Back to Journals » OncoTargets and Therapy » Volume 14
Circular RNA circSEC24A Promotes Cutaneous Squamous Cell Carcinoma Progression by Regulating miR-1193/MAP3K9 Axis
Received 5 August 2020
Accepted for publication 7 December 2020
Published 22 January 2021 Volume 2021:14 Pages 653—666
DOI https://doi.org/10.2147/OTT.S275691
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Federico Perche
Xiaoyan Lu, Quan Gan, Caibin Gan
Department of Dermatological, Xinxiang Central Hospital, Xinxiang City, Henan Province, People’s Republic of China
Correspondence: Xiaoyan Lu
Department of Dermatological, Xinxiang Central Hospital, No. 56 Jinsui Avenue, Xinxiang City, Henan Province 453000, People’s Republic of China
Tel +86-0373-2052459
Email [email protected]
Background: Circular RNAs (circRNAs) have been increasingly demonstrated to play critical roles in cancer progression. However, the biological functions and underlying mechanism of circRNA SEC24 homolog A, COPII coat complex component (circSEC24A) in cutaneous squamous cell carcinoma (CSCC) have not been well elucidated yet.
Methods: The expression levels of circSEC24A, microRNA-1193 (miR-1193) and mitogen-activated protein kinase kinase kinase 9 (MAP3K9) were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and colony formation assay were used to assess cell proliferation ability. Flow cytometry and transwell assay were utilized to detect cell apoptosis and migration and invasion. Glycolytic metabolism was examined via the measurement of lactate production, glucose consumption, extracellular acidification rate (ECAR), hexokinase 2 (HK2) and Lactate dehydrogenase A (LDHA) expression. The interaction between miR-1193 and circSEC24A or MAP3K9 was predicted by starBase v2.0 and verified by dual-luciferase reporter, RNA Immunoprecipitation (RIP) and pull-down assay assays. The mice xenograft model was established to investigate the roles of circSEC24A in vivo.
Results: CircSEC24A and MAP3K9 were upregulated and miR-1193 was downregulated in CSCC tissues and cells. CircSEC24A knockdown inhibited the progression of CSCC cells by inhibiting cell proliferation, migration, invasion, and glycolysis and inducing apoptosis. Moreover, miR-1193 was a direct target of circSEC24A and its downregulation reversed the inhibitory effect of circSEC24A knockdown on the progression of CSCC cells. Furthermore, MAP3K9 was a downstream target of miR-1193 and its upregulation attenuated the anti-cancer role of miR-1193 in CSCC cells. Additionally, circSEC24A acted as a molecular sponge of miR-1193 to regulate MAP3K9 expression. Furthermore, interference of circSEC24A repressed tumor growth via upregulating miR-1193 and downregulating MAP3K9.
Conclusion: CircSEC24A interference suppressed the progression of CSCC by regulating miR-1193/MAP3K9 axis, which might be a promising strategy for CSCC treatment.
Keywords: cutaneous squamous cell carcinoma, circSEC24A, miR-1193, MAP3K9
Introduction
Cutaneous squamous cell carcinoma (CSCC) is the second most common skin cancer leading to 10–20% of skin cancer-related deaths.1 Although outstanding advances have been made in the treatment of CSCC in recent decades, the 5-year overall survival of CSCC patients is still unsatisfactory (25%-50%) due to tumor aggressive and distal metastasis.2,3 Hence, it is crucial to understand the molecular mechanism of CSCC progression and find effective therapeutic strategies.
Circular RNAs (circRNAs) are a special type of non-coding RNAs that form a continuous covalently closed loop without 5ʹ-end cup and 3ʹ-end ploy A tail (unlike lncRNA).4 Currently, growing evidence has suggested that circRNAs are more stable and difficult to degrade in response to the RNA exonuclease due to their closed-loop structures.5 CircRNAs have been confirmed to be related to the occurrence and development of many diseases, including cancers.6,7 For instance, circRNA circAGFG1 facilitated the progression of triple-negative breast cancer by regulating miR-195-5p/CCNE1 axis.8 Moreover, circRNA circPDSS1 promoted gastric cancer progression via regulating miR-186-5p/NEK2 axis.9 CircRNA SEC24 homolog A, COPII coat complex component (circSEC24A; also known as hsa_circ_0003528, chr5:134032815–134044578) is derived from back-splicing of SEC24A transcript and has been suggested to be upregulated in CSCC.10 Nevertheless, the exact roles and regulatory mechanism of circSEC24A in CSCC have not been reported.
It is well known that circRNAs usually serve as competing endogenous RNAs (ceRNAs) to suppress the functions of their target microRNAs (miRNAs) through binding with miRNAs.11 MiRNAs usually bind to the 3ʹ-untranslated regions (3ʹ-UTR) of target mRNAs to inhibit target gene expression and can act as tumor facilitator or inhibitor in many types of cancer.12 Hence, circRNAs binding to miRNAs can regulate target gene expression and form a regulatory circRNA-miRNA-mRNA network. A previous study showed that miR-1193 was downregulated in CSCC and can inhibit the progression of CSCC.13 Moreover, mitogen-activated protein kinase kinase kinase 9 (MAP3K9) has been demonstrated to act as a tumor-promoting gene in CSCC.14 Interestingly, starBase v2.0 predicted that circSEC24A and MAP3K9 shared the complementary binding sequence for miR-1193, which prompted us to assume the ceRNA network of circSEC24A/miR-1193/MAP3K9.
In this research, we analyzed the levels of circSEC24A, miR-1193 and MAP3K9 in CSCC tissues and cells. Additionally, we investigated their biological functions in the progression of CSCC by gain- and loss-of-function experiments. Furthermore, the potential regulatory network of circSEC24A/miR-1193/MAP3K9 axis was also explored in CSCC cells. We aimed to identify promising therapeutic targets for treatment of CSCC and explore a novel mechanism for better understanding the pathogenesis of CSCC.
Materials and Methods
Specimens Collection
The clinical specimens including CSCC tissues (n=55) and adjacent normal tissues (n=55) were provided by the patients who had undergone surgery at Xinxiang Central Hospital. These patients were divided into early pathological stages I+II (n=23) and advanced pathological stages III+IV (n=32) based on histopathological evaluation. These patients did not receive radiotherapy or chemotherapy prior to surgical resection. These samples were harvested, promptly frozen in liquid nitrogen and then kept in a refrigerator at −80°C after surgical resection. Informed consent had acquired from each patient. This research was authorized by the Research Ethics Committee of Xinxiang Central Hospital.
Cell Culture and Transfection
Human keratinocyte cell line (HaCaT) was obtained from CLS Cell Line Service GmbH (Eppelheim, Germany), and two CSCC cell lines (SCC13 and Colo16) were bought from COBIOER (Nanjing, China) and cultured in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA) at 37°C under a humidified atmosphere containing 5% CO2.
The small interfering RNAs (siRNA) against circSEC24A (si-circSEC24A) and corresponding control (si-NC), miR-1193 mimic (miR-1193) and corresponding control (miR-NC), miR-1193 inhibitor (anti-miR-1193) and corresponding control (anti-miR-NC), MAP3K9 overexpression vector (MAP3K9) and corresponding control (vector) were obtained from Genecreat (Wuhan, China). Lentivirus-mediated short hairpin RNA (shRNA) targeting circSEC24A (sh-circSEC24A) and corresponding control (sh-NC) were constructed by RiboBio (Guangzhou, China). SCC13 and Colo16 cells were seeded into seeded in six-well plates (5 × 105 cells/well) and then transfected with oligonucleotide (50 nM miRNA mimic/inhibitor and 20 nM siRNA) or plasmid (2 μg) using the Lipofectamine® 3000 reagent (Invitrogen) when cell confluence reached 60–70%. The cells were collected for subsequent experimentation following 24 h of transfection at 37°C.
GSE Microarray Analysis
CircRNA expression profiles of CSCC (GSE74758; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) were obtained from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo). In the study, punch biopsies from patients with CSCC (n=3) and non-lesional skin (n=3) were included. Microarray based circRNA expression profiles were acquired using Arraystar circRNA Arrays V. 2.0.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA form cells (HaCaT, SCC13 and Colo16) and tissues (human tumor, human normal, and mice xenograft model tumor) was extracted with Trizol reagent (Invitrogen). Next, complementary DNA (cDNA) was synthesized by One Step PrimeScript cDNA Synthesis Kit (Takara, Shiga, Japan) and microRNA First-Strand cDNA Synthesis Kit (Sangon Biotech, Shanghai, China). Then, qRT-PCR reactions were performed using the Premix Ex Taq II kit (Takara) on 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). In this study, primers used for amplification were listed as follows: circSEC24A (Forward, 5ʹ-GCTCTCCTTAAACAGGATATACACAA-3ʹ; Reverse, 5ʹ-TGTCCACTGAGAAGGAATAAGTCA-3ʹ); SEC24A (Forward, 5ʹ-TTCCTTCTCAGTGGACAGTATTC-3ʹ; Reverse, 5ʹ-GCTGATGATGGTAAGAGGGATAG-3ʹ); miR-1193 (Forward, 5ʹ-ACACTCCAGCTGGGATAGACCGGTGACGTGC-3ʹ; Reverse, 5ʹ-CTCAACTGGTGTCGTGGA-3ʹ); MAP3K9 (Forward, 5ʹ-ATGCAAGATTTGGCTGTCCTTC-3ʹ; Reverse, 5ʹ-TGGTAGTAATATGAGAACTAC-3ʹ); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Forward, 5ʹ-GGAGAAGCTGAGTCATGGGT-3ʹ; Reverse, 5ʹ-GGTGGGAGATCTGGTTTCCG-3ʹ), U6 (Forward, 5ʹ-GTGCGTGTCGTGGAGTCG-3ʹ; Reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ). The relative expression of circSEC24A, SEC24A, MAP3K9, and miR-1193 was evaluated with 2-ΔΔCt method, and the expression of circSEC24A, SEC24A and MAP3K9 was normalized by GAPDH and miR-1193 level was normalized by U6.
RNase R Treatment
In order to analyze the stability of circSEC24A and its linear isoform. Total RNA (2 μg) was incubated using the RNase R (3 U/μg, Epicentre Technologies, Madison, WI, USA) for 30 min at 37°C. After that, qRT-PCR was conducted to test the expression levels of circSEC24A and SEC24A.
Cell Viability Assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) analysis was utilized for measuring cell viability. Briefly, SCC13 and Colo16 cell suspension (100 µL, 2×103 cells) were seeded in the 96-well plates overnight. After transfection at different times, MTT reagent (20 μL, 5 mg/mL, Sangon Biotech) was added to per well using a pasteur pipette (20 μL), followed by incubation for 3–4 h. After removing cultured medium, methyl sulfoxide solution (DMSO; 150 μL) was added to each well using a pasteur pipette (200 μL). Finally, a microplate reader (Bio-Teck, Winooski, VT, USA) was applied to examine the absorbance at 490 nm.
Colony Formation Assay
Briefly, SCC13 and Colo16 cells (300 cells/well) suspended in 2 mL of complete medium (DMEM containing 10% FBS) were seeded into each well of six-well plates for 24 h and then transfected with si-circSEC24A, si-circSEC24A + anti-miR-1193, miR-1193, miR-1193 + MAP3K9, or their corresponding controls. The medium was changed with fresh medium (DMEM containing 10% FBS) after transfection for 48 h and updated every 3 days. After 2 weeks later, the cells were carefully washed with pre-cold phosphate-buffered saline (PBS) and fixed using the 4% paraformaldehyde for 30 min at 4°C, followed by staining with 0.1% crystal violet for 2 h at room temperature. Next, the cells were again washed with PBS until per well was clear. Finally, the number of colonies containing at least 50 cells was counted by a microscope (Olympus, Tokyo, Japan).
Cell Apoptosis Assay
According to the recommendations, Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Sangon Biotech) was utilized for detecting cell apoptosis. In brief, SCC13 and Colo16 cells suspended in 2 mL of complete medium (DMEM containing 10% FBS) were seeded into 6-well plates with 5×105 cells/well. After 48 h of incubation, cells were harvested, followed by staining with Annexin V-FITC and PI. Next, a flow cytometry (Partec AG, Arlesheim, Switzerland) was employed for measuring apoptotic cells.
Transwell Assay
Transwell chamber (Corning Incorporation, Corning, NY, USA) was pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA) for evaluating cell invasion capacity and the uncoated membrane was employed to assess cell migration. Briefly, SCC13 and Colo16 cells (2×104 cells/well) suspended in serum-free medium (DMEM, 200 µL) were placed in the top chambers, while the bottom chamber was filled with 600 µL of 10% FBS medium. Following incubation for 24 h, cells remained on the top surface of the membrane were carefully wiped off by a cotton wool, while cells passed through the filter were fixed using paraformaldehyde (4% for 15 min), followed by staining with crystal violet (0.1% for 1 h). Lastly, an inverted microscope (100× magnification) was applied to count and photograph the migrated and invaded cells.
Measurement of Lactate Production and Glucose Consumption
SCC13 and Colo16 cells (5×105 cells/well) suspended in 2 mL of complete medium (DMEM containing 10% FBS) were seeded into six-well plates for 24 h and transfected with si-circSEC24A, si-circSEC24A + anti-miR-1193, miR-1193, miR-1193 + MAP3K9, or their matched controls. Cell culture media were collected 48 h after the transfection. Based on the manufacturer’s instructions, the lactate production and glucose consumption were examined by lactate assay kit (BioVision, Mountain View, CA, USA) and glucose assay kit (Sigma-Aldrich, St. Louis, MO, USA), respectively. Finally, the glucose consumption or lactate production was determined using a standard calibration curve and normalized to total protein.
Extracellular Acidification Rate (ECAR) Assay
ECAR was determined by a Seahorse Bioscience XF24 extracellular flux analyzer (Seahorse Bioscience, North Billerica, MA, USA). Briefly, SCC13 and Colo16 cells (2×104 cells/well) were seeded into XF microplate, followed by the measurement of baseline. At the indicated time points, glucose, oxidative phosphorylation inhibitor oligomycin (OM) and 2glycolytic inhibitor (2-DG) were then injected into per well, respectively, as manufacturer’s recommendation.
Western Blot Assay
Cells (HaCaT, SCC13 and Colo16) and tissues (human tumor, human normal, and mice xenograft model tumor) were lysed using RIPA lysis buffer (Solarbio, Beijing, China) for extracting the total protein. BCA protein assay kit (Solarbio) was employed to evaluate protein concentration, and then an equal amount (40 µg) of protein was resolved by separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by being transferred onto polyvinylidene fluoride membranes. These membranes were blocked using the 5% non-fat milk (Sangon Biotech) before being incubated with primary antibody against hexokinase 2 (HK2) (1:8000, ab227198, Abcam, Cambridge, UK), Lactate dehydrogenase A (LDHA) (1:2000, ab125683, Abcam), MAP3K9 (1:1000, ab154506, Abcam) or GAPDH (1:1000, ab37168, Abcam) for 12 h at 4°C. Next, membranes were then continuously incubated with secondary antibody (1:4000, ab205718, Abcam). At last, the protein signal was visualized using the enhanced chemiluminescence reagent (Solarbio). The protein levels were normalized by GAPDH, and ImageJ software was employed to assess the bands density.
Bioinformatics Analysis and Dual-Luciferase Reporter Assay
The potential complementary sequence of miR-1193 and circSEC24A or MAP3K9 was predicted by starbase v2.0 (http://starbase.sysu.edu.cn/). The wild-type (WT) luciferase reporter plasmids of circSEC24A and MAP3K9 3ʹ-UTR were obtained by inserting the sequences of circSEC24A or MAP3K9 3ʹ-UTR containing the binding sequence for miR-1193 into pmirGLO luciferase reporter vector (Promega, Madison, WI, USA). The mutant (MUT) luciferase reporter plasmids of circSEC24A and MAP3K9 3ʹ-UTR were obtained by mutating the seed sites of miR-1193. SCC13 and Colo16 cells were co-transfected with the constructed reporter plasmid (WT or MUT) and miR-1193 or miR-NC for 48 h. At last, dual-Luciferase Reporter Assay System (Promega) was applied for measuring the firefly and Renilla luciferase activities.
RNA Immunoprecipitation (RIP) Assay
RIP experiment was performed using EZ-Magna RIP Kit (Millipore, Bedford, MA, USA) to validate the interaction between circSEC24A and miR-1193. In brief, SCC13 and Colo16 cells were lysed by complete RIP lysis buffer. After that, cell lysate (100 µL) was incubated by RIP buffer supplemented with magnetic beads conjugated with human anti-Argonaute2 (AGO2) or anti-immunoglobulin G (IgG) antibody. Subsequently, proteinase K was employed for separating the RNA-protein complexes from beads. Next, immunoprecipitated RNA was extracted, and qRT-PCR was conducted to detect the expression of miR-1193 and circSEC24A.
RNA Pull-Down Assay
Biotin labeled miR-1193 or circSEC24A (bio-miR-1193 or bio-circSEC24A) and negative control (bio-NC) were generated from RiboBio and incubated with cell lysates, followed by incubation with streptavidin beads (Invitrogen). Then, the bound RNAs were eluted and used for detecting the enrichment of circSEC24A and MAP3K9 by qRT-PCR.
In vivo Tumor Growth Assay
SCC13 cells were transfected with sh-circSEC24A or sh-NC. SCC13 cells with stable knockdown or control cells (5×106) were injected subcutaneously into BALB/c nude mice (n=5/group, female, 5–6 weeks, Huafukang, Beijing, China). The animal experiments obtained approval from the Animal Care and Use Committee of Xinxiang Central Hospital, and performed according to the guidelines for laboratory animal welfare (GB T 35892–2018). Tumor volume was detected at the indicated times and calculated following the equation: volume = (length× width2)/2. Mice were killed following injection for 4 weeks, and the formed tumors were excised, weighed and harvested for further analysis.
Statistical Analysis
All data were shown as the mean ± standard deviation (SD) from at least three independent experiments. Statistical data analysis was performed with GraphPad Prism 6.0. Student’s t-test or a one-way analysis of variance (ANOVA) was utilized to analyze significant differences between different groups. Kaplan-Meier method was used to generate survival curve. Statistical significance was considered when P< 0.05.
Results
CircSEC24A Was Upregulated in CSCC Tissues and Cells
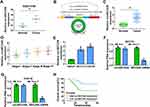
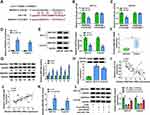
Firstly, we analyzed GSE74758 from the GEO database to explore the relative expression of circSEC24A in CSCC tissues and normal tissues. The data showed that the circSEC24A expression was enhanced in CSCC tissues compared to normal tissues (Figure 1A). CircSEC24A (hsa_circ_0003528) is generated from exons 14–18 of SEC24A gene and is located on chromosome 5, whose spliced mature sequence length is 741 bp (Figure 1B). To further confirm the expression of circSEC24A in CSCC tissues and cells, qRT-PCR was conducted. The results presented that circSEC24A was overexpressed in CSCC tissues (n=55) relative to normal tissues (n=55) (Figure 1C). Moreover, circSEC24A expression in patients with pathological stages I, II, III, and IV stages were presented in Figure 1D, and we found that circSEC24A expression was positively related to the pathological stage. Similarly, circSEC24A level was increased in CSCC cells (SCC13 and Colo16) cells compared with that in human keratinocyte cell line (HaCaT) (Figure 1E). Moreover, to verify that circSEC24A was a circular RNA, SCC13 and Colo16 cells were treated with RNase R. According to the qRT-PCR analysis, compared with the linear SEC24A, circSEC24A was resistant to RNase R in SCC13 and Colo16 cells (Figure 1F and G), suggesting that circSEC24A had a loop structure. Moreover, we further divided 55 CSCC patients into high circSEC24A (n=28) and low circSEC24A (n=27) expression groups according to median value of circSEC24A expression. As displayed in Figure 1H, patients with high circSEC24A level had shorter overall survival than those with low circSEC24A level (P=0.0089). To summarize, circSEC24A was confirmed to be a circular RNA and might be a prognosis marker.
Knockdown of circSEC24A Inhibited the Progression of CSCC Cells
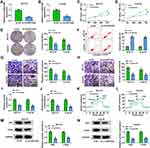
To explore the biological roles of the circSEC24A in CSCC cells, we performed loss-of-function experiments by transfection of si-circSEC24A in SCC13 and Colo16 cells. As displayed in Figure 2A and B, qRT-PCR results revealed that the expression of circSEC24A was decreased in SCC13 and Colo16 cells transfected with si-circSEC24A compared to those cells transfection with si-NC, suggesting that si-circSEC24A was successfully introduced into SCC13 and Colo16 cells. MTT and colony formation analyses showed that circSEC24A knockdown reduced cell viability and the number of colonies in SCC13 and Colo16 cells (Figure 2C–E), suggesting that circSEC24A downregulation inhibited SCC13 and Colo16 cell proliferation. In addition, flow cytometry results manifested that circSEC24A interference accelerated SCC13 and Colo16 cell apoptosis (Figure 2F). Moreover, wound healing assay (Supplementary material and methods) and transwell assay demonstrated that migration and invasion of SCC13 and Colo16 cells were suppressed by circSEC24A silence (Figure 2G and H, Supplementary Figure 1A and 1B). Glycolysis has been reported to exert the positive role in the progression of CSCC cells,15 hence, we examine whether circSEC24A had a role in glycolysis in CSCC cells. We observed that knockdown of circSEC24A inhibited lactate production and glucose consumption in SCC13 and Colo16 cells (Figure 2I and J). ECAR approximated the glycolysis flux rate.16 The results showed that circSEC24A silence decreased glycolysis rate and glycolytic capacity (Figure 2K and L), indicating that circSEC24A knockdown decreased the ECAR in SCC13 and Colo16 cells. Next, glycolytic enzymes (HK2 and LDHA) were analyzed by Western blot in SCC13 and Colo16 cells. The results indicated that circSEC24A downregulation suppressed the protein levels of HK2 and LDHA (Figure 2M and N). All these findings suggested that circSEC24A knockdown could inhibit proliferation, migration, invasion and glycolysis but induced apoptosis in CSCC cells.
CircSEC24A Served as a Sponge of miR-1193 in CSCC Cells
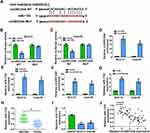
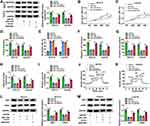
Previous study reported that circRNAs could act as a molecular sponge to interact with miRNAs,17 so the potential target miRNAs of circSEC24A were predicted by starbase v2.0. We selected 5 miRNAs (miR-1193, miR-217, miR-192-5p, miR-215-5p, and miR-381-3p) with relatively high prediction scores. Next, we explored the effect of circSEC24A on expression of potential target miRNAs. We found that the expression of miR-1193 increased most significantly after knockdown circSEC24A (Supplementary Figure 1C and 1D). Therefore, miR-1193 was selected for subsequent studies. The results presented that circSEC24A had binding sequence with miR-1193 (Figure 3A), indicating that circSEC24A might act as a sponge of miR-1193. To confirm the direct binding between circSEC24A and miR-1193, dual-luciferase reporter, RIP and RNA pull-down assays were performed. Results from dual-luciferase reporter assay suggested that miR-1193 overexpression markedly reduced the luciferase activity of circSEC24A WT, but had no obvious impact on the luciferase activity of circSEC24A MUT (Figure 3B and C). RIP assay indicated that the levels of circSEC24A and miR-1193 were strikingly enriched in AGO2 group in contrast to IgG control group (Figure 3D and E). Moreover, RNA pull-down assay showed that the enrichment of circSEC24A was increased after treatment with bio-miR-1193 in SCC13 and Colo16 cells compared with bio-NC group (Figure 3F), suggesting that miR-1193 could pull down circSEC24A. Subsequently, the effect of circSEC24A on miR-1193 expression was explored. We found that silencing circSEC24A promoted the expression of miR-1193 in SCC13 and Colo16 cells (Figure 3G). Moreover, we observed that the level of miR-1193 was significantly lower in CSCC tissues and cells than in normal tissues and cells (Figure 3H and I). Furthermore, we observed that circSEC24A expression was negatively correlated with miR-1193 level in CSCC tissues (Figure 3J). Collectively, these data suggested that miR-1193 was a direct target of circSEC24A in CSCC cells.
Downregulation of circSEC24A Suppressed the Progression of CSCC Cells by Sponging miR-1193
To explore whether circSEC24A exerted its biological functions via sponging miR-1193, SCC13 and Colo16 cells were transfected with si-NC, si-circSEC24A, si-circSEC24A + anti-miR-NC, or si-circSEC24A + anti-miR-1193. The results of qRT-PCR showed that the expression of miR-1193 was enhanced after transfection with si-circSEC24A in SCC13 and Colo16 cells, while this effect was reversed by transfection of anti-miR-1193 (Figure 4A and B). Moreover, knockdown of miR-1193 partially abolished the inhibitory effects of si-circSEC24A on cell viability and colony formation ability (Figure 4C–E). Meanwhile, miR-1193 silence abated the anti-migration, anti-invasion and pro-apoptosis effects caused by si-circSEC24A (Figure 4F–H). Furthermore, the suppressive effects of circSEC24A downregulation on lactate production, glucose consumption, ECAR, HK2 and LDHA expression were mitigated by downregulating miR-1193 (Figure 4I–N). Collectively, these findings demonstrated that circSEC24A regulated proliferation, migration, invasion, apoptosis, and glycolysis by sponging miR-1193.
MAP3K9 Was a Downstream Target of miR-1193 in CSCC Cells
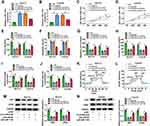
MiRNAs were well known to play critical roles in a variety of cancers via suppressing their target genes. We next used bioinformatics tool (starbase v2.0) to predict potential target genes of miR-1193. We selected 5 mRNAs (MAP3K9, SOX4, RAB23, PTEN, and FOSL1) with relatively high prediction scores. Subsequently, the effect of miR-1193 on expression of potential target mRNAs was explored. We found that the expression of MAP3K9 decreased most significantly by overexpression of miR-1193 (Supplementary Figure 1E and 1F). As presented in Figure 5A, 3ʹ-UTR of MAP3K9 had several binding sites within miR-1193, suggesting that MAP3K9 might be a target of miR-1193. Subsequently, the targeting relationship between MAP3K9 and miR-1193 was identified using the dual-luciferase reporter and RNA pull-down assays. We found that forced expression of miR-1193 inhibited the luciferase activity of MAP3K9 3ʹ-UTR-WT, while the luciferase activity of MAP3K9 3ʹ-UTR-MUT was unaffected by transfection of miR-1193 (Figure 5B and C). RNA pull-down assay showed that bio-miR-1193 led to higher MAP3K9 enrichment than treatment of bio-NC in SCC13 and Colo16 cells (Figure 5D). Moreover, we observed that the protein expression of MAP3K9 was reduced in SCC13 and Colo16 cells transfected with miR-1193 compared with those cells transfected with miR-NC (Figure 5E), suggesting that MAP3K9 expression was negatively regulated by miR-1193. The results from qRT-PCR and Western blot indicated that the mRNA and protein expression of MAP3K9 were enhanced in CSCC tissues compared to normal tissues (Figure 5F and G). Likewise, the protein level of MAP3K9 was also increased in CSCC cells (SCC13 and Colo16) in contrast to HaCaT cells (Figure 5H). Meanwhile, we observed a negative correlation between MAP3K9 mRNA expression and miR-1193 expression in CSCC tissues and a positive correlation between MAP3K9 mRNA expression and circSEC24A expression in CSCC tissues (Figure 5I and J). RNA pull-down assay demonstrated that MAP3K9 expression was enriched by bio-circSEC24A (Figure 5K), indicating that MAP3K9 could interact with circSEC24A in SCC13 and Colo16 cells. Next, we further elucidated the relationships among circSEC24A, miR-1193 and MAP3K9 in CSCC cells. The results showed that MAP3K9 protein expression was inhibited by downregulating circSEC24A, while knockdown of miR-1193 rescued the effect (Figure 5L), suggesting that circSEC24A positively modulated MAP3K9 expression by sponging miR-1193. Altogether, these data proved that circSEC24A acted as a molecular sponge of miR-1193 to regulate the expression of MAP3K9.
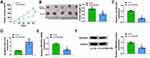
Overexpression of miR-1193 Suppressed the Progression of CSCC Cells by Targeting MAP3K9
To explore whether the effect of miR-1193 on the progression of CSCC was mediated by MAP3K9, SCC13 and Colo16 cells were transfected with miR-NC, miR-1193, miR-1193 + vector, or miR-1193 + MAP3K9. Western blot assay indicated that the protein expression of MAP3K9 was reduced after transfection with miR-1193, which could be reversed by addition of MAP3K9 (Figure 6A). Moreover, overexpression of miR-1193 inhibited cell viability and colony formation in SCC13 and Colo16 cells, while these effects were abated by upregulation of MAP3K9 (Figure 6B–D). In addition, enforced expression of miR-1193 facilitated apoptosis and limited migration and invasion of SCC13 and Colo16 cells, which was restored by overexpression of MAP3K9 (Figure 6E–G). Besides, restoration of miR-1193 inhibited lactate production, glucose consumption, ECAR, HK2 and LDHA expression, while these effects were also reversed by upregulating MAP3K9 (Figure 6H–M). Taken together, these findings demonstrated that miR-1193 exerted its biological roles in CSCC cells by targeting MAP3K9.
Interference of MAP3K9 Restrained the Tumor Growth by Regulating miR-1193 and MAP3K9 Expression
To assess the effect of circSEC24A on CSCC in vivo, sh-NC- or sh-circSEC24A-transfected SCC13 cells were introduced into nude mice. The results demonstrated that silence of circSEC24A significantly decreased tumor volume and weight when compared with the sh-NC group (Figure 7A and B). Moreover, our results indicated that interference of circSEC24A significantly downregulated circSEC24A expression and upregulated miR-1193 expression in tumor tissues (Figure 7C and D). Additionally, we found that knockdown of circSEC24A decreased the mRNA and protein expression of MAP3K9 in tumor tissues (Figure 7E and F). Thus, these data suggested that silencing circSEC24A inhibited tumor growth through upregulating miR-1193 and downregulating MAP3K9.
Discussion
CSCC incidence and mortality increase year by year.18 In recent years, dysregulation of circRNA has been suggested to be associated with the initiation and development of diverse cancers, including CSCC.19 In this work, we showed that circSEC24A interference repressed the progression of CSCC via regulating miR-1193/MAP3K9 axis.
Previous studies have suggested that circRNAs are abundant in eukaryotic cells and recognized as significant prognostic biomarkers for tumors due to their abundance and stability.4,20 Although the exact roles of the majority of most circRNAs in CSCC remain undefined, some reports suggested that certain circRNAs could influence the occurrence and development of CSCC. An et al reported that circ_0070934 upregulation facilitated growth and invasion of CSCC cells via sponging miR-1238 and miR-1247-5p.21 Sand et al demonstrated that 143 circRNAs were upregulated, including circSEC24A, and 179 circRNAs were downregulated in CSCC.10 However, the exact functions of circSEC24A in CSCC are still unclear. Here, we also observed that circSEC24A level was elevated in CSCC tissues and cells. Functionally, circSEC24A silence limited the progression of CSCC cells through inhibiting proliferation, migration, invasion and glycolysis and promoting apoptosis in CSCC cells. These findings suggested that circSEC24A acted as an oncogene in CSCC.
Emerging evidence has elaborated that circRNAs can act as miRNA sponges to exert a variety of biological effects as well as repress the activity of miRNAs and modulate their downstream target genes expression in various cancers.4,22 So, starBase v2.0 software was applied to predict the target miRNAs of circSEC24A, and the results indicated that circSEC24A might bind with miR-1193, which was verified through a series of experiments in CSCC cells. Previous reports proved that miR-1193 served as an anti-oncogene in diverse tumors. For instance, Li et al stated that miR-1193 suppressed breast cancer cell growth and metastasis via targeting IGF2BP2.23 Moreover, Chen et al clarified that miR-1193 repressed the progression of glioma cells through regulating IGF2BP2 expression.24 Shen et al showed that miR-1193 limited the growth and invasion of human T-cell leukemia cells via modulating TM9SF3.25 More importantly, Wang et al proved that low expression of miR-1193 was observed in CSCC cells, and it was sponged by LINC00963 and inhibited the progression of CSCC cells by targeting SOX4.13 In line with this research, we uncovered that miR-1193 level was also declined in CSCC tissues and cells. Moreover, downregulation of miR-1193 reversed the suppressive impact of si-circSEC24A on the progression of CSCC. Altogether, these findings indicated that circSEC24A silence suppressed the progression of CSCC cells by upregulation of miR-1193.
Increasing studies have implied that miRNAs can participate in various biological and pathological processes by regulating target genes expression.26 Therefore, we screened the mRNAs that contained complementary sequences of miR-1193 by starBase v2.0 software. The data showed that MAP3K9 might bind with miR-1193. Next, MAP3K9 was demonstrated as a direct target of miR-1193. MAP3K9 has been found to serve as a tumor promoter in serval cancers, including renal cancer,27 pancreatic cancer28 and esophagus squamous cell carcinoma.29 Moreover, Luo et al reported that miR-148a significantly inhibited the progression of CSCC cells by targeting MAP3K9.14 In this research, we uncovered that MAP3K9 expression was elevated in CSCC tissues and cells. Moreover, rescue experiments indicated that MAP3K9 could abate the anti-cancer role of miR-1193 in CSCC cells. Additionally, circSEC24A positively regulated MAP3K9 expression by sponging miR-1193. In vivo experiments demonstrated that circSEC24A interference restrained tumor growth via upregulation of miR-1193 and downregulation of MAP3K9. Collectively, these results suggested that circSEC24A downregulation limited the progression of CSCC by regulating miR-1193/MAP3K9 axis.
In conclusion, we discovered that circSEC24A and MAP3K9 were overexpressed whilst miR-1193 was lowly expressed in CSCC tissues and cells. Moreover, circSEC24A silence repressed the progression of CSCC by upregulating miR-1193 and downregulating MAP3K9. Our study might contribute to a better understanding of the underlying mechanism of CSCC progression and offer a promising circRNA-targeted therapy for CSCC.
Abbreviations
circSEC24A, circRNA SEC24 homolog A; CSCC, cutaneous squamous cell carcinoma; miR-1193, microRNA-1193; MAP3K9, mitogen-activated protein kinase kinase kinase 9; ECAR, extracellular acidification rate; HK2, hexokinase 2; LDHA, Lactate dehydrogenase A.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Ratushny V, Gober MD, Hick R, Ridky TW, Seykora JT. From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. 2012;122(2):464–472. doi:10.1172/JCI57415
2. Keyal U, Bhatta AK, Zhang G, Wang XL. Present and future perspectives of photodynamic therapy for cutaneous squamous cell carcinoma. J Am Acad Dermatol. 2019;80(3):765–773. doi:10.1016/j.jaad.2018.10.042
3. Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344(13):975–983. doi:10.1056/NEJM200103293441306
4. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333. doi:10.1038/nature11928
5. Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
6. Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8(12):353.
7. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
8. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):1–19. doi:10.1186/s12943-018-0933-7
9. Ouyang Y, Li Y, Huang Y, et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234(7):10458–10469. doi:10.1002/jcp.27714
10. Sand M, Bechara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83(3):210–218. doi:10.1016/j.jdermsci.2016.05.012
11. Bach D-H, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
12. Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161.
13. Wang J, Li C, Xu L, Yang C, Zhang X. MiR-1193 was sponged by LINC00963 and inhibited cutaneous squamous cell carcinoma progression by targeting SOX4. Pathology. 2019;215(10):152600. doi:10.1016/j.prp.2019.152600
14. Luo Q, Li W, Zhao T, Tian X, Liu Y, Zhang X. Role of miR-148a in cutaneous squamous cell carcinoma by repression of MAPK pathway. Arch Biochem Biophys. 2015;583:47–54. doi:10.1016/j.abb.2015.07.022
15. McCoy DE, Feo T, Harvey TA, et al. Structural absorption by barbule microstructures of super black bird of paradise feathers. Nat Commun. 2018;9(1):1–17. doi:10.1038/s41467-017-02088-w
16. Dranka BP, Benavides GA, Diers AR, et al. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 2011;51(9):1621–1635. doi:10.1016/j.freeradbiomed.2011.08.005
17. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
18. Samarasinghe V, Madan V, Lear JT. Management of high-risk squamous cell carcinoma of the skin. Expert Rev Anticancer Ther. 2011;11(5):763–769. doi:10.1586/era.11.36
19. Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. doi:10.1186/s12943-017-0598-7
20. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
21. An X, Liu X, Ma G, Li C. Upregulated circular RNA circ_0070934 facilitates cutaneous squamous cell carcinoma cell growth and invasion by sponging miR-1238 and miR-1247–5p. Biochem Biophys Res Commun. 2019;513(2):380–385. doi:10.1016/j.bbrc.2019.04.017
22. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
23. Li X, Li Y, Lu H. MiR-1193 suppresses proliferation and invasion of human breast cancer cells through directly targeting IGF2BP2. Oncol Res Featuring Preclin Clin Cancer Therapeutics. 2017;25(4):579–585. doi:10.3727/97818823455816X14760504645779
24. Chen Z, Wu C, Li X, Zhang X, Huang L. miR-1193 Inhibits Glioma Cells Proliferation, Migration, and Invasion via Targeting IGF2BP2. J Biomater Tissue Eng. 2018;8(11):1558–1565. doi:10.1166/jbt.2018.1904
25. Shen L, Du X, Ma H, Mei S. miR-1193 suppresses the proliferation and invasion of human T-cell leukemia cells through directly targeting the transmembrane 9 superfamily 3 (TM9SF3). Oncol Res Featuring Preclin Clin Cancer Therapeutics. 2017;25(9):1643–1651. doi:10.3727/096504017X14908284471361
26. Felekkis K, Touvana E, Stefanou C, Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14(4):236.
27. Nie F, Liu T, Zhong L, et al. MicroRNA-148b enhances proliferation and apoptosis in human renal cancer cells via directly targeting MAP3K9. Mol Med Report. 2016;13(1):83–90. doi:10.3892/mmr.2015.4555
28. Xia J, Cao T, Ma C, et al. miR-7 suppresses tumor progression by directly targeting MAP3K9 in pancreatic cancer. Molecular Therapy - Nucleic Acids. 2018;13:121–132. doi:10.1016/j.omtn.2018.08.012
29. Zhang B, Yu T, Yu Z, Yang X. MicroRNA-148a regulates the MAPK/ERK signaling pathway and suppresses the development of esophagus squamous cell carcinoma via targeting MAP3K9. Eur Rev Med Pharmacol Sci. 2019;23(15):6497–6504. doi:10.26355/eurrev_201908_18533
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.