Back to Journals » Cancer Management and Research » Volume 13
CircPRDM2 Contributes to Doxorubicin Resistance of Osteosarcoma by Elevating EZH2 via Sponging miR-760
Authors Yuan J, Liu Y, Zhang Q, Ren Z, Li G, Tian R
Received 1 December 2020
Accepted for publication 8 May 2021
Published 2 June 2021 Volume 2021:13 Pages 4433—4445
DOI https://doi.org/10.2147/CMAR.S295147
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Jianjun Yuan,* Yan Liu,* Quan Zhang, Zhishuai Ren, Guang Li, Rong Tian
Department of Spine Surgery, Tianjin Union Medical Center, Tianjin, 300121, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Rong Tian
Department of Spine Surgery, Tianjin Union Medical Center, No. 190, Jieyuan Road, Hongqiao District, Tianjin, 300121, People’s Republic of China
Tel +86-022-27557253
Email [email protected]
Background: Circular RNAs (circRNAs) are implicated in the chemoresistance of human cancers. However, the functions of circRNA PR/SET domain 2 (circPRDM2) in the resistance of osteosarcoma (OS) to doxorubicin (DXR) are unknown.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) assay was conducted to determine the levels of circPRDM2, microRNA-760 (miR-760) and enhancer of zeste homolog 2 (EZH2). RNase R assay was used to analyze the characteristics of circPRDM2. IC50 of DXR was estimated by Cell Counting Kit-8 (CCK-8) assay. Colony formation assay was performed for cell colony formation ability. Wound-healing assay and transwell assay were utilized for cell migration and invasion. Flow cytometry analysis was conducted for cell apoptosis. Western blot assay was employed for protein levels. Dual-luciferase reporter assay, RNA immunoprecipitation (RIP) assay and RNA pull-down assay were adopted to analyze the relationships among circPRDM2, miR-760 and EZH2. Murine xenograft model assay was utilized to explore DXR resistance in vivo.
Results: CircPRDM2 level was enhanced in DXR-resistant OS tissues and cells. CircPRDM2 deficiency inhibited IC50 of DXR, colony formation, migration and invasion and facilitated apoptosis in DXR-resistant OS cells in vitro. CircPRDM2 was identified as the sponge for miR-760. MiR-760 inhibition reversed the inhibitory effects of circPRDM2 knockdown on DXR resistance and cell progression in DXR-resistant OS cells. Moreover, EZH2 was identified as the target gene of miR-760 and EZH2 overexpression abolished miR-760-mediated impacts on DXR sensitivity and malignant behaviors in DXR-resistant OS cells. Also, circPRDM2 silencing improved DXR sensitivity in vivo.
Conclusion: Our study demonstrated the role of circPRDM2/miR-760/EZH2 axis in enhancing DXR resistance.
Keywords: OS, DXR, circPRDM2, miR-760, EZH2
Introduction
Osteosarcoma (OS) is a common primary bone malignancy, mainly occurring in children and adolescents.1,2 Due to advancements in surgery and chemotherapy, the overall survival of OS has been significantly improved over the past decades.3,4 Doxorubicin (DXR) is one of the first-line chemotherapeutic drugs for OS; however, the acquisition of drug resistance limited its clinical efficacy.5,6 Thus, a better understanding of the mechanisms behind OS chemoresistance is critical to discover more effective treatment methods for OS.
Circular RNAs (circRNAs) are a series of covalently closed non-coding RNA molecules, which are more stable than linear RNA.7 CircRNAs can act as the sponges for microRNAs (miRNAs) to regulate gene expression at the transcriptional or post-transcriptional level.8,9 Many studies have suggested that circRNAs are linked to tumor progression and drug sensitivity in human cancers, including OS. For example, circ_0003496 promoted the DXR resistance and malignancy of OS by decoying miR-370 and elevating KLF12.10 Circ_0000073 enhanced OS cell growth and motility and improved methotrexate resistance of OS by the modulation of miR-145-5p/NRAS axis or miR-151-3p/NRAS axis.11 Hierarchical cluster analysis and heat map analysis showed that circRNA PR/SET domain 2 (circPRDM2) (also named as circ_0005986) level was elevated in OS tissues, indicating the potential role of circPRDM2 in OS development.12 However, the biological roles of circPRDM2 in the carcinogenesis and chemoresistance of OS are unacknowledged.
MiRNAs are small non-coding RNAs that exert essential functions in cancer biology and chemoresistance by regulating target gene expression.13,14 As a member of miRNAs, miR-760 has been verified to be associated with the chemoresistance of human cancers. For instance, Wang et al confirmed that miR-760 repressed colorectal cancer (CRC) progression and 5-FU resistance by targeting DCP1A.15 Yang et al unraveled that miR-760 was able to decrease gemcitabine resistance in pancreatic cancer by binding to ITGB1.16 In OS, Yin et al demonstrated that miR-760 served as a tumor inhibitor via interacting with HDGF.17 Even so, the exploration of miR-760 on drug resistance remains dismal.
Enhancer of zeste homolog 2 (EZH2) belongs to the histone methyltransferase family and plays a vital role in tumor carcinogenesis.18 Moreover, EZH2 was reported to exert vital functions in the carcinogenesis and chemosensitivity of OS.19 In this study, by analyzing bioinformatics prediction softwares starbase and DianaTools=microT_CDS, we found that miR-760 shared the binding sites with circPRDM2 and EZH2. However, the relationships among circPRDM2, miR-760 and EZH2 are unclear.
In the research, the expression profiles of circPRDM2, miR-760 and EZH2 in chemoresistant OS tissues and cells were determined. Moreover, their functional functions in the development and chemoresistance of OS were investigated.
Materials and Methods
Tissue Acquisition
In total, 43 OS tissue specimens and adjacent nontumor tissue specimens were acquired from OS patients (including 18 chemoresistant and 13 chemosensitive patients) who received doxorubicin (DXR)-based chemotherapy before surgery at Tianjin Union Medical Center. The chemoresistant and chemosensitivity patients were classified according to the Huvos scoring system.20,21 The specimens were preserved at −80°C until use. The work obtained approval from the Ethics Committee of Tianjin Union Medical Center. Written informed consents were offered by the patients or their guardians.
Cell Culture
OS cells (KHOS and MG63) and osteoblast cells (hFOB1.19) acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA) were incubated at 37°C in an incubator containing 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Gibco). The DXR-resistant OS cells (KHOS/DXR and MG63/DXR) were generated by exposing KHOS and MG63 cells to serially increased doses of DXR (Solarbio, Beijing, China). To retain the DXR-resistant phenotype, 1 μg/mL DXR (Solarbio) was supplemented into the culture media.
Cell Transfection
CircPRDM2 small interfering RNA (si-circPRDM2#1 and si-circPRDM2#2) and si-NC, miR-760 mimics (miR-760) and miR-NC, miR-760 inhibitors (in-miR-760) and in-miR-NC, EZH2 overexpression plasmid (EZH2) and pcDNA control plasmid, circPRDM2 short hairpin RNA (sh-circPRDM2) and sh-NC were synthesized by GenePharma (Shanghai, China) and then transfected into cells utilizing Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ instructions.
RNA Isolation, RNase R Treatment and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
Total RNA was extracted through the usage of RNAiso Plus (Takara, Dalian, China). RNase R treatment was performed on total RNA using RNase R (3 U/μg; Epicenter Biotechnologies, Madison, WI, USA) for 20 min at 37°C. Next, cDNAs were synthesized utilizing PrimeScript™ RT reagent Kit (Takara) or All-in-One™ miRNA First-Strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA). QRT-PCR was then executed with SYBR Premix Ex Taq II (Takara). The relative expression was computed through the 2−ΔΔCt strategy with GAPDH or U6 as the negative control. The primers were as follows: circPRDM2: 5ʹ-ATTTGGGATGGATGTGCATT-3ʹ and R: 5ʹ-CAACAGCAGAAGGGAAAAGC-3ʹ); miR-760: (F: 5ʹ-CGGCTCTGGGTCTGTGGGGA-3ʹ and R: 5ʹ-CTCTACAGCTATATTGCCAGCCA-3ʹ); EZH2: (F: 5ʹ-GTACACGGGGATAGAGAATGTGG-3ʹ and R: 5ʹ-GGTGGGCGGCTTTCTTTATCA-3ʹ); GAPDH: (F: 5ʹ-GGGAGCCAAAAGGGTCAT-3ʹ and R: 5ʹ-GAGTCCTTCCACGATACCAA-3ʹ); U6: (F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
Cell Counting Kit-8 (CCK-8) Assay
The 50% inhibitory concentration (IC50) of DXR was assessed by CCK-8 assay (Boster, Wuhan, China). In brief, the transfected KHOS/DXR and MG63/DXR cells were seeded into 96-well plates with 5×103 cells/well and cultured with different doses of DXR (Solarbio) for 48 h. Next, 10 µL CCK-8 was supplemented into each well and maintained for a further 3 h. The absorption was measured at 450 nm with a microplate reader (Potenov, Beijing, China). IC50 was defined as the concentration of chemotherapeutic drugs that repressed cell proliferation by 50%.
Colony Formation Assay
DXR-resistant OS cells with various transfections were plated into 6-well plates. The media were changed every 3 days. After 2 weeks, the colonies were fixed in 4% paraformaldehyde (Sangon, Shanghai, China) followed by crystal violet (Sangon) staining for 15 min. At last, the colony number was counted using a microscope (Olympus, Tokyo, Japan).
Wound-Healing Assay
The transfected DXR-resistant OS cells were cultured in 6-well plates with 5×105 cells/well for 24 h. Then, the wounds were made using a pipette tip. After 24 h, the images were captured with a microscope (Olympus). The scratch area was measured to evaluate cell migration ability.
Transwell Assay
To test cell invasion and migration, the transwell chambers (Corning Incorporated, Corning, NY, USA) coated with or without Matrigel (Corning Incorporated) were used, respectively. The transfected KHOS/DXR and MG63/DXR cells (2×104) were suspended in 200 μL serum-free medium and then plated into the upper chambers. The lower chambers were added with DMEM (Gibco) containing 10% FBS (Gibco). 24 h later, the cells that invaded/migrated into the lower chambers were stained with crystal violet (Sangon) for 15 min and quantified under a microscope (100×; Olympus).
Flow Cytometry Analysis
The transfected DXR-resistant OS cells were harvested and washed with PBS (Solarbio). Next, the cells were suspended and stained with Annexin V-fluorescein isothiocyanate (FITC; Beyotime, Shanghai, China) and propidium iodide (PI; Beyotime) for 15 min in the dark according to the protocols. The apoptotic cells were analyzed with FACScan® flow cytometry (BD Biosciences, San Jose, CA, USA).
Western Blot Assay
The tissues and cells were lysed in RIPA buffer (Beyotime) to extract the total protein. The extracted proteins were examined with a BCA Protein Quantification Kit (Beyotime). Next, 10% sodium dodecyl sulfonate-polyacrylamide gel (Solarbio) was employed to separate the proteins, which were then blotted onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After 1 h of blockage in 5% skim milk, the membranes were probed with primary antibodies overnight at 4°C and then cultivated with the relevant secondary antibody (ab6728; Abcam, Cambridge, MA, USA) at indoor temperature for 2 h. The bands were visualized utilizing an ECL kit (Beyotime). All primary antibodies including multidrug resistance protein 1 (MRP1; ab32574), P-glycoprotein (P-gp; ab170904), lung-resistance-related protein (LPR; ab97311), EZH2 (ab186006) and β-actin (ab8224) were purchased from Abcam.
Dual-Luciferase Reporter Assay
The luciferase reporter vectors (circPRDM2 WT, circPRDM2 MUT, EZH2 3ʹUTR WT and EZH2 3ʹUTR MUT) were generated by cloning the sequences of circPRDM2 or EZH2 3ʹUTR including the wild-type or mutant miR-760 binding sites into pmirGLO plasmid (Promega, Fitchburg, WI, USA). Then, KHOS/DXR and MG63/DXR cells were co-transfected with the generated plasmids and miR-760/miR-NC and cultured for 48 h. Finally, the luciferase activity was detected with dual-Luciferase Reporter Assay Kit (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP assay was done utilizing Magna RNA-binding protein immunoprecipitation kit (Millipore) in line with the protocols. In brief, KHOS/DXR and MG63/DXR cell extracts were prepared and inoculated with magnetic beads coated with Anti-immunoglobulin G (Anti-IgG) or Anti-Argonaute-2 (Anti-AGO2). The enrichment of miR-760 and circPRDM2 was detected with the aforementioned qRT-PCR assay after RNA was isolated from RNA-protein complexes.
RNA Pull-Down Assay
The Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for RNA pull-down experiment. In brief, biotinylated circPRDM2 (Bio-circPRDM2 WT), biotinylated mutant circPRDM2 (Bio-circPRDM2 MUT), biotinylated EZH2 (Bio-EZH2 WT), biotinylated mutant EZH2 (Bio-EZH2 MUT) or biotinylated NC (Bio-NC) was transfected into KHOS/DXR and MG63/DXR cells and cultivated for 48 h. Next, cell lysates were inoculated with streptavidin-coated magnetic beads (Invitrogen). At last, the enrichment of miR-760 in co-precipitated RNAs was examined with qRT-PCR analysis.
Murine Xenograft Model
The BALB/c nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) were divided into 4 groups (n=5/group). About 2×106 MG63/DXR cells with sh-NC or sh-circPRDM2 transfection were resuspended in PBS (Solarbio) and then injected into the right back of nude mice. One week later, the mice were administrated with 2 mg/kg DXR (Solarbio) or PBS (Solarbio) once a week. The tumor volume was examined every week. The mice were sacrificed and the xenografted tumors were weighted after 4 weeks. The animal experiments were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of “National Institutes of Health” and approved by the Ethics Committee of Animal Research of Tianjin Union Medical Center.
Statistical Analysis
The data obtained from three independent experiments were analyzed by GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA) and expressed as mean ± standard deviation. The differences were estimated with Student’s t-test or one-way analysis of variance. Survival curve of patients was generated by Kaplan-Meier plot and analyzed by Log rank test. P<0.05 indicated a statistically significant difference.
Results
CircPRDM2 Was Upregulated in DXR-Resistant OS Tissues and Cells
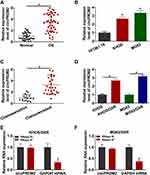
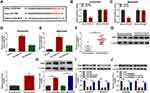
As shown in Figure 1A and B, circPRDM2 level was markedly increased in OS tissues and cells compared to normal tissues and cells. According to circPRDM2 expression in tumor tissues, the patients with OS were divided to 2 groups: High circPRDM2 expression group (n=22) and Low circPRDM2 expression group (n=21). We found that the patients in the high circPRDM2 expression group had worse overall survival and disease free survival than patients in the low circPRDM2 expression group (Figure S1A and B). Then, to explore the functions of circPRDM2 in the chemoresistance of OS, the expression of circPRDM2 in DXR-resistant OS tissues was detected by qRT-PCR assay. The results exhibited that circPRDM2 level was elevated in DXR-resistant OS tissues compared to DXR-sensitive OS tissues (Figure 1C). Next, we established DXR resistant OS cells (KHOS/DXR and MG63/DXR cells). CCK-8 assay showed that IC50 of DXR was markedly increased in KHOS/DXR and MG63/DXR cells compared to KHOS and MG63 cells, indicating the acquisition of DXR resistance of KHOS/DXR and MG63/DXR cells (Figure S2A and B). Moreover, it was found that circPRDM2 was highly expressed in KHOS/DXR and MG63/DXR cells relative to KHOS and MG63 cells (Figure 1D). In addition, we found that circPRDM2 was resistant to RNase R treatment, while GAPDH was evidently digested by RNase R, indicating that circPRDM2 possessed a ring structure (Figure 1E and F). All these results suggested that circPRDM2 might play a role in the chemoresistance of OS.
CircPRDM2 Knockdown Inhibited the Resistance of DXR-Resistant OS Cells to DXR
To explore the exact roles of circPRDM2 in DXR resistance of OS, KHOS/DXR and MG63/DXR cells with circPRDM2 silencing were constructed by transfecting si-circPRDM2#1 or si-circPRDM2#2 into KHOS/DXR and MG63/DXR cells. As a result, the transfection of si-circPRDM2#1 or si-circPRDM2#2 markedly reduced circPRDM2 expression in KHOS/DXR and MG63/DXR cells compared to si-NC control groups (Figure 2A). IC50 determination showed that circPRDM2 knockdown reduced DXR resistance in KHOS/DXR and MG63/DXR cells (Figure 2B). As illustrated by cell colony formation assay, the colony formation capacity of KHOS/DXR and MG63/DXR cells was repressed following circPRDM2 silencing compared to control groups (Figure 2C). The results of wound-healing assay indicated that KHOS/DXR and MG63/DXR cells transfected with si-circPRDM2#1 or si-circPRDM2#2 had a wider scratch wound after 24 h in comparison with si-NC control groups, indicating cell migration ability was repressed after circPRDM2 knockdown (Figure 2D). Transwell assay showed that circPRDM2 silencing restrained the migration and invasion of KHOS/DXR and MG63/DXR cells (Figure 2E and F). Flow cytometry analysis demonstrated that circPRDM2 deficiency facilitated cell apoptosis in KHOS/DXR and MG63/DXR cells compared to control groups (Figure 2G). Moreover, our results showed that silencing of circPRDM2 decreased the levels of drug resistance-related proteins (MRP1, P-gp and LRP) in KHOS/DXR and MG63/DXR cells compared to control groups (Figure 2H and I). Collectively, circPRDM2 knockdown inhibited the resistance of DXR-resistant OS cells to DXR.
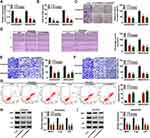
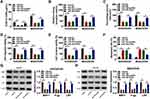
CircPRDM2 Directly Interacted with miR-760
In order to explore the underlying mechanism of circPRDM2 in DXR resistance of OS cells, starbase was used to analyze the target of circPRDM2. Bioinformatics analysis showed that circPRDM2 harbored the binding sites of miR-760 (Figure 3A). As displayed in Figure 3B, miR-760 transfection increased miR-760 expression and in-miR-760 transfection decreased miR-760 expression in KHOS/DXR and MG63/DXR cells. Then, dual-luciferase reporter assay, RIP assay and RNA pull-down assay were performed to verify the interaction between circPRDM2 and miR-760. Dual-luciferase reporter assay showed that miR-760 overexpression evidently reduced the luciferase activity of circPRDM2 WT groups instead of circPRDM2 MUT groups in KHOS/DXR and MG63/DXR cells (Figure 3C and D). RIP assay showed that miR-760 and circPRDM2 were markedly enriched in Anti-AGO2 pellets compared to Anti-IgG groups (Figure 3E and F). Moreover, RNA pull-down assay exhibited that miR-760 was notably pulled down by Bio-circPRDM2 WT but not the mutated oligos, further confirming the combination between circPRDM2 and miR-760 (Figure 3G and H). Indeed, miR-760 was weakly expressed in DXR-resistant OS tissues and cells compared to corresponding normal tissues and cells (Figure 3I and J). Furthermore, we observed that circPRDM2 silencing elevated the expression of miR-760 in KHOS/DXR and MG63/DXR cells, but miR-760 inhibition reversed the effect (Figure 3K). Taken together, circPRDM2 directly targeted miR-760 to negatively regulate miR-760 expression.
CircPRDM2 Knockdown Suppressed DXR Resistance in DXR-Resistant OS Cells by Targeting miR-760
We then investigated whether circPRDM2 could alter DXR resistance in DXR-resistant OS cells by targeting miR-760. As exhibited in Figure 4A, circPRDM2 knockdown reduced IC50 of DXR in KHOS/DXR and MG63/DXR cells, while miR-760 inhibition reversed the effect. The results of colony formation assay indicated that miR-760 downregulation ameliorated the inhibitory effect of circPRDM2 silencing on the colony formation ability of KHOS/DXR and MG63/DXR cells (Figure 4B). As illustrated by wound-healing assay and transwell assay, circPRDM2 deficiency restrained the migration and invasion of KHOS/DXR and MG63/DXR cells, whereas these effects were abrogated by reducing miR-760 (Figure 4C–E). Our results also showed that circPRDM2 knockdown-induced apoptosis of KHOS/DXR and MG63/DXR cells was abated by the downregulation of miR-760 (Figure 4F). Additionally, circPRDM2 silencing decreased the protein levels of MRP1, P-gp and LRP in KHOS/DXR and MG63/DXR cells, with miR-760 inhibition reversed the impacts (Figure 4G and H). These findings suggested that circPRDM2 knockdown enhanced DXR sensitivity of DXR-resistant OS cells to DXR by sponging miR-760.
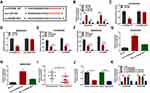
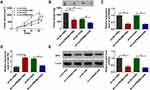
EZH2 Was the Direct Target Gene of miR-760
To further clarify the underlying mechanism of circPRDM2/miR-760 axis in the chemoresistance of OS, we performed bioinformatics analysis using DianaTools=microT_CDS and found that miR-760 contained the binding sites of EZH2 (Figure 5A). As demonstrated by dual-luciferase reporter assay, miR-760 elevation led to a notable suppression in the luciferase activity of EZH2 3ʹUTR WT in KHOS/DXR and MG63/DXR cells, but the luciferase activity in EZH2 3ʹUTR MUT groups was not affected (Figure 5B and C). RNA pull-down assay showed that miR-760 level was remarkably pulled down by Bio-EZH2 WT in KHOS/DXR and MG63/DXR cells compared to Bio-NC and Bio-EZH2 MUT groups (Figure 5D and E). These results demonstrated the interaction between miR-760 and EZH2. Thereafter, we found that EZH2 mRNA and protein levels were elevated in DXR-resistant OS tissues compared to DXR-sensitive OS tissues (Figure 5F and G). Moreover, compared to KHOS and MG63 cells, EZH2 protein level was enhanced in KHOS/DXR and MG63/DXR cells (Figure 5H). MiR-760 overexpression led to a marked reduction in EZH2 protein level in KHOS/DXR and MG63/DXR cells, while this effect was weakened by the elevation of EZH2 (Figure 5I). Of note, circPRDM2 silencing decreased the protein level of EZH2 in KHOS/DXR and MG63/DXR cells, while miR-760 inhibition reversed the impact (Figure 5J). To summarize, circPRDM2 positively regulated EZH2 expression by targeting miR-760.
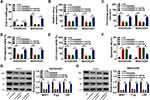
Overexpression of miR-760 Promoted the Sensitivity of DXR-Resistant OS Cells to DXR by Interacting with EZH2
As shown in Figure 6A, miR-760 overexpression suppressed DXR resistance in KHOS/DXR and MG63/DXR cells, while this effect was ameliorated after EZH2 elevation. Then, we found that the colony formation of KHOS/DXR and MG63/DXR cells was repressed by the overexpression of miR-760, with EZH2 upregulation reversed this impact (Figure 6B). Wound healing assay and transwell assay indicated that miR-760 overexpression restrained the migration and invasion of KHOS/DXR and MG63/DXR cells, but EZH2 elevation abated the effects (Figure 6C–E). Flow cytometry analysis showed that KHOS/DXR and MG63/DXR cells with miR-760 overexpression showed a promotion in cell apoptosis, whereas this effect was overturned by elevating EZH2 (Figure 6F). Moreover, overexpression of miR-760 reduced MRP1, P-gp and LRP protein levels in KHOS/DXR and MG63/DXR cells, while EZH2 upregulation reversed the impacts (Figure 6G and H). These results indicated that miR-760 suppressed DXR resistance in DXR-resistant OS cells by targeting EZH2.
CircPRDM2 Knockdown Inhibited DXR Resistance of OS in vivo
To determine the function of circPRDM2 in DXR resistance in vivo, the murine xenograft model was established. MG63/DXR cells transfected with sh-NC or sh-circPRDM2 were injected into the mice and treated with PBS or DXR every week. It was found that circPRDM2 knockdown and DR treatment remarkably repressed tumor volume and tumor weight (Figure 7A and B). The results of qRT-PCR assay showed that circPRDM2 level was reduced and miR-760 level was elevated in the tumor tissues harvested from sh-circPRDM2 groups compared to sh-NC groups (Figure 7C and D). Moreover, we found that EZH2 protein level was decreased in the tumor tissues harvested from sh-circPRDM2 groups compared to sh-NC groups (Figure 7E). Collectively, circPRDM2 knockdown enhanced DXR sensitivity in vivo.
Discussion
Chemoresistance is one of the main obstacles in cancer treatment.6 Clarifying the mechanisms behind the development of drug resistance will guide us to make more effective and specific efforts for cancer therapy. Currently, the involvement of circRNAs in tumor progression and chemoresistance has been gradually identified with the development of high-throughput sequencing and bioinformatics technology. Even so, the functions of circRNAs in the chemoresistance of OS are largely unknown. In the present study, we clarified the effects and related mechanisms of circPRDM2 in DXR resistance of OS. Our results demonstrated that circPRDM2 contributed to the resistance of OS to DXR via miR-760/EZH2 axis.
Diverse circRNAs, such as circ_001569,22 circ_0004674,23 circ_000307424 and circ_0003496,10 have been reported to play vital roles in the drug resistance of OS. Previous research indicated that circ_0005986 suppressed the malignancy of hepatocellular carcinoma by sponging miR-129-5p.25 Liu et al claimed that circPRDM2 was abnormally upregulated in OS patients.12 Herein, circPRDM2 was found to be abnormally upregulated in OS tissues and chemoresistant OS tissues. Thus, we speculated that circPRDM2 might play a role in the chemoresistance of OS. Thereafter, we performed experiments to explore the roles of circPRDM2 in the chemoresistance of OS. High expression of circPRDM2 in OS patients was related to poor prognosis as well as tumor recurrence and metastasis, indicating that circPRDM2 might be a diagnostic and prognostic marker for OS. Functionally, circPRDM2 silencing inhibited DXR resistance, cell growth and motility, as well as accelerated cell apoptosis in DXR-resistant OS cells in vitro. The multidrug resistance (MDR)-related proteins are related to the resistance of OS to drugs.26 Thus, we measured the protein levels of MRP1, P-gp and LRP in DXR-resistant OS cells. It was found that circPRDM2 deficiency led to a remarkable reduction in MRP1, P-gp and LRP levels in DXR-resistant OS cells. Furthermore, we demonstrated that circPRDM2 downregulation enhanced DXR sensitivity in vivo. Cancer stem cells (CSCs) are relatively rare for several tumor types and have the capabilities of self-renewal and differentiation.27 Previous studies showed that CSCs facilitated tumorigenesis and chemoresistance in human cancers, including OS.28–31 However, whether circPRDM2 can regulate the properties of CSCs to participate in tumor formation and drug resistance in OS remains unclear.
Afterward, the underlying mechanism of circPRDM2 in regulating DXR resistance of OS was investigated. We found that circPRDM2 served as the sponge for miR-760, which directly targeted EZH2. Moreover, circPRDM2 positively modulated EZH2 expression in DXR-resistant OS cells by decoying miR-760. MiR-760 has been verified to act as a tumor suppressor in diverse cancers, such as CRC,32 glioma,33 gastric cancer,34 as well as OS.17 Moreover, miR-760 could regulate the chemoresistance of cancers through lncRNA KCNQ1OT1/miR-760/PPP1R1B axis,35 miR-760/ITGB1 axis16 and circ_0007031/miR-760/DCP1A axis.15 However, it remains unclear about the effects and regulatory mechanisms of miR-760 in DXR resistance in OS. Our data indicated that miR-760 downregulation abated the suppressive roles of circPRDM2 knockdown in DXR resistance and cell malignant characteristics in DXR-resistant OS cells, indicating circPRDM2 enhanced DXR resistance by sponging miR-760. EZH2 is an epigenetic regulator and has been reported to be targeted by miR-138 to participate in tumor progression and chemoresistance in OS.19 However, the relationship between miR-760 and EZH2 was investigated for the first time. Herein, we demonstrated that miR-760 overexpression improved DXR sensitivity and repressed cell progression in DXR-resistant OS cells, while EZH2 elevation reversed the impacts.
In conclusion, circPRDM2 contributed to the resistance of OS to DXR through regulating miR-760 and EZH2. The study deepened our understandings on the incidence of DXR resistance in OS and might provide new opportunities for the therapeutic interventions of OS.
Data Sharing Statement
The underlying data is available from the corresponding author upon reasonable request.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Ther. 2017;4(1):25–43. doi:10.1007/s40744-016-0050-2
2. Chen L, Zou X, Wang Y, Mao Y, Zhou L. Central nervous system tumors: a single center pathology review of 34,140 cases over 60 years. BMC Clin Pathol. 2013;13(1):14. doi:10.1186/1472-6890-13-14
3. Federman N, Bernthal N, Eilber FC, Tap WD. The multidisciplinary management of osteosarcoma. Curr Treat Options Oncol. 2009;10(1–2):82–93. doi:10.1007/s11864-009-0087-3
4. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. doi:10.1080/14737140.2018.1413939
5. Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol. 2014;16:15–23. doi:10.1016/j.coph.2014.02.002
6. Chou AJ, Gorlick R. Chemotherapy resistance in osteosarcoma: current challenges and future directions. Expert Rev Anticancer Ther. 2006;6(7):1075–1085. doi:10.1586/14737140.6.7.1075
7. Das A, Gorospe M, Panda AC. The coding potential of circRNAs. Aging (Albany NY). 2018;10(9):2228–2229. doi:10.18632/aging.101554
8. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
9. Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32(7):923–925. doi:10.1038/emboj.2013.53
10. Xie C, Liang G, Xu Y, Lin E. Circular RNA hsa_circ_0003496 contributes to tumorigenesis and chemoresistance in osteosarcoma through targeting (microRNA) miR-370/kruppel-like factor 12 axis. Cancer Manag Res. 2020;12:8229–8240. doi:10.2147/CMAR.S253969
11. Li X, Liu Y, Zhang X, et al. Circular RNA hsa_circ_0000073 contributes to osteosarcoma cell proliferation, migration, invasion and methotrexate resistance by sponging miR-145-5p and miR-151-3p and upregulating NRAS. Aging (Albany NY). 2020;12(14):14157–14173. doi:10.18632/aging.103423
12. Liu X, Zhong Y, Li J, Shan A. Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget. 2017;8(70):114829–114838. doi:10.18632/oncotarget.22162
13. Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–587. doi:10.1016/j.molmed.2006.10.006
14. Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer–a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013
15. Wang Y, Wang H, Zhang J, et al. Circ_0007031 serves as a sponge of miR-760 to regulate the growth and chemoradiotherapy resistance of colorectal cancer via regulating DCP1A. Cancer Manag Res. 2020;12:8465–8479. doi:10.2147/CMAR.S254815
16. Yang D, Hu Z, Xu J, et al. MiR-760 enhances sensitivity of pancreatic cancer cells to gemcitabine through modulating Integrin beta1. Biosci Rep. 2019;39(11):BSR20192358. doi:10.1042/BSR20192358
17. Yin R, Liu J, Zhao D, Wang F. Long non-coding RNA ASB16-AS1 functions as a miR-760 sponge to facilitate the malignant phenotype of osteosarcoma by increasing HDGF expression. Onco Targets Ther. 2020;13:2261–2274. doi:10.2147/OTT.S240022
18. Tsang DP, Cheng AS. Epigenetic regulation of signaling pathways in cancer: role of the histone methyltransferase EZH2. J Gastroenterol Hepatol. 2011;26(1):19–27. doi:10.1111/j.1440-1746.2010.06447.x
19. Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One. 2016;11(3):e0150026. doi:10.1371/journal.pone.0150026
20. Ferrari S, Ruggieri P, Cefalo G, et al. Neoadjuvant chemotherapy with methotrexate, cisplatin, and doxorubicin with or without ifosfamide in nonmetastatic osteosarcoma of the extremity: an Italian sarcoma group trial ISG/OS-1. J Clin Oncol. 2012;30(17):2112–2118. doi:10.1200/JCO.2011.38.4420
21. Vijayanarasimha D, Nayanar SK, Vikram S, Patil VM, Babu S, Satheesan B. Clinico-pathological study of limb salvage surgery for osteosarcoma: experience in a rural cancer center. Indian J Surg Oncol. 2017;8(2):136–141. doi:10.1007/s13193-016-0547-8
22. Zhang H, Yan J, Lang X, Zhuang Y. Expression of circ_001569 is upregulated in osteosarcoma and promotes cell proliferation and cisplatin resistance by activating the Wnt/beta-catenin signaling pathway. Oncol Lett. 2018;16(5):5856–5862. doi:10.3892/ol.2018.9410
23. Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin Z, Jian-Ping H, Tai-Cheng Z. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing. Epigenomics. 2018;10(10):1327–1346. doi:10.2217/epi-2018-0023
24. Lei S, Xiang L. Up-regulation of circRNA hsa_circ_0003074 expression is a reliable diagnostic and prognostic biomarker in patients with osteosarcoma. Cancer Manag Res. 2020;12:9315–9325. doi:10.2147/CMAR.S262093
25. Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8(27):43878–43888. doi:10.18632/oncotarget.16709
26. Li S, Sun W, Wang H, Zuo D, Hua Y, Cai Z. Research progress on the multidrug resistance mechanisms of osteosarcoma chemotherapy and reversal. Tumour Biol. 2015;36(3):1329–1338. doi:10.1007/s13277-015-3181-0
27. Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi:10.1016/j.stem.2007.08.014
28. Li B, Lu Y, Wang H, et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother. 2016;79:93–101. doi:10.1016/j.biopha.2016.01.045
29. Wang K, Hu YB, Zhao Y, Ye C. LncRNA ANRIL regulates ovarian cancer progression and tumor stem cell-like characteristics via miR-324-5p/ran axis. Onco Targets Ther. 2021;14:565–576. doi:10.2147/OTT.S273614
30. Qian L, Liu F, Chu Y, et al. MicroRNA-200c nanoparticles sensitized gastric cancer cells to radiotherapy by regulating PD-L1 expression and EMT. Cancer Manag Res. 2020;12:12215–12223. doi:10.2147/CMAR.S279978
31. Cheng M, Duan PG, Gao ZZ, Dai M. MicroRNA487b3p inhibits osteosarcoma chemoresistance and metastasis by targeting ALDH1A3. Oncol Rep. 2020;44(6):2691–2700. doi:10.3892/or.2020.7814
32. Cao L, Liu Y, Wang D, et al. MiR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J Exp Clin Cancer Res. 2018;37(1):83. doi:10.1186/s13046-018-0757-8
33. Li Q, Lu J, Xia J, Wen M, Wang C. Long non-coding RNA LOC730100 enhances proliferation and invasion of glioma cells through competitively sponging miR-760 from FOXA1 mRNA. Biochem Biophys Res Commun. 2019;512(3):558–563. doi:10.1016/j.bbrc.2019.03.124
34. Liu W, Li Y, Feng S, Guan Y, Cao Y. MicroRNA-760 inhibits cell viability and migration through down-regulating BST2 in gastric cancer. J Biochem. 2020;168(2):159–170. doi:10.1093/jb/mvaa031
35. Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med. 2019;23(6):3808–3823. doi:10.1111/jcmm.14071
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.