis of cerebrospinal fluid. Chin J Neurol. 2023;56(5):526–531.
21. Jiang JJ, Yang Y, Yin ZT, Zhao H, Huo YH, Bu H. Diagnosis of brain metastasis by copy number variation analysis and ctDNA based on cerebrospinal fluid metagenome next-generation sequencing: a case report. Chin J Nerv Ment Dis. 2023;49(3):155–159.
22. Su J, Han X, Xu X, et al. Simultaneous detection of pathogens and tumors in patients with suspected infections by next-generation sequencing. Front Cell Infect Microbiol. 2022;12:892087. doi:10.3389/fcimb.2022.892087
23. Lin P, Chen Y, Xu J, et al. A multicenter-retrospective cohort study of chromosome instability in lung cancer: clinical characteristics and prognosis of patients harboring chromosomal instability detected by metagenomic next-generation sequencing. J Thorac Dis. 2023;15(1):112–122. doi:10.21037/jtd-22-1732
24. Respiratory branch of Chinese medical association. Guidelines for the diagnosis and treatment of community-acquired pneumonia in Chinese adults (2016). Chin J Tuberc respir Dis. 2016;39(4):253–279.
25. Infectious Diseases Group, Respiratory Branch of Chinese Medical Association. Guidelines for the diagnosis and treatment of hospital-acquired pneumonia and ventilator-associated pneumonia in Chinese adults (2018). Chin J Tuberc respir Dis. 2018;41(4):255–280.
26. Ren MX, Pan L, Li J. Metagenomic next-generation sequencing of pathogenic microorganism in BALF from patients with abnormal lung imaging. Int J Respir. 2021;41(18):1398–1403.
27. Huang M, Lin Y, Chen X, Wu D. The value of gene chip detection of bronchoalveolar lavage fluid in the diagnosis of nontuberculous mycobacterial lung disease. Ann Palliat Med. 2021;10(6):6438–6445. doi:10.21037/apm-21-1205
28. Zhang R, Zhuang Y, Xiao ZH, et al. Diagnosis and surveillance of neonatal infections by metagenomic next-Generation sequencing. Front Microbiol. 2022;13:855988. doi:10.3389/fmicb.2022.855988
29. Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42. doi:10.1093/nar/gks1195
30. Luan Y, Hu H, Liu C, et al. A proof-of-concept study of an automated solution for clinical metagenomic next-generation sequencing. J Appl Microbiol. 2021;131(2):1007–1016. doi:10.1111/jam.15003
31. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi:10.1038/s41564-018-0349-6
32. Huang J, Jiang E, Yang D, et al. Metagenomic next-generation sequencing versus traditional pathogen detection in the diagnosis of peripheral pulmonary infectious lesions. Infect Drug Resist. 2020;13:567–576. doi:10.2147/IDR.S235182
33. Xie SS, Li X, Li P, et al. Diagnostic value and clinical application of metagenomic next-generation sequencing technology in infected patients. Int J Respir. 2020;40(9):641–646.
34. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–40.
35. Zhou H, Ouyang C, Han X, et al. Metagenomic sequencing with spiked-in internal control to monitor cellularity and diagnosis of pneumonia. J Infect. 2022;84(1):e13–e17. doi:10.1016/j.jinf.2021.09.018.
36. Lin P, Chen Y, Su S, et al. Diagnostic value of metagenomic next-generation sequencing of bronchoalveolar lavage fluid for the diagnosis of suspected pneumonia in immunocompromised patients. BMC Infect Dis. 2022;22(1):416. doi:10.1186/s12879-022-07381-8.
37. Wang J, Han Y, Feng J Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. 2019;19(1):252. doi:10.1186/s12890-019-1022-4
38. Ke CH, Hang HQ, Yang TJ, et al. Value of bronchoalveolar lavage fluid test by high-throughput sequencing technology in etiological diagnosis of infantile pneumonia. J Chin Pract Diagn Ther. 2020;34(4):398–400.
39. Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):162ra154. doi:10.1126/scitranslmed.3004742
40. Smatti MK, Yassine HM, AbuOdeh R, et al. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS ONE. 2017;12(12):e0189033. doi:10.1371/journal.pone.0189033
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.