Back to Journals » OncoTargets and Therapy » Volume 14
Circular RNAs Regulate Glucose Metabolism in Cancer Cells
Authors Ji X, Sun W, Lv C, Huang J, Zhang H
Received 20 April 2021
Accepted for publication 18 June 2021
Published 2 July 2021 Volume 2021:14 Pages 4005—4021
DOI https://doi.org/10.2147/OTT.S316597
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Xiaoyu Ji, Wei Sun, Chengzhou Lv, Jiapeng Huang, Hao Zhang
Department of Thyroid Surgery, The First Affiliated Hospital of China Medical University, Shenyang, Liaoning, 110001, People’s Republic of China
Correspondence: Hao Zhang Email [email protected]
Abstract: Circular RNAs (circRNAs) were originally thought to result from RNA splicing errors. However, it has been shown that circRNAs can regulate cancer onset and progression in various ways. They can regulate cancer cell proliferation, differentiation, invasion, and metastasis. Moreover, they modulate glucose metabolism in cancer cells through different mechanisms such as directly regulating glycolytic enzymes and glucose transporter (GLUT) or indirectly regulating signal transduction pathways. In this review, we elucidate on the role of circRNAs in regulating glucose metabolism in cancer cells, which partly explains the pathogenesis of malignant tumors, and provides new therapeutic targets or new diagnostic and prognostic markers for human cancers.
Keywords: circRNAs, glucose metabolism, Warburg effect, signaling pathway, targeted therapy
Background
Glucose metabolism is one of the most basic life properties. Under normoxia, most differentiated cells convert glucose into carbon dioxide and acetyl coenzyme A through oxidative phosphorylation to maintain energy metabolism. Besides, under anoxic conditions, glucose is directly reduced to lactic acid through glycolysis. In the 1920s, Otto Heinrich Warburg discovered that tumor cells exhibit unique reprogramming phenotypes,1–3 a phenomenon he named the “Warburg effect”.4,5 One of the main characteristic of Warburg effect is that, despite sufficient oxygen supply, cancer cells still produce energy through glycolysis instead of relying on mitochondrial oxidative phosphorylation, leading to elevated glucose uptake as well as ATP and lactic acid accumulation in cancer cells.4,6 Although more ATP can be produced by oxidative phosphorylation, precursors or intermediates produced during glycolysis by the pentose phosphate pathway, the hexosamine pathway, and the serine/glycine biosynthesis pathway can provide carbon sources for various biosynthesis reactions, thereby meeting the needs for rapid DNA replication.7–11 Glycolysis produces less reactive oxygen species (ROS) when compared to mitochondrial oxidative phosphorylation, which can induce tumor cell apoptosis or senescence under oxygen stress.12 In addition, the lactic acid produced by glycolysis can lower the pH of the extracellular matrix (ECM).13 An acidic microenvironment promotes tumor cell invasion, metastasis, and enhances their resistance to radiotherapy.14,15 Therefore, it is beneficial for tumor cells to rely on glycolysis to meet their energy needs because the Warburg effect is an optimized way through which tumor cells stimulate growth by cell stress.
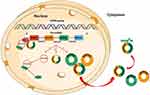
Circular RNAs (circRNAs) were first discovered in ribonucleic acid (RNA) viruses in 1976,16 followed by their discovery in form of endogenous RNA splicing products in eukaryotes in 1979.17 circRNAs were originally thought to result from RNA splicing errors, however, this theory was negated by the discovery of a large number of circRNAs in mammalian cells using RNA sequencing (RNA-seq) technology and bioinformatics. Salzman et al18 confirmed that circRNA is the main transcript for a variety of human cell types. Jack et al19 identified more than 25,000 circRNAs in human fibroblasts, implying that they are stable, conservative, and non-random products of RNA splicing, which may be associated with complementary ALU repeats in adjacent introns. These findings reveal dynamic expression patterns of circRNAs in various developmental stages and physiological conditions. For example, they serve as scaffolds in the assembly of protein complexes,20,21 and are also involved in; the isolation of proteins from their natural subcellular localization,22,23 regulating the expression of parental genes,24–27 regulating other splicing processes,28,29 RNA-protein interactions,30 and act as microRNA (miRNA) sponges31–33 (Figure 1).
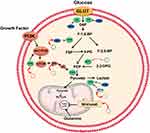
The most direct strategy for manipulating glucose metabolism is by affecting metabolic enzymes or kinases. Some signaling pathways, which play an important role in glucose metabolism can also be manipulated. Alterations in mRNA and protein levels are associated with glucose metabolism reprogramming in tumor cells.34–36 This implies that glycolytic enzymes and signaling pathways may be targets for cancer treatment.34,37 (Figure 2 and Figure 3).
Therefore, this review focuses on the mechanisms through which circRNAs regulate glucose metabolism, with the aim of elucidating on the complex cancer metabolism regulatory networks and providing a better theoretical basis for clinical diagnosis and treatment.
Table 1 summarizes circRNAs and their targets in the regulation of glucose metabolism in cancer.
 |
Table 1 CircRNAs and Their Targets in the Regulation of Glucose Metabolism in Cancer |
CircRNAs Regulate the Enzymes, Regulatory Molecules, and Oncogenes Involved in Glucose Metabolism in Cancer
CircRNAs Regulate Glucose Uptake by Altering the Expression of Glucose Transporter (GLUT)
Glucose transporter (GLUT) is a membrane protein that regulates cellular glucose metabolism. Under normal physiological conditions, GLUT promotes passive glucose transport by quickly transporting glucose from capillaries to cells. Among the 14 identified GLUT subtypes, GLUT1, GLUT3, and GLUT4 are up-regulated in malignant tumor cells, implying that they accelerate glucose transport in malignant tumors.38,39 This feature has been used in positron emission tomography for non-invasive diagnostic imaging of human cancer using radiolabeled glucose analogues and computer tomography.40
CircRNAs are involved in the regulation of GLUT1 expression. Circ100290, as a competitive endogenous RNA (ceRNA), elevates GLUT1 expression through mir-378a, thereby promoting glycolysis, proliferation, and invasion of oral squamous cell carcinoma cells.41 Qi et al42 found that circ0072995, as a type of carcinogenic circular RNA, induces malignant phenotype of cells through the mir-149-5p/SHMT2 axis. Therefore, circ0072995 enhances glucose uptake and lactic acid production, and promotes anaerobic glycolysis in breast cancer by promoting GLUT expression. circ0007534 significantly regulates protein levels of glycolysis-related genes (GULT1) in colon cancer through the mir-613/SLC25A22 axis, thereby affecting the efficiency of glycolysis and colon cancer cell progression.43 Propofol is the most widely used intravenous anesthetic in clinical surgery.44,45 In addition to various anesthetic effects, propofol exhibits anti-cancer affects46,47 against breast cancer48 and stomach cancer.49 Zhao et al50 found that, propofol regulates protein levels of GLUT1 by regulating circTADA2A expression through the mir-455 - 3p/FOXM1 axis, which suppresses glucose uptake by cancer cells, lactic acid production, and extracellular acidification, and inhibits lung cancer cell proliferation. Xiong et al51 reported that in non-small cell lung cancer cells (NSCLC), circMYLK is a molecular sponge for miR-195-5p while glucose transporter member 3 (GLUT3) is the target gene of miR-195-5p. Suppression of GLUT3 reduces lactic acid production in cancer cells. The decrease in lactic acid production was attributed to a decreased efficiency of aerobic glycolysis or the conversion of pyruvate to acetyl-CoA (Figures 2 and 4).
 |
Figure 2 CircRNAs regulate the molecules involved in glucose metabolism in cancer. CircRNAs regulate glucose uptake and glycolytic flux by modulating GLUTs and glycolic enzymes. |
CircRNA Affects Glycolysis by Regulating Enzymes or Kinases
Hexokinase (HK) is the first rate-limiting enzyme in the glycolytic pathway and is associated with cancer progression.52 It transforms glucose into 6-phosphate-glucose and promotes glucose transport into cells by glucose transporter 1 (GLUT1). In addition to the widely expressed HK1, cancer cells also overexpress HK2, which can enhance glycolysis.53
Expression of circPVT1 in highly invasive oral squamous cell carcinoma (OSCC) cells was found to be significantly elevated, and was associated with increased HK2 expression and suppressed mir-106a-5p expression.54 In vitro, HK2 was shown to be a direct target of mir-106a-5p while circPVT1 reversely regulated mir-106a-5p at the transcriptional level. These findings show that mir-106a-5p is a key factor between circPVT1 and HK2. In nasopharyngeal carcinoma and prostate cancer cells, the circRNA of the ZNF609 gene promotes cancer cell occurrence and development by up-regulating HK2 expression. Knockout of the HRAS gene inhibited HK2 expression and suppressed glucose consumption, lactate production, and ATP levels in nasopharyngeal carcinoma cells.55 Besides, the HK2 gene is the target of mir-501-3p in prostate cancer cells. Transfection of mir-501-3p can inhibit the expression of the HK2 protein, promote glycolysis, and improve radiation resistance of prostate cancer cells.56 In addition to the above-mentioned circRNA, circRNF20 occurs in breast cancer,57 circENO1 in lung adenocarcinoma,57 and circNFIX in glioma.58 These circRNAs can affect HK2 expression and promote cancer cell progression (Figures 2 and 4).
Pyruvate kinase (PK) is the last rate-limiting enzyme in glycolysis. Its activity can be affected by metamorphism and covalent modification. PK converts phosphoenolpyruvic acid (PEP) to pyruvic acid, where the first and second steps consume ATP while the last step produces ATP. Currently, four PK isoenzymes have been identified: M, K, L, and R types. The aberrant expression of pyruvate kinase M2 (PKM2) is very common in tumor cells.59 It is a splicing isoform of a dimeric pyruvate kinase involved in aerobic glycolysis, and it determines the proportion of carbon derived from glucose during energy production through glycolysis.60 Dysregulated circRNAs in cancer influence PKM2 expression. In liver cancer, mir-338 - 3p was found to be down-regulated despite PKM2 overexpression.61 This can be attributed to elevated expressions of circular RNA MAT2B, which acts as a molecular sponge for capturing mir-338-3p. Therefore, the MAT2B/mir-338-3p axis promotes glycolysis and liver cancer progression under hypoxic conditions by regulating PKM2 expression. Furthermore, circ0006168 regulates the mir-384/RBBP7 axis by activating the S6K/S6 pathway, and regulates PK protein levels in order to promote esophageal cancer cell growth, migration, invasion, and glycolysis.62 Hou et al63 knocked out circ0025033 in ovarian cancer cells and found that glucose consumption and lactic acid production were inhibited while si-circ0025033 transfection significantly suppressed PKM2 expression when compared to si-NC transfection (Figures 2 and 4).
Lactate dehydrogenase A (LDHA) is one of the subunits of lactate dehydrogenase (LDH) isoenzyme, which was first identified as one of the glycolytic genes induced by the myc gene.64 Its expression levels are associated with survival outcomes of various cancers, and it modulates tumor cell proliferation and invasion.45,65 Moreover, it can catalyze reversible conversion of pyruvate and lactic acid, resulting in lactic acid accumulation, which leads to tumor microenvironment acidification and enhanced tumor invasion. Secreted lactic acid can also be absorbed by adjacent tumor-associated stromal cells, ultimately producing pyruvate.66,67 CircRNAs modulate glycolysis by regulating the expression of the LDHA gene. Long et al68 reported that circPOSTN elevates the expression of LDHA in glioma cells. The underlying mechanism involves lactic acid production regulation through the circPOSTN/mir-361-5p /TPX2 axis, which provides an acidic environment for cells and eventually, promotes glioma cell proliferation and invasion. circABCB10 knockout in breast cancer cells negatively regulates glycolysis through the mir-223 - 3P/PFN axis, thereby reducing glucose consumption and lactic acid production, suppressing LDH-A activity and ATP level, promoting infrared treatment sensitivity, and providing a new direction for breast cancer radiotherapy69 (Figure 2).
The pyruvate dehydrogenase complex (PDC) is essential in metabolic homeostasis and is regulated by pyruvate dehydrogenase kinases (PDKs). The PDKs control PDC activity by phosphorylating its specific serine residues and, subsequently, deactivating the system if present in excess. There are four known isoforms of PDK’s (PDK1, PDK2, PDK3, and PDK4), that have different binding affinities to the complex.70 The isozymes bind phosphates to specific serine residues present within the α-subunits of the E1 of the complex.71,72 Hu et al73 reported that Circ-CNST regulates PDK1 expression by sponging miR-578 and glucose consumption. Moreover, lactate production and ATP/ADP ratios were found to be suppressed in Circ-CNST upregulated osteosarcoma cells. Xie et al74 knocked out Circ_0002711 in ovarian cancer cells and found that they could significantly inhibit glucose consumption, lactic acid production and PDK1 expression by modulating the miR-1244/ROCK1 axis. In gliomas, CircRNA EPHB4 modulates significantly promoting glucose consumption, lactate production and PDK1 expression levels by sponging miR-637.75 In Hepatocellular carcinoma, Circ0091579 enhances PDK2 protein expression under hypoxic stress conditions and promotes glucose uptake as well as lactic acid production.76 Moreover, PDK4 protein expression is enhanced by CircCCDC66 in thyroid cancer77 (Figures 3 and 4).
 |
Figure 4 The ceRNA network of circRNA-miRNA-mRNA. Spherical indicate circRNA, diamond represent miRNA, and rectangles indicate mRNA. |
CircRNA Affects Glycolysis by Regulating Oncogenes
Molecular mechanisms of metabolic abnormalities are associated with oncogene activation or tumor suppressor loss, eventually leading to elevated expression levels of hypoxia inducible factor 1α (HIF-1α) or the c-Myc oncogene. Studies64,78,79 have shown that aberrant regulation of the MYC oncogene is common during tumorigenesis. Notably, c-Myc is a transcription factor that is encoded by the MYC oncogene. Under normal oxygen conditions, regulation of the glycolytic genes by c-Myc promotes glucose metabolism as well as cell growth and proliferation.
Kim et al80 reported that dysregulated hypoxia-inducible factor-1 (HIF-1) can cooperate with dysregulated c-Myc to induce catalase hexose kinase 2 (the first key enzyme of glycolysis) and inactivate lactate dehydrogenase, thereby suppressing mitochondrial respiration and promoting glycolysis. TXNIP is an effective negative regulator of glucose uptake and aerobic glycolysis.81 Shen et al82 found that mRNA and protein expression levels of TXMP were significantly elevated in osteosarcoma cells with circECE1 knock out when compared to control cells. This indicates that TXNIP is mainly regulated by circECE1 transcription. Moreover, circECE1 interacts with c-Myc to prevent spop-induced c-Myc ubiquitination and degradation, and activates the c-Myc-TXNIP signal-regulated Warburg effect. There is a feedback loop between c-Myc and circECE1 - TXNIP, which regulates c-Myc protein expression and glucose metabolism. Circ-ACACA regulates the PI3K/PKB signaling pathway in non-small cell lung cancer (NSCLC) cells by interacting with MIR-1183, and suppresses protein expression levels of c-Myc, MMP9, and GLUT-1 genes in tumor cells, thereby promoting glycolysis.83
Enolase (ENO1) is a glycolytic enzyme that acts as a metabolic tumor promoter of the Warburg effect in cancer cells. It plays a vital role in aerobic glycolysis by converting 2-phosphoglyceride into phosphoenolpyruvate.84–86 In lung adenocarcinoma, ENO1 is the main target of miR-22-3p, and miR-22-3p transfection can suppress ENO1 protein expression. Therefore, circRNA of the ENO1 gene elevates protein expression levels of ENO1 as well as promote glycolysis and tumor progression of lung adenocarcinoma by up-regulating miR-22 - 3p.87
Phosphate isomerase (PI) is a cytoplasmic enzyme responsible for catalyzing mutual conversions between glucose-6-phosphatase and fructose-6-phosphatase. Therefore, it plays a key role in the glycolytic pathway.88 Phosphoglycerate kinase 1 (PGK1) contributes to ATP production and is involved in tumor occurrence and development.89 In addition, overexpression or knockdown of p200 CUX1 (CUX1) elevated or suppressed expression levels of p110 CUX1, ENO1, glucose-6-phosphate isomerase (GPI), or PGK1.90 The binding of circ-CUX1 to the RRM region of EWSR1 leads to EWSR1-mediated MAZ reverse activation, implying that the circ-CUX1/ EWSR1/MAZ axis promotes aerobic glycolysis and tumor progression (Figures 3 and 4).
CircRNAs Affect Glucose Metabolism by Regulating Cancer-Associated Signal Pathways
The HIF Signaling Pathway
Hypoxia inducible factor (HIF) is a nuclear transcription factor produced by cancer cells that have adapted to hypoxic environments. HIF1 is an oxygen-sensitive transcription complex composed of the oxygen regulatory subunit (HIF-1a) and the constitutive expression subunit (HIF-1b). Under hypoxia stress and PI3K activation, HIF-1a combines with the HIF-1b dimer to form active HIF-1 complexes, which regulate the transcription of various genes by binding hypoxia response elements.91 In a hypoxic environment, most tumor cells carry out aerobic glycolysis, thereby promoting the expression of glucose transporters, which increase glucose uptake by activating HIF-1.92 Moreover, it increases the expression of glycolytic enzymes,93,94 inhibits oxidative phosphorylation,95 and up-regulates LDHA, resulting in an acidic tumor microenvironment.93,96,97 In breast cancer, circDENND4C and HIF-1a form a positive feedback loop, which contributes to the Warburg effect. Under hypoxic stress conditions, elevated HK2, MMP9, and MMP2 protein expressions promote glucose uptake and lactic acid production. In the hypoxia-inducible chain, circDENND4C, a direct transcription target of HIF-1 was shown to promote glycolysis by elevating HK2, MMP9, and MMP2 levels. However, this effect was reversed by ectopic expressions of HIF-1a, indicating that the function of circDENND4C was dependent on HIF-1.98 Xu et al99 showed that circNRIP1 sponged miR-138-5p through HIF-1α dependent glucose metabolism, thereby maintaining the resistance of gastric carcinoma (GC) cells to 5-FU under hypoxia. Therefore, the combination of targeted circNRIP1 and 5-FU can significantly improve the prognosis of GC patients (Figures 3 and 4).
The Wnt/Snail Signaling Pathway
Wnt signaling regulates embryonic development and its imbalance is closely associated with the occurrence of many malignant tumors (including breast and colon cancers). Moreover, it induces snail-dependent epithelial-mesenchymal transition (EMT), which is the main cause of tumor invasion and metastasis.100–104
Metabolic changes can control the EMT process and trigger tumor malignancies, commonly referred to as the Warburg effect.105–107 Su et al108 revealed that Wnt inhibits mitochondrial respiration by inhibiting cytochrome C oxidase and promotes glycolysis by inducing pyruvate carboxylase (a key peroxidase). This process depends on the β-catendon/T cytokine 4/Snail signaling pathway. Inhibition of E-cadherin inhibits mitochondrial respiration and stimulates glycolysis through snail activation, implying that EMT contributes to Wnt/snail-mediated regulation of mitochondrial respiration and glucose metabolism. FBP1 is one of the gluconeogenesis-related restriction enzymes that modulates glucose metabolism.109 Pan et al111 identified a novel protein encoded by circFNDC3B, which inhibits tumor progression and epithelial-mesenchymal transition in colon cancer by alleviating the inhibitory effect of snail on the FBP1 gene. Therefore, it has been postulated that circFNDC3B plays a tumor suppressor role through the snail/FBP1/EMT axis.110 Han et al111 found that hsa_circ_0072387 expression was significantly downregulated while miR-503-5p was upregulated in OSCC cells and tissues. The gain of hsa_circ_0072387 or knockdown of miR-503-5p was shown to suppress OSCC cell proliferation, migration and invasion, EMT, as well as glycolysis. hsa_circ_0072387 targeted miR-503-5p and inversely regulated it expression. Moreover, upregulation of miR-503-5p partially reversed the tumor suppressive effects of hsa_circ_0072387 on OSCC cells (Figure 3).
The PI3K/AKT/mTOR Signaling Pathway
The phosphoinositide 3-kinase (PI3K) signaling pathway is involved in glucose metabolism. PI3K indirectly elevates the expression of GLUTs and enzymes by modulating Akt and mammalian target of rapamycin (mTOR). Akt-related metabolic factors include apoptosis-related kinases and GLUTs. Activation of Akt can elevate cellular ATP production and oxygen consumption.112 Akt regulates glycolysis and can elevate the expression of GLUTs113 and glycolytic enzymes such as HK2, PKM2112,114,115 or inhibit mitochondrial oxidative phosphorylation116,117 or activate mTORC1, which in turn elevates HIF-1 levels.118,119
CircHIPK3 is upregulated in lung cancer and promotes glucose uptake and utilization during aerobic glycolysis by enhancing Akt phosphorylation and activating the mTOR signaling pathway, resulting in the upregulation of HK2.120 Besides, Yalan et al found that CircRHOBTB3 plays a suppressor role and inhibits tumorigenesis by inactivating the PI3K/AKT pathway in ovarian cancer. Lentivectors for short hairpin RNA (shRNA) against circRHOBTB3 (sh-circRHOBTB3) or pcDNA-circRHOBTB3 were used to downregulate or upregulate circRHOBTB3 expression in an animal tumor model. It was found that the protein expressions of GLUT1, HK2 and LDHA were altered.121(Figure 3).
Therapeutic Potential of CircRNA in Targeted Cancer Treatment
High rates of aerobic glycolysis in cancer cells are important for proliferation. Although oxidative phosphorylation is more beneficial for ATP production, high ATP production rates but at low efficiencies is uncommon in cancer cells. Instead, cancer cells may benefit from elevated levels of glycolysis intermediates such as nucleotides, amino acids, lipids, and NADPH.122
Identifying key nodes in the pathway network that regulates aerobic glycolysis can lead to the discovery of new targets for anti-tumor therapy. Among the various enzymes involved in the glycolysis cascade, GLUT-1, HK2, LDHA, and PKM2 are potential targets because they are overexpressed in cancer cells.123–126 In addition, targeting the Warburg effect through precision medicine, a multi-pronged approach, may be an effective anti-cancer treatment strategy. For example, the Warburg effect was reversed in lung adenocarcinoma cells by inhibiting the EDFR signaling pathway.127 Another treatment strategy involves combining targeted glycolysis drugs and mTOR inhibitors to prevent metabolic reprogramming from inducing cancer cell apoptosis.128 For instance, the combination of acarbinidine (AICAR) and methotrexate (glucosamine) reversed the Warburg effect in MCF-7 breast cancer cells.129 Although single drug therapy may induce resistance, combined therapy can induce AMPK and FOX1 expression, resulting in increased mitochondrial oxidative phosphorylation and reduced glycolysis. These metabolic changes indicate that the anti-Warburg effect prevents G1/S and G2/M transitions and slows cell cycle progression. A previous study performed on kidney and ganglion cell tumors reported germline mutations of succinate dehydrogenase and fumarate hydratase in the TCA cycle.130 The role of one of these mutations is to activate HIF-1α-regulated glucose metabolism. Collectively, these findings imply that targeting glucose metabolism-related genes or pathways has great potentials in cancer treatment.
Interactions between circRNAs and key transcription factors or metabolic enzymes involved in glycolysis can effectively regulate glucose metabolism and promote tumor progression. In addition to these key molecules, other metabolic pathways are also crucial for glucose metabolism in cancer, especially the PI3K/AKT/ mTOR and the AMPK pathways. Considering glucose metabolism reprogramming, targeting circRNAs may have an important impact on cancer cell invasion and proliferation. Therefore, the circRNAs associated with aerobic glycolysis may be diagnostic and prognostic biomarkers for reprogramming glucose metabolism. Moreover, elucidation of circRNA-mediated glucose metabolism regulation in tumor cells may inform the development of circRNA inhibitors and prevent tumor progression.
Therefore, circRNA, as a regulator of metabolites, is more likely to provide novel targets for cancer treatment. However, there are many challenges associated with these methods. First, cells can recruit other glycolytic enzymes to promote glycolysis despite enzyme inhibitors being highly specific. Second, key target pathways must be identified. However, this may be difficult to achieve because of the phenotypic and functional heterogeneity between cancer and a single tumor. Third, it is necessary to clarify the toxic effects of inhibiting glycolytic enzymes in normal cells since aerobic glycolysis is a key process in immune and stem cells. Fluorodeoxyglucose positron emission tomography (FDG-PET) has previously been used to measure glucose metabolism, detect cancer, and predict prognosis.131 Current methods, including positron emission tomography (PET), autoradiography and magnetic resonance imaging (MRI) can measure the primary metabolic rate of glucose. However, these methods are limited by the fact that they cannot distinguish between markers and intermediates.
Conclusion
Implementation of integrated treatment strategies for patients based on their genetic backgrounds is becoming a reality in cancer treatment. Advances in systematic biology, such as reconstruction of genome-scale metabolic models (GEMs), can enhance the accuracy of systematic assessment of cancer cell type-specific metabolic profiles.132–134 Transgenic bio-integromics data can reveal the biomarkers and anti-metabolites of potential specific patients and cancer types. Furthermore, reprogrammed glucose metabolism is a recently identified marker for cancer cells. Therefore, elucidation of the roles of circRNAs in glucose metabolism and their mechanisms will help in development of new strategies for controlling abnormal metabolic phenotypes and for inhibiting the Warburg effect.
Abbreviations
circRNA, Circular RNA; RNA-seq, RNA sequencing; miRNA, microRNA; ceRNA, competitive endogenous RNA; GLUT, glucose transporter; ROS, reactive oxygen species; ECM, extracellular matrix; HK, Hexokinase; OSCC, oral squamous cell carcinoma; PK, Pyruvate kinase; PEP, phosphoenolpyruvic acid; PKM2, pyruvate kinase M2; LDH, lactate dehydrogenase; HIF, Hypoxia induced factor; NSCLC, non-small-cell lung cancer; PDC, pyruvate dehydrogenase complex; PDKs, pyruvate dehydrogenase kinases; PI, phosphate isomerase; PGK1, Phosphoglycerate kinase 1; GPI, glucose-6-phosphate isomerase; GC, Gastric Carcinoma; EMT, epithelial-mesenchymal transition; FDG-PET, Fluorodeoxyglucose positron emission tomography; PET, positron emission tomography; MRI, magnetic resonance imaging; GEM, genome-scale metabolic model.
Ethical Approval
Ethical approval was waived because human participants or animals were not involved in any way.
Consent for Publication
All authors read and approved the final manuscript for publication.
Authors’ Statement
We hereby confirm that neither the manuscript nor any part of it has been published or is being considered for publication elsewhere. We acknowledge that all authors sufficiently participated in the study and take responsibility for its content.
Funding
This study was financially supported by: The National Natural Science Foundation of China (grant number 81902726); The Project funded by China Postdoctoral Science Foundation (grant number 2018M641739); and Natural Science Foundation of Liaoning Province (grant number 20180530090).
Disclosure
All authors have no conflicts of interest to declare.
References
1. Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi:10.1038/nrc2981
2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674.
3. van Staa TP, Patel D, Gallagher AM, et al. Glucose-lowering agents and the patterns of risk for cancer: a study with the General Practice Research Database and secondary care data. Diabetologia. 2012;55(3):654–665. doi:10.1007/s00125-011-2390-3
4. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi:10.1126/science.123.3191.309
5. Warburg O. The chemical constitution of respiration ferment. Science. 1928;68(1767):437–443. doi:10.1126/science.68.1767.437
6. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8(6):519–530. doi:10.1085/jgp.8.6.519
7. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
8. DeBerardinis RJ, et al., Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis.
9. Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi:10.1038/nrd3504
10. DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148(6):1132–1144. doi:10.1016/j.cell.2012.02.032
11. Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27(1):441–464. doi:10.1146/annurev-cellbio-092910-154237
12. Wang Y, Wu Y, Wang Y, et al. Bacillus amyloliquefaciens SC06 alleviates the oxidative stress of IPEC-1 via modulating Nrf2/Keap1 signaling pathway and decreasing ROS production. Appl Microbiol Biotechnol. 2017;101(7):3015–3026. doi:10.1007/s00253-016-8032-4
13. Held-Warmkessel J, Dell DD. Lactic acidosis in patients with cancer. Clin J Oncol Nurs. 2014;18(5):592–594. doi:10.1188/14.CJON.592-594
14. Peppicelli S, Bianchini F, Calorini L. Extracellular acidity a “Reappreciated” trait of tumor environment driving malignancy: perspectives in diagnosis and therapy. Cancer Metastasis Rev. 2014;33(2–3):823–832.
15. Shiraishi T, Verdone JE, Huang J, et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget. 2015;6(1):130–143. doi:10.18632/oncotarget.2766
16. Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
17. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi:10.1038/280339a0
18. Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi:10.1371/journal.pone.0030733
19. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
20. Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi:10.1038/cdd.2016.133
21. Abbaszadeh-Goudarzi K, Radbakhsh S, Pourhanifeh MH, et al. Circular RNA and diabetes: epigenetic regulator with diagnostic role. Curr Mol Med. 2020;20(7):516–526. doi:10.2174/1566524020666200129142106
22. Armakola M, Higgins MJ, Figley MD, et al. Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet. 2012;44(12):1302–1309. doi:10.1038/ng.2434
23. Borran S, Ahmadi G, Rezaei S, et al. Circular RNAs: new players in thyroid cancer. Pathol Res Pract. 2020;216(10):153217. doi:10.1016/j.prp.2020.153217
24. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
25. Zhang Y, Zhang X-O, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi:10.1016/j.molcel.2013.08.017
26. Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5(1):12453. doi:10.1038/srep12453
27. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi:10.18632/oncotarget.3469
28. Ashwal-Fluss R, Meyer M, Pamudurti N, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
29. Nahand JS, Jamshidi S, Hamblin MR, et al. Circular RNAs: new epigenetic signatures in viral infections. Front Microbiol. 2020;11:1853. doi:10.3389/fmicb.2020.01853
30. Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi:10.1093/nar/gkw027
31. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
32. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
33. Shabaninejad Z, Vafadar A, Movahedpour A, et al. Circular RNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12(1):84. doi:10.1186/s13048-019-0558-5
34. Zhao M, Fan J, Liu Y, et al. Oncogenic role of the TP53-induced glycolysis and apoptosis regulator in nasopharyngeal carcinoma through NF-κB pathway modulation. Int J Oncol. 2016;48(2):756–764. doi:10.3892/ijo.2015.3297
35. Faubert B, Vincent EE, Griss T, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci U S A. 2014;111(7):2554–2559. doi:10.1073/pnas.1312570111
36. Mirzaei H, Hamblin MR. Regulation of glycolysis by non-coding rnas in cancer: switching on the Warburg effect. Mol Ther Oncolytics. 2020;19:218–239.
37. Panisova E, Kery M, Kopacek J, et al. Enhanced metabolism as a common feature of cancer plasticity. Neoplasma. 2016;63(6):836–845. doi:10.4149/neo_2016_602
38. Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34(2–3):121–138. doi:10.1016/j.mam.2012.07.001
39. Fan C, Tang Y, Wang J, et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16(1):130. doi:10.1186/s12943-017-0699-3
40. Greco M, Crippa F, Agresti R, et al. Axillary lymph node staging in breast cancer by 2-fluoro-2-deoxy-D-glucose-positron emission tomography: clinical evaluation and alternative management. J Natl Cancer Inst. 2001;93(8):630–635. doi:10.1093/jnci/93.8.630
41. Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol. 2019;234(11):19130–19140. doi:10.1002/jcp.28692
42. Qi C, Qin X, Zhou Z, et al. Circ_0072995 promotes cell carcinogenesis via up-regulating miR-149-5p-mediated SHMT2 in breast cancer. Cancer Manag Res. 2020;12:11169–11181. doi:10.2147/CMAR.S272274
43. Ding DY, Wang D, Shu ZB. Hsa_circ_0007534 knockdown represses the development of colorectal cancer cells through regulating miR-613/SLC25A22 axis. Eur Rev Med Pharmacol Sci. 2021;25(2):570.
44. Chidambaran V, Costandi A, D’Mello A. Propofol: a review of its role in pediatric anesthesia and sedation. CNS Drugs. 2015;29(7):543–563.
45. Oren-Ziv A, Hoppenstein D, Shles A, et al. Sedation methods for intra-articular corticosteroid injections in Juvenile Idiopathic Arthritis: a review. Pediatr Rheumatol Online J. 2015;13(1):28. doi:10.1186/s12969-015-0021-0
46. Kushida A, Inada T, Shingu K. Enhancement of antitumor immunity after propofol treatment in mice. Immunopharmacol Immunotoxicol. 2020;Volume 13(3):5293–5307. doi:10.1080/08923970701675085
47. Song J, et al. Mini profile of potential anticancer properties of propofol. PLoS One. 2021;19(1):e114440. doi:10.1371/journal.pone.0114440
48. Yu B, et al. Propofol induces apoptosis of breast cancer cells by downregulation of miR-24 signal pathway. Cancer Biomark. 2018;21(3):513–519. doi:10.3233/CBM-170234
49. Zhang W, et al. Propofol inhibits proliferation, migration and invasion of gastric cancer cells by up-regulating microRNA-195. Int J Biol Macromol. 2018;120(Pt A):975–984. doi:10.1016/j.ijbiomac.2018.08.173
50. Zhao H, Wei H, He J, et al. Propofol disrupts cell carcinogenesis and aerobic glycolysis by regulating circTADA2A/miR-455-3p/FOXM1 axis in lung cancer. Cell Cycle. 2020;19(19):2538–2552. doi:10.1080/15384101.2020.1810393
51. Xiong S, et al. <p>Circular RNA MYLK promotes glycolysis and proliferation of non-small cell lung cancer cells by sponging miR-195-5p and increasing glucose transporter Member 3 expression. Cancer Manag Res. 2020;19:5469–5478. doi:10.2147/CMAR.S257386
52. Tan VP, Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11(6):963–964. doi:10.1080/15548627.2015.1042195
53. Heneberg P. Redox Regulation of Hexokinases. Antioxid Redox Signal. 2019;30(3):415–442. doi:10.1089/ars.2017.7255
54. Zhu X, Du J, Gu Z. Circ-PVT1/miR-106a-5p/HK2 axis regulates cell growth, metastasis and glycolytic metabolism of oral squamous cell carcinoma. Mol Cell Biochem. 2020;474(1–2):147–158. doi:10.1007/s11010-020-03840-5
55. Liu Z, Liu F, Wang F, et al. CircZNF609 promotes cell proliferation, migration, invasion, and glycolysis in nasopharyngeal carcinoma through regulating HRAS via miR-338-3p. Mol Cell Biochem. 2021;476(1):175–186. doi:10.1007/s11010-020-03894-5
56. Du S, et al. Circ-ZNF609 accelerates the radioresistance of prostate cancer cells by promoting the glycolytic metabolism through miR-501-3p/HK2 axis. Cancer Manag Res. 2020;12:7487–7499. doi:10.2147/CMAR.S257441
57. Cao L, et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 2020;11(2):145. doi:10.1038/s41419-020-2336-0
58. Ding C, Wu Z, You H, et al. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res. 2019;38(1):506. doi:10.1186/s13046-019-1483-6
59. Li Q, et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37(9):12503–12512. doi:10.1007/s13277-016-5113-z
60. Liang F, et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49(3):1046–1056. doi:10.3892/ijo.2016.3599
61. Li Q, Pan X, Zhu D, et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi:10.1002/hep.30671
62. Xie ZF, et al. Circular RNA hsa_circ_0006168 contributes to cell proliferation, migration and invasion in esophageal cancer by regulating miR-384/RBBP7 axis via activation of S6K/S6 pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):151–163. doi:10.26355/eurrev_202001_19906
63. Hou W, Zhang Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract. 2021;217:153275. doi:10.1016/j.prp.2020.153275
64. Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15(21):6479–6483. doi:10.1158/1078-0432.CCR-09-0889
65. Wulaningsih W, et al. Serum lactate dehydrogenase and survival following cancer diagnosis. Br J Cancer. 2015;113(9):1389–1396. doi:10.1038/bjc.2015.361
66. Fukumura D, Xu L, Chen Y, et al. Hypoxia and acidosis independently up-regulate vascular endothelial growth factor transcription in brain tumors in vivo. Cancer Res. 2001;61(16):6020–6024.
67. Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71(22):6921–6925. doi:10.1158/0008-5472.CAN-11-1457
68. Long N, et al. CircPOSTN/miR-361-5p/TPX2 axis regulates cell growth, apoptosis and aerobic glycolysis in glioma cells. Cancer Cell Int. 2020;20:374. doi:10.1186/s12935-020-01454-x
69. Zhao Y, et al. Circle RNA circABCB10 modulates PFN2 to promote breast cancer progression, as well as aggravate radioresistance through facilitating glycolytic metabolism via miR-223-3p. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2019.3389
70. Patel MS, Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans. 2006;34(Pt 2):217–222. doi:10.1042/BST0340217
71. Gudi R, et al. Diversity of the pyruvate dehydrogenase kinase gene family in humans. J Biol Chem. 1995;270(48):28989–28994. doi:10.1074/jbc.270.48.28989
72. Sugden MC, Holness MJ. Interactive regulation of the pyruvate dehydrogenase complex and the carnitine palmitoyltransferase system. FASEB j. 1994;8(1):54–61. doi:10.1096/fasebj.8.1.8299890
73. Hu R, Chen S, Yan J. Blocking circ-CNST suppresses malignant behaviors of osteosarcoma cells and inhibits glycolysis through circ-CNST-miR-578-LDHA/PDK1 ceRNA networks. J Orthop Surg Res. 2021;16(1):300. doi:10.1186/s13018-021-02427-0
74. Xie W, et al. Circ_0002711 knockdown suppresses cell growth and aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian cancer. J Biosci. 2021;46:1.
75. Jin C, et al. CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10. Mol Oncol. 2021;15(2):596–622. doi:10.1002/1878-0261.12830
76. Shu J, Du J, Wang F, et al. Circ_0091579 enhances the malignancy of hepatocellular carcinoma via miR-1287/PDK2 axis. Open Life Sci. 2021;16(1):69–83. doi:10.1515/biol-2021-0009
77. Ren H, Song Z, Chao C, et al. circCCDC66 promotes thyroid cancer cell proliferation, migratory and invasive abilities and glycolysis through the miR-211-5p/PDK4 axis. Oncol Lett. 2021;21(5):416. doi:10.3892/ol.2021.12677
78. Kelly B, O’Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25(7):771–784. doi:10.1038/cr.2015.68
79. Masui K, Tanaka K, Akhavan D, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 2013;18(5):726–739. doi:10.1016/j.cmet.2013.09.013
80. Kim J-W, Gao P, Liu Y-C, et al. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27(21):7381–7393. doi:10.1128/MCB.00440-07
81. Sullivan WJ, Mullen PJ, Schmid EW, et al. Extracellular matrix remodeling regulates glucose metabolism through TXNIP destabilization. Cell. 2018;175(1):117–132.e21. doi:10.1016/j.cell.2018.08.017
82. Shen S, Yao T, Xu Y, et al. CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol Cancer. 2020;19(1):151. doi:10.1186/s12943-020-01269-4
83. Wu W, Xi W, Li H, et al. Circular RNA circ‑ACACA regulates proliferation, migration and glycolysis in non‑small‑cell lung carcinoma via miR‑1183 and PI3K/PKB pathway. Int J Mol Med. 2020;45(6):1814–1824. doi:10.3892/ijmm.2020.4549
84. Qian X, Xu W, Xu J, et al. Enolase 1 stimulates glycolysis to promote chemoresistance in gastric cancer. Oncotarget. 2017;8(29):47691–47708. doi:10.18632/oncotarget.17868
85. Capello M, Ferri-Borgogno S, Riganti C, et al. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget. 2016;7(5):5598–5612. doi:10.18632/oncotarget.6798
86. Chen S, Zhang Y, Wang H, et al. WW domain-binding protein 2 acts as an oncogene by modulating the activity of the glycolytic enzyme ENO1 in glioma. Cell Death Dis. 2018;9(3):347. doi:10.1038/s41419-018-0376-5
87. Zhou J, Zhang S, Chen Z, et al. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10(12):885. doi:10.1038/s41419-019-2127-7
88. Zdralevic M, Marchiq I, de Padua MMC, et al. Metabolic plasiticy in cancers-distinct role of glycolytic enzymes GPI, LDHs or membrane transporters MCTs. Front Oncol. 2017;7:313. doi:10.3389/fonc.2017.00313
89. Li X, Jiang Y, Meisenhelder J, et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol Cell. 2016;61(5):705–719. doi:10.1016/j.molcel.2016.02.009
90. Li H, Yang F, Hu A, et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol Med. 2019;11(12):e10835. doi:10.15252/emmm.201910835
91. Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001;11(3):293–299. doi:10.1016/S0959-437X(00)00193-3
92. Starska K, Forma E, Jóźwiak P, et al. Gene and protein expression of glucose transporter 1 and glucose transporter 3 in human laryngeal cancer-the relationship with regulatory hypoxia-inducible factor-1alpha expression, tumor invasiveness, and patient prognosis. Tumour Biol. 2015;36(4):2309–2321. doi:10.1007/s13277-014-2838-4
93. Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12(2):149–162. doi:10.1101/gad.12.2.149
94. Hussien R, Brooks GA. Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines. Physiol Genomics. 2011;43(5):255–264. doi:10.1152/physiolgenomics.00177.2010
95. Kim J-W, Tchernyshyov I, Semenza GL, et al. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi:10.1016/j.cmet.2006.02.002
96. Hayashi M, et al. Induction of glucose transporter 1 expression through hypoxia-inducible factor 1α under hypoxic conditions in trophoblast-derived cells. J Endocrinol. 2017;8(29):47691–47708. doi:10.1677/joe.1.05599
97. Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO j. 1998;17(11):3005–3015.
98. Liang G, Liu Z, Tan L, et al. HIF1alpha-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37(8):4337–4343. doi:10.21873/anticanres.11827
99. Xu G, et al. Circular RNA circNRIP1 Sponges microRNA-138-5p to maintain hypoxia-induced resistance to 5-fluorouracil through HIF-1α-dependent glucose metabolism in gastric carcinoma. Cancer Manag Res. 2020;12:2789–2802. doi:10.2147/CMAR.S246272
100. Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8(5):387–398. doi:10.1038/nrc2389
101. Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi:10.1038/nature03319
102. Bagherian A, Mardani R, Roudi B, et al. Combination therapy with nanomicellar-curcumin and temozolomide for in vitro therapy of glioblastoma multiforme via Wnt signaling pathways. J Mol Neurosci. 2020;70(10):1471–1483. doi:10.1007/s12031-020-01639-z
103. Yousefi F, Shabaninejad Z, Vakili S, et al. TGF-β and WNT signaling pathways in cardiac fibrosis: non-coding RNAs come into focus. Cell Commun Signal. 2020;18(1):87. doi:10.1186/s12964-020-00555-4
104. Ghasemi F, Shafiee M, Banikazemi Z, et al. Curcumin inhibits NF-kB and Wnt/β-catenin pathways in cervical cancer cells. Pathol Res Pract. 2019;215(10):152556. doi:10.1016/j.prp.2019.152556
105. Halldorsson S, Rohatgi N, Magnusdottir M, et al. Metabolic re-wiring of isogenic breast epithelial cell lines following epithelial to mesenchymal transition. Cancer Lett. 2017;396:117–129. doi:10.1016/j.canlet.2017.03.019
106. Lunetti P, Di Giacomo M, Vergara D, et al. Metabolic reprogramming in breast cancer results in distinct mitochondrial bioenergetics between luminal and basal subtypes. Febs j. 2019;286(4):688–709. doi:10.1111/febs.14756
107. Peppicelli S, Toti A, Giannoni E, et al. Metformin is also effective on lactic acidosis-exposed melanoma cells switched to oxidative phosphorylation. Cell Cycle. 2016;15(14):1908–1918. doi:10.1080/15384101.2016.1191706
108. Lee SY, Jeon HM, Ju MK, et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res. 2012;72(14):3607–3617. doi:10.1158/0008-5472.CAN-12-0006
109. Liu G-M, Li Q, Zhang P-F, et al. Restoration of FBP1 suppressed Snail-induced epithelial to mesenchymal transition in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1132. doi:10.1038/s41419-018-1165-x
110. Pan Z, Cai J, Lin J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19(1):71. doi:10.1186/s12943-020-01179-5
111. Han L, Cheng J, Li A. hsa_circ_0072387 suppresses proliferation, metastasis, and glycolysis of oral squamous cell carcinoma cells by downregulating miR-503-5p. Cancer Biother Radiopharm. 2021;36(1):84–94. doi:10.1089/cbr.2019.3371
112. Gottlob K, et al. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15(11):1406–1418. doi:10.1101/gad.889901
113. van Dam EM, Govers R, James DE. Akt activation is required at a late stage of insulin-induced GLUT4 translocation to the plasma membrane. Mol Endocrinol. 2005;19(4):1067–1077. doi:10.1210/me.2004-0413
114. Salani B, Ravera S, Amaro A, et al. IGF1 regulates PKM2 function through Akt phosphorylation. Cell Cycle. 2015;14(10):1559–1567. doi:10.1080/15384101.2015.1026490
115. Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab. 2002;13(10):444–451. doi:10.1016/S1043-2760(02)00662-8
116. Miyamoto S, Murphy AN, Brown JH. Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ. 2008;15(3):521–529. doi:10.1038/sj.cdd.4402285
117. Robey RB, Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25(34):4683–4696. doi:10.1038/sj.onc.1209595
118. Majumder PK, Febbo PG, Bikoff R, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10(6):594–601. doi:10.1038/nm1052
119. Skeen JE, Bhaskar PT, Chen -C-C, et al. Akt deficiency impairs normal cell proliferation and suppresses oncogenesis in a p53-independent and mTORC1-dependent manner. Cancer Cell. 2006;10(4):269–280. doi:10.1016/j.ccr.2006.08.022
120. Gu F, Zhang J, Yan L, et al. CircHIPK3/miR-381-3p axis modulates proliferation, migration, and glycolysis of lung cancer cells by regulating the AKT/mTOR signaling pathway. Open Life Sci. 2020;15(1):683–695. doi:10.1515/biol-2020-0070
121. Yalan S, Yanfang L, He C, et al. Circular RNA circRHOBTB3 inhibits ovarian cancer progression through PI3K/AKT signaling pathway. Panminerva Med. 2020. doi:10.23736/S0031-0808.20.03957-9
122. Vander Heiden MG, Lunt SY, Dayton TL, et al. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2011;76:325–334. doi:10.1101/sqb.2012.76.010900
123. Wolf A, Agnihotri S, Micallef J, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. doi:10.1084/jem.20101470
124. Bluemlein K, Grüning N-M, Feichtinger RG, et al. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2(5):393–400. doi:10.18632/oncotarget.278
125. Chan DA, Sutphin PD, Nguyen P, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3(94):94ra70. doi:10.1126/scitranslmed.3002394
126. Le A, Cooper CR, Gouw AM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107(5):2037–2042. doi:10.1073/pnas.0914433107
127. Janssen HLA, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi:10.1056/NEJMoa1209026
128. Pusapati RV, Daemen A, Wilson C, et al. mTORC1-dependent metabolic reprogramming underlies escape from glycolysis addiction in cancer cells. Cancer Cell. 2016;29(4):548–562. doi:10.1016/j.ccell.2016.02.018
129. Fodor T, Szántó M, Abdul-Rahman O, et al. Combined treatment of MCF-7 cells with AICAR and methotrexate, arrests cell cycle and reverses warburg metabolism through AMP-Activated Protein Kinase (AMPK) and FOXO1. PLoS One. 2016;11(2):e0150232. doi:10.1371/journal.pone.0150232
130. Baysal BE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287(5454):848–851. doi:10.1126/science.287.5454.848
131. Ben-Haim S, Ell P. 18F-FDG PET and PET/CT in the evaluation of cancer treatment response. J Nucl Med. 2009;50(1):88–99. doi:10.2967/jnumed.108.054205
132. Folger O, Jerby L, Frezza C, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7(1):501. doi:10.1038/msb.2011.35
133. Frezza C, Zheng L, Folger O, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477(7363):225–228. doi:10.1038/nature10363
134. Agren R, Mardinoglu A, Asplund A, et al. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10(3):721. doi:10.1002/msb.145122
135. Yi G, Wang D, Han J, et al. circKLHL24 blocks breast cancer development by regulating the miR-1204/ALX4 Network. Cancer Biother Radiopharm. 2021. doi:10.1089/cbr.2020.3992
136. Li C, Liu H, Niu Q, et al. <p>Circ_0000376, a Novel circRNA, promotes the progression of non-small cell lung cancer through regulating the miR-1182/NOVA2 network. Cancer Manag Res. 2020;12:7635–7647. doi:10.2147/CMAR.S258340
137. Ma J, Qi G, Li L, Novel Serum A. Exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307. doi:10.2147/OTT.S243214
138. Dong W, Zhao L, Zhang S, et al. Circ-KIAA0907 inhibits the progression of oral squamous cell carcinoma by regulating the miR-96-5p/UNC13C axis. World J Surg Oncol. 2021;19(1):75. doi:10.1186/s12957-021-02184-8
139. Guo J, Su Y, Zhang M. Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p. Cancer Cell Int. 2020;20(1):407. doi:10.1186/s12935-020-01501-7
140. Hu B, Xian Z, Zou Q, et al. CircFAT1 Suppresses Colorectal Cancer Development Through Regulating miR-520b/UHRF1 Axis or miR-302c-3p/UHRF1 Axis. Cancer Biother Radiopharm. 2021;36(1):45–57. doi:10.1089/cbr.2019.3291
141. Zhang ZJ, Zhang Y-H, Qin X-J, et al. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur Rev Med Pharmacol Sci. 2020;24(5):2387–2400. doi:10.26355/eurrev_202003_20506
142. Zhang X, Wang S, Lin G, et al. Down-regulation of circ-PTN suppresses cell proliferation, invasion and glycolysis in glioma by regulating miR-432-5p/RAB10 axis. Neurosci Lett. 2020;735:135153. doi:10.1016/j.neulet.2020.135153
143. Chen Y, et al. Silencing of circular RNA circPDE5A suppresses neuroblastoma progression by targeting the miR-362-5p/NOL4L axis. Int J Neurosci. 2021:1–11.
144. Liu J, Li X, Yue L, et al. Circ_0105346 knockdown inhibits osteosarcoma development via regulating miR-1182/WNT7B axis. Cancer Manag Res. 2021;13:521–535. doi:10.2147/CMAR.S281430
145. Chen W, Li Y, Zhong J, et al. circ-PRKCI targets miR-1294 and miR-186-5p by downregulating FOXK1 expression to suppress glycolysis in hepatocellular carcinoma. Mol Med Rep. 2021;23(6):6. doi:10.3892/mmr.2021.12103
146. Peng L, Chen G, Zhu Z, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8(1):808–818. doi:10.18632/oncotarget.13656
147. Qiu L, Zheng L, Gan C, et al. circBICD2 targets miR-149-5p/IGF2BP1 axis to regulate oral squamous cell carcinoma progression. J Oral Pathol Med. 2020. doi:10.1111/jop.13156
148. Zheng Z, Ma X, Li H. Circular RNA circMDM2 accelerates the glycolysis of oral squamous cell carcinoma by targeting miR-532-3p/HK2. J Cell Mol Med. 2020;24(13):7531–7537. doi:10.1111/jcmm.15380
149. Xing Z, Wang X, Liu J, et al. Hsa_circ_0069094 accelerates cell malignancy and glycolysis through regulating the miR-591/HK2 axis in breast cancer. Cell Signal. 2021;79:109878. doi:10.1016/j.cellsig.2020.109878
150. Dou D, Ren X, Han M, et al. Circ_0008039 supports breast cancer cell proliferation, migration, invasion, and glycolysis by regulating the miR-140-3p/SKA2 axis. Mol Oncol. 2021;15(2):697–709. doi:10.1002/1878-0261.12862
151. Zang H, Li Y, Zhang X, et al. <p>Knockdown of circRAD18 Mitigates Breast Cancer Progression through the Regulation of miR-613/HK2 Axis. Cancer Manag Res. 2020;12:3661–3672. doi:10.2147/CMAR.S243300
152. Lu J, Zhu Y, Qin Y, et al. <p>CircNFIX Acts as a miR-212-3p sponge to enhance the malignant progression of non-small cell lung cancer by up-regulating ADAM10. Cancer Manag Res. 2020;12:9577–9587. doi:10.2147/CMAR.S272309
153. Shi Q, Ji T, Ma Z, et al. Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother Radiopharm. 2021. doi:10.1089/cbr.2020.4490
154. Ding Z, Guo L, Deng Z, et al. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19(3):269–279. doi:10.1016/j.aohep.2020.01.002
155. Fang Q, Liu H, Zhou A, et al. <p>Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manag Res. 2020;12:6849–6860. doi:10.2147/CMAR.S253510
156. Li Y, Zang H, Zhang X, et al. <p>Exosomal Circ-ZNF652 promotes cell proliferation, migration, invasion and glycolysis in hepatocellular carcinoma via miR-29a-3p/GUCD1 axis. Cancer Manag Res. 2020;12:7739–7751. doi:10.2147/CMAR.S259424
157. Liu W, Yin C, Liu Y. Circular RNA circ_0091579 promotes hepatocellular carcinoma proliferation, migration, invasion, and glycolysis through miR-490-5p/CASC3 axis. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2019.3472
158. Liu Y, Li R, Wang X, et al. <p>CircTTBK2 contributes to the progression of glioma through regulating miR-145-5p/CPEB4 axis. Cancer Manag Res. 2020;12:8183–8195. doi:10.2147/CMAR.S263586
159. Zhang K, Hu H, Xu J, et al. Circ_0001421 facilitates glycolysis and lung cancer development by regulating miR-4677-3p/CDCA3. Diagn Pathol. 2020;15(1):133. doi:10.1186/s13000-020-01048-1
160. Che H, Ding H, Jia X. circ_0080145 enhances imatinib resistance of chronic myeloid leukemia by regulating miR-326/PPFIA1 axis. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3600
161. Jia Y, Liu M, Wang S. CircRNA hsa_circRNA_0001776 inhibits proliferation and promotes apoptosis in endometrial cancer via downregulating LRIG2 by sponging miR-182. Cancer Cell Int. 2020;20(1):412. doi:10.1186/s12935-020-01437-y
162. Fang X, Bai Y, Zhang L, et al. Silencing circSLAMF6 represses cell glycolysis, migration, and invasion by regulating the miR-204-5p/MYH9 axis in gastric cancer under hypoxia. Biosci Rep. 2020;40(6):6. doi:10.1042/BSR20201275
163. Li Z, Fu Y, Ouyang W, et al. Circ_0016347 promotes osteosarcoma progression by regulating miR-1225-3p/KCNH1 axis. Cancer Biother Radiopharm. 2021. doi:10.1089/cbr.2019.3349
164. Zhang Y, Zhao L, Yang S, et al. CircCDKN2B-AS1 interacts with IMP3 to stabilize hexokinase 2 mRNA and facilitate cervical squamous cell carcinoma aerobic glycolysis progression. J Exp Clin Cancer Res. 2020;39(1):281. doi:10.1186/s13046-020-01793-7
165. Zhang Y, Shi Z, Li Z, et al. <p>Circ_0057553/miR-515-5p regulates prostate cancer cell proliferation, apoptosis, migration, invasion and aerobic glycolysis by targeting YES1. Onco Targets Ther. 2020;13:11289–11299. doi:10.2147/OTT.S272294
166. Tian S, Han G, Lu L, et al. Circ-FOXM1 contributes to cell proliferation, invasion, and glycolysis and represses apoptosis in melanoma by regulating miR-143-3p/FLOT2 axis. World J Surg Oncol. 2020;18(1):56. doi:10.1186/s12957-020-01832-9
167. Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi:10.1002/1878-0261.12629
168. Huo S, Dou D. Circ_0056285 regulates proliferation, apoptosis and glycolysis of osteosarcoma cells via miR-1244/TRIM44 axis. Cancer Manag Res. 2021;13:1257–1270. doi:10.2147/CMAR.S290645
169. Tai G, Zhang M, Liu F. Circ_0000735 enhances the proliferation, metastasis and glycolysis of non-small cell lung cancer by regulating the miR-635/FAM83F axis. Exp Lung Res. 2021;47(3):136–148. doi:10.1080/01902148.2021.1881188
170. Fang K, Chen X, Qiu F, et al. Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of non-small-cell lung cancer through the miR-638/FAM83F axis. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2019.3534
171. Ding C, Xi G, Wang G, et al. Exosomal Circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS axis. Front Genet. 2020;11:962. doi:10.3389/fgene.2020.00962
172. Wang B, Lu Y, Feng E. hsa_circ_0001610 knockdown modulates miR-646-STAT3 axis to suppress endometrial carcinoma progression. J Gene Med. 2021:e3337.
173. Lu X, Gan Q, Gan C. Circular RNA circSEC24A promotes cutaneous squamous cell carcinoma progression by regulating miR-1193/MAP3K9 axis. Onco Targets Ther. 2021;14:653–666. doi:10.2147/OTT.S275691
174. Ma L, Li H, Lin Y, et al. CircDUSP16 Contributes to Cell Development in Esophageal Squamous Cell Carcinoma by Regulating miR-497-5p/TKTL1 Axis. J Surg Res. 2021;260:64–75. doi:10.1016/j.jss.2020.11.052
175. Wei Y, Zhang Y, Meng Q, et al. Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder cancer cell growth through activation of LDHA. Am J Transl Res. 2019;11(11):6838–6849.
176. Jin C, Dong D, Yang Z, et al. CircMYC regulates glycolysis and cell proliferation in melanoma. Cell Biochem Biophys. 2020;78(1):77–88. doi:10.1007/s12013-019-00895-0
177. Li Y, Zang H, Zhang X, et al. <p>circ_0136666 facilitates the progression of colorectal cancer via miR-383/CREB1 axis. Cancer Manag Res. 2020;12:6795–6806. doi:10.2147/CMAR.S251952
178. Zhang Y, Tang K, Chen L, et al. <p>Exosomal CircGDI2 suppresses oral squamous cell carcinoma progression through the regulation of MiR-424-5p/SCAI axis. Cancer Manag Res. 2020;12:7501–7514. doi:10.2147/CMAR.S255687
179. Ju C, Zhou J, Miao H, et al. Bupivacaine suppresses the progression of gastric cancer through regulating circ_0000376/miR-145-5p axis. BMC Anesthesiol. 2020;20(1):275. doi:10.1186/s12871-020-01179-4
180. Wu M, Sun T, Xing L. Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR-184 to regulate HAMP in Hepatocellular Carcinoma. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3779
181. Zhang X, Li J, Feng Q. CircRNA circYY1 (hsa_circ_0101187) modulates cell glycolysis and malignancy through regulating YY1 expression by sponging miR-769-3p in breast cancer. Cancer Manag Res. 2021;13:1145–1158. doi:10.2147/CMAR.S289172
182. Dong C, Fan B, Ren Z, et al. CircSMARCA5 facilitates the progression of prostate cancer through miR-432/PDCD10 axis. Cancer Biother Radiopharm. 2021;36(1):70–83. doi:10.1089/cbr.2019.3490
183. Liu Y, Chen G, Wang B, et al. Silencing circRNA protein kinase C iota (circ-PRKCI) suppresses cell progression and glycolysis of human papillary thyroid cancer through circ-PRKCI/miR-335/E2F3 ceRNA axis. Endocr J. 2021. doi:10.1507/endocrj.EJ20-0726
184. Wang L, Li H, Qiao Q, et al. Circular RNA circSEMA5A promotes bladder cancer progression by upregulating ENO1 and SEMA5A expression. Aging (Albany NY). 2020;12(21):21674–21686. doi:10.18632/aging.103971
185. Liu J, Liu H, Zeng Q, et al. Circular RNA circ-MAT2B facilitates glycolysis and growth of gastric cancer through regulating the miR-515-5p/HIF-1α axis. Cancer Cell Int. 2020;20(1):171. doi:10.1186/s12935-020-01256-1
186. Chen Z, Wang F, Xiong Y, et al. CircZFR functions as a sponge of miR-578 to promote breast cancer progression by regulating HIF1A expression. Cancer Cell Int. 2020;20(1):400. doi:10.1186/s12935-020-01492-5
187. Guan Y, Cao Z, Du J, et al. Circular RNA circPITX1 knockdown inhibits glycolysis to enhance radiosensitivity of glioma cells by miR-329-3p/NEK2 axis. Cancer Cell Int. 2020;20(1):80. doi:10.1186/s12935-020-01169-z
188. Yue L, Wang G, Zhu M. CircRNA SEPT9 contributes to malignant behaviors of glioma cells via miR-432-5p-mediated regulation of LASP1. Brain Res. 2021;1766:147501. doi:10.1016/j.brainres.2021.147501
189. Liu A, Xu J. Circ_03955 promotes pancreatic cancer tumorigenesis and Warburg effect by targeting the miR-3662/HIF-1α axis. Clin Transl Oncol. 2021. doi:10.1007/s12094-021-02599-5
190. Xu Y, Jiang T, Wu C, et al. CircAKT3 inhibits glycolysis balance in lung cancer cells by regulating miR-516b-5p/STAT3 to inhibit cisplatin sensitivity. Biotechnol Lett. 2020;42(7):1123–1135. doi:10.1007/s10529-020-02846-9
191. Shangguan H, Feng H, Lv D, et al. Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis. 2020;11(6):437. doi:10.1038/s41419-020-2635-5
192. Liu Z, Wang R, Zhu G. Circ_0035483 functions as a tumor promoter in renal cell carcinoma via the miR-31-5p-Mediated HMGA1 upregulation. Cancer Manag Res. 2021;13:693–706. doi:10.2147/CMAR.S282806
193. Lin L, Li N, Hu X, et al. Identification of circ_0085616 as an Upregulated and Oncogenic Circular RNA in Cervical Cancer Via the miR-503-5p-Mediated ATXN7L3 Activation. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3865
194. Gao Y, Ma H, Gao Y, et al. <p>CircRNA Circ_0001721 Promotes the Progression of Osteosarcoma Through miR-372-3p/MAPK7 Axis. Cancer Manag Res. 2020;12:8287–8302. doi:10.2147/CMAR.S244527
195. He S, Yang J, Jiang S, et al. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20(1):404. doi:10.1186/s12935-020-01496-1
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.