Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
Circ_0040929 Serves as Promising Biomarker and Potential Target for Chronic Obstructive Pulmonary Disease
Authors Miao Y, Wu J, Wu R, Wang E, Wang J
Received 1 March 2022
Accepted for publication 4 July 2022
Published 7 September 2022 Volume 2022:17 Pages 2079—2092
DOI https://doi.org/10.2147/COPD.S364553
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Yi Miao,1 Junfang Wu,1 Runmiao Wu,1 Enguang Wang,2 Jing Wang3
1Department of Respiratory Medicine, Shaanxi Provincial People’s Hospital, Xi’an City, 710068, People’s Republic of China; 2Department of Respiratory and Critical Care, the Fifth Affiliated Hospital of Xinjiang Medical University, Urumqi City, 830000, People’s Republic of China; 3Department of Clinical Laboratory, Shaanxi Provincial People’s Hospital, Xi’an City, 710068, People’s Republic of China
Correspondence: Junfang Wu, Department of Respiratory Medicine, Shaanxi Provincial People’s Hospital, No. 256, West Youyi Road, Beilin District, Xi’an City, Shaanxi Province, People’s Republic of China, Tel +86 29-85251331, Email [email protected]
Background: Circular RNAs (circRNAs) can act as essential regulators in many diseases, including chronic obstructive pulmonary disease (COPD). We aimed to explore the role and underlying mechanism of circ_0040929 in COPD.
Methods: A cellular model of COPD was constructed by treating human bronchial epithelial cells (16HBE) with cigarette smoke extract (CSE). The levels of circ_0040929, microRNA-515-5p (miR-515-5p) and insulin-like growth factor-binding protein 3 (IGFBP3) were measured by quantitative real-time PCR. Cell proliferation was assessed by Cell Counting Kit-8 and 5-ethynyl-2’-deoxyuridine assays. Cell apoptosis was evaluated by flow cytometry. Protein expression was measured using Western blot assay. The levels of inflammatory factors and airway remodeling were assayed via enzyme-linked immunosorbent assay. The interaction between miR-515-5p and circ_0040929/IGFBP3 was confirmed by dual-luciferase reporter, RNA pull-down and RNA immunoprecipitation assays. Exosomes were detected using transmission electron microscopy.
Results: Circ_0040929 expression and IGFBP3 expression were upregulated in the serum of smokers (n = 22) compared to non-smokers (n = 22) and more significantly upregulated in the serum of COPD patients (n = 22). However, miR-515-5p expression was decreased in the serum of smokers compared to non-smokers and further reduced in the serum of COPD. Circ_0040929 knockdown attenuated CSE-induced cell injury by increasing proliferation and reducing apoptosis, inflammation, and airway remodeling in 16HBE cells. MiR-515-5p was a direct target of circ_0040929, and miR-515-5p inhibition reversed the effect of circ_0040929 knockdown in CSE-treated 16HBE cells. IGFBP3 was a direct target of miR-515-5p, and miR-515-5p overexpression alleviated CSE-induced cell injury via targeting IGFBP3. Moreover, circ_0040929 regulated IGFBP3 expression by targeting miR-515-5p. Importantly, circ_0040929 was upregulated in serum exosomes from COPD patients.
Conclusion: Circ_0040929 played a promoting role in CSE-induced COPD by regulating miR-515-5p/IGFBP3 axis, suggesting that it might be a novel potential target for COPD treatment.
Keywords: chronic obstructive pulmonary disease, circ_0040929, miR-515-5p, IGFBP3
Introduction
Chronic obstructive pulmonary disease (COPD) is a progressive inflammatory lung disease and is one of the leading causes of death worldwide.1 COPD is characterized by airway limitation and abnormal inflammatory response in lungs caused by multiple risk factors, including cigarette smoke (CS).2 CS can induce airway inflammation in COPD, and the excessive ROS induced by CS can damage the lungs and promote COPD pathogenesis. So far, CS is the most important risk factor for COPD, and smoking cessation can slow down COPD development at the early diagnosed phase.3,4 CS may directly damage pulmonary endothelial cells, which then leads to increased apoptosis and reduced epithelial barrier formation, ultimately resulting in a continuous inflammatory response in lung tissues.5–7 Currently, the effective and specific treatments for COPD have not been fully discovered. Hence, it is essential to study the underlying mechanism of COPD for better prevention and treatment of COPD.
Circular RNA (circRNA) has a unique covalent closed-loop without 5’-end cap and 3’-end poly A tail.8 Due to its closed-loop structure, circRNA is more stable and can avoid degradation by RNases.9 Increasing studies have shown that circRNAs can participate in diverse pathological and physiological processes, including cell proliferation, apoptosis and inflammation.10,11 Many studies have demonstrated that the abnormal expression of circRNAs is tightly associated with multiple human diseases, including COPD.12 For example, circ-RBMS1 might be a potential target for COPD, whose knockdown could alleviate cigarette smoke extract (CSE)-induced human bronchial epithelial cell (16HBE) injury.13 Also, circ-HACE1 could aggravate CSE-induced 16HBE cell apoptosis, inflammatory response and oxidative stress.14 Circ_0040929 is derived from ankyrin repeat domain 11 (ANKRD11) gene and located at chr16:89371613–89383486. A former study revealed that circ_0040929 was overexpressed in CSE-treated human small airway epithelial cells.15 However, the functional role and regulatory mechanism of circ_0040929 in COPD have not been investigated.
MicroRNAs (miRNAs) are a large class of short (~22 nt) noncoding RNAs that can regulate gene expression.16 Recently, the connection between circRNAs and miRNAs has attracted extensive attention. In terms of mechanism, circRNAs can sponge certain miRNAs and prevent the degradation of messenger RNAs targeted by these miRNAs.17 Many miRNAs are reported to be implicated in various diseases, including COPD.18,19 A previous report showed that miR-515-5p could reduce epithelial cell epithelial–mesenchymal transition (EMT) in CS-induced airway remodeling.20 Moreover, insulin-like growth factor-binding protein 3 (IGFBP3) was overexpressed in alveolar epithelial cells treated with CSE, which might be a target for the therapy or prevention of COPD.21 Interestingly, online bioinformatics database showed that both circ_0040929 and IGFBP3 had complementary-binding sequence for miR-515-5p. Therefore, we assumed that circ_0040929/miR-515-5p/IGFBP3 axis might participate in COPD progression.
In this paper, CSE-treated 16HBE cells were used as an in vitro model of COPD. In addition, we investigated the biological functions of circ_0040929, miR-515-5p, IGFBP3 in cell model of COPD. Furthermore, the interactions among circ_0040929, miR-515-5p, and IGFBP3 were explored to reveal an underlying mechanism of circ_0040929 in COPD. We aimed to offer a promising avenue for the treatment of COPD.
Materials and Methods
Serum Collection
Blood samples from COPD patients (n = 22), smokers (n = 22, without COPD), and non-smokers (n = 22, without COPD) were collected at Shaanxi Provincial People’s Hospital. Patients with COPD were diagnosed in accordance with the global Initiative for Chronic Obstructive Pulmonary Disease criteria, and all subjects had no other respiratory diseases. Blood samples were then centrifugated (3000 rpm, 10 min). Next, the supernatant was put in a clean tube and then centrifuged (1500 rpm, 30 min). At last, the final supernatant was kept in a refrigerator at −80°C. The participants provided written informed consent. Demographic and clinicopathological characteristics of the subjects in this study are listed in Table 1. This research had acquired approval from the Research Ethics Committee of Shaanxi Provincial People’s Hospital.
 |
Table 1 Demographic and Clinicopathological Characteristics of the Subjects in This Study |
Preparation of CSE
CSE preparation was performed following the methods described previously.22 Briefly, the smoke from 10 cigarettes was bubbled through 30 mL of RPMI-1640 (Invitrogen, Carlsbad, CA, USA). The resulting suspension was filtered using a 0.22 μm cellulose membrane for removing large particles and the bacteria, which was considered as 100% CSE. This CSE was diluted with medium for obtaining the concentrations of 0.5%, 1%, 2%, and 4% CSE.
Cell Culture and Transfection
16HBE cells purchased from Procell (Wuhan, China) was cultured in RPMI-1640 (Invitrogen) that contained 10% fetal bovine serum (FBS; Invitrogen) in an incubator at 37°C with 5% CO2. For CSE treatment, 16HBE cells were stimulated with various doses (0%, 1%, 2%, and 4%) of CSE, or CSE (2%) for indicated times (0 h, 6 h, 12 h, 24 h, and 36 h).
Small interfering RNA (siRNA) against circ_0040929 (si-circ_0040929) and corresponding control (si-NC), miR-515-5p mimics or inhibitor (miR-515-5p or anti-miR-515-5p) and corresponding control (miR-NC or anti-NC), IGFBP3 overexpression vector (IGFBP3) and corresponding control (vector) were all provided by Genechem (Shanghai, China). The sequences were as follows: si-circ_0040929 (5’-GACACAGCAGACGGTTGATGA-3’); si-NC (5’-GCGCGATAGCGCGAATATA-3’); miR-515-5p mimic (5’-UUCUCCAAAAGAAAGCACUUUCUG-3’); miR-NC (5’-ACUCUAUCUGCACGCUGACUU-3’); miR-1253 inhibitor (5’-CAGAAAGUGCUUUCUUUUGGAGAA-3’); anti-miR-NC (5’-CAGUACUUUUGUGUAGUACAA-3’). Lipofectamine 3000 Reagent (Invitrogen) was employed to transfect the oligonucleotide and/or vector into 16HBE cells.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA from cells was isolated utilizing TRIzol (Invitrogen). The total RNA from the serum (250 μL/sample) was isolated using a TRIzol LS Reagent (Invitrogen). Next, the purity and concentration of RNA samples were detected by the measurement of the absorbance at 260 and 280 nm using NanoDrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). OD260/OD280 ratio ranged from 18 to 2.1 could be classified as a qualified sample. 1 μg of RNA was then reversed into cDNA with Primescript RT Reagent (TaKaRa, Kusatsu, Japan) for analysis of circRNA and mRNA or miScript II RT kit (Invitrogen) for detection of miRNA. Thereafter, qRT-PCR reaction was manipulated on the ABI-7500 Real-time PCR machine (Applied Biosystems, Foster City, CA, USA) with SYBR Green PCR Master Mix (Invitrogen). The RNA levels were calculated using the 2−ΔΔCt method. U6 (for miRNA) and β-actin (for circRNA and mRNA) were used as an internal inference. Primer information was listed: circ_0040929 (sense: 5’-GAGCAGAAGGACTCGGACAC-3’ and anti-sense: 5’-CTTCCTGCTGTGGTGCTTTA-3’); ANKRD11 (sense: 5’-CCTAGATGACGACACGCCTTTG-3’ and anti-sense: 5’-GTCTCGCCTTTCCTGTTGCTCT-3’); miR-515-5p (sense: 5’-GGGTTCTCCAAAAGAAAGC-3’ and anti-sense: 5’-CAGTGCGTGTCGTGGAGT-3’); IGFBP3 (sense: 5’-TTGCACAAAAGACTGCCAAG-3’ and anti-sense: 5’-CAACATGTGGTGAGCATTCC-3’); GAPDH (sense: 5’- GTCTCCTCTGACTTCAACAGCG-3’ and anti-sense: 5’- ACCACCCTGTTGCTGTAGCCAA-3’); β-actin (sense: 5’-CTCCATCCTGGCCTCGCTGT-3’ and anti-sense: 5’-GCTGTCACCTTCACCGTTCC-3’); U6 (sense: 5’-CTCGCTTCGGCAGCACATATACT-3’ and anti-sense: 5’-ACGCTTCACGAATTTGCGTGTC-3’). All experiments were repeated three times.
RNase R and Actinomycin D Treatment
RNase R treatment was used for degrading linear RNAs. RNA (2 μg) was treated with or without RNase R (3 U/μg) (Seebio, Shanghai, China) for half an hour. Next, circ_0040929 and ANKRD11 levels were assessed by qRT-PCR. All experiments were repeated three times.
For Actinomycin D assay, cells were exposed Actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) to block transcription. Thereafter, qRT-PCR was used for measuring circ_0040929 and ANKRD11 expression. All experiments were repeated three times.
Subcellular Fraction Assay
The fractions of nuclear and cytoplasmic were isolated with PARIS Kit (Invitrogen) for detecting the cellular localization of circ_0040929. Afterwards, the expression of GAPDH (as the controls for cytoplasmic transcript), U6 (as the controls for nuclear transcript) and circ_0040929 was examined through qRT-PCR. All experiments were repeated three times.
Cell Proliferation Assays
Cell Counting Kit-8 (CCK-8) purchased from Dojindo (Kumamoto, Japan) was employed for analyzing cell viability. Briefly, cell suspension (100 μL) was seeded into 96-well plates. Following the treatment, 10 μL of CCK-8 reagent was placed into each well. After 2–3 h, a microplate reader (BioTeck, Winooski, VT, USA) was employed for examining the absorbance of per well at 450 nm. All experiments were repeated three times.
For 5-ethynyl-20-deoxyuridine (EdU) assay, EdU kit (KeyGene, Nanjing, China) was used to test cell proliferation and DNA synthesis. In short, we seeded cells into a 24-well plate. After treatment, the cells were incubated with EdU solution (50 μM) for 2 h. After fixing with paraformaldehyde (4%), cells were treated with Triton-X-100 (0.5%), followed by staining with Click-It reaction in a dark place for 0.5 h. The nucleic acids were stained with DAPI, followed by analysis under fluorescence microscope (Leica, Wetzlar, Germany) at 200× magnification. All experiments were repeated three times.
Flow Cytometry Analysis
After treatment, 16HBE cells were harvested by centrifugation and then suspended in binding buffer (0.4 mL), followed by staining with Annexin V-FITC and PI (BD Biosciences, San Diego, CA, USA) in the darkness for 20–30 min. At last, cell apoptosis was monitored via a flow cytometer (BD Biosciences). All experiments were repeated three times.
Western Blot Assay
RIPA lysis buffer (KeyGene) was used for extracting total proteins. After measurement of protein concentration, proteins were subjected to SDS-PAGE, followed by transferring onto the PVDF membranes (Millipore, Billerica, MA, USA). After blockage using 5% skim milk (Beyotime, Shanghai, China), these membranes were immunoblotted with primary antibodies at 4°C for 10–14 h. Lastly, the combined signals were detected by an enhanced chemiluminescence kit (KeyGene) after incubation of corresponding secondary antibody. The antibodies including proliferating cell nuclear antigen (PCNA; 1:500, ab18197), Bcl-2 (1:500, ab196495), Bax (1:500, ab32503), Cleaved Caspase 3 (1:500, ab32042), IGFBP3 (1:1000, ab193910), β-actin (1:5000, ab227387), CD63 (1:1000, ab118307), TSG101 (1:500, ab30871), and HRP-conjugated IgG anti-rabbit (1:4000, ab205718) were commercially acquired from Abcam (Cambridge, MA, USA). All experiments were repeated three times.
Enzyme-Linked Immunosorbent Assay (ELISA)
The levels of factors related to inflammatory response (including IL-6, IL-1β and TNF-α), airway remodeling (including α-SMA, collagen I) and IGFBP3 in cell supernatant were determined by ELISA (Abcam). The data were presented in terms of pg/mL. All experiments were repeated three times.
Dual-Luciferase Reporter Assay
The candidate target miRNAs of circ_0040929 were predicted using circBANK (http://www.circbank.cn/), starbase (http://starbase.sysu.edu.cn/), and circinteractome (https://circinteractome.nia.nih.gov/). Starbase was employed to predict the candidate target genes for miR-515-5p. To generate wild-type luciferase reporter vector (circ_0040929-WT or IGFBP3-3’UTR-WT), the fragments of circ_0040929 or IGFBP3 3’UTR harboring miR-515-5p binding sequence were synthesized and introduced into the pmirGLO Dual-luciferase vectors (GenePharma, Shanghai, China). The mutant luciferase reporter vector (circ_0040929-MUT or IGFBP3-3’UTR-MUT) was created via mutating the binding sequence. After that, 16HBE cells were co-introduced with the corresponding reporter vector and miR-515-5p/miR-NC. Next, Dual Luciferase Reporter Gene Assay Kit (LMAI Bio, Shanghai, China) was applied for the measurement of luciferase activities. All experiments were repeated three times.
RNA Pull-Down Assay
Biotin-labeled negative control (Bio-NC) and biotin-labeled miR-515-5p (Bio-miR-515-5p) (RiboBio, Guangzhou, China) were introduced into 16HBE cells, respectively. After incubation for 48 h, cells were collected and lysed. Following the addition of streptavidin beads (Invitrogen) at 4°C for 2 h, the pulled down RNAs were tested via qRT-PCR. All experiments were repeated three times.
RNA Immunoprecipitation (RIP) Assay
RIP experiment was conducted with Magna RIP Kit (Millipore). In short, cells were lysed by complete RIP lysis buffer. Next, cell lysate was incubated with magnetic beads which linked with Anti-IgG antibody (as negative control) or anti-Argonaute2 (Anti-Ago2). After digestion with protease K, the extracted RNA was further used for qRT-PCR for detecting circ_0040929 and miR-515-5p enrichment. All experiments were repeated three times.
Transmission Electron Microscopy (TEM)
ExoQuick precipitation kit (System Biosciences, Mountain View, CA, USA) was utilized for isolating exosomes from serum samples. Next, isolated exosomes were suspended in PBS (Beyotime), followed by fixing in glutaraldehyde (2.5%, pH 7.2). After that, fixed exosomes were then put on a carbon-coated copper grid and subsequently stained by phosphotungstic acid solution (pH = 7.0) and dried at 65°C. Next, JEM-1400 (JEOL, Akishima, Japan) was utilized to observe exosome morphology and ultrastructure at 100 kV.
Nanoparticle Tracking Analysis (NTA)
To observe exosome size distribution and concentration, NTA was carried out using a NanoSight NS300 (NanoSight, Amesbury, UK). Isolated exosome samples were diluted in PBS and injected into the NanoSight sample pool, and five recordings of 30 sec each were captured. NTA 3.0 software (NanoSight) was used to analyze the data.
Statistical Analysis
Data from at least 3 independent experiments were presented as the mean ± standard deviation and analyzed by GraphPad Prism version 6.0 software (GraphPad Software, La Jolla, CA, USA). Difference comparison depended on Student’s t-test and one-way analysis of variance. The correlation between circ_0040929 and miR-515-5p in the serum of COPD patients was performed by Spearman correlation coefficient. P<0.05 was regarded as statistically significant.
Results
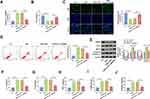
Circ_0040929 is Upregulated in COPD Patients and CSE-Treated 16HBE Cells
The expression of circ_0040929 in the serum of COPD patients, smokers, and non-smokers was examined by qRT-PCR. As shown in Figure 1A, circ_0040929 expression was higher in the serum of smokers than that in non-smokers, and its expression was the highest in the serum of COPD patients. According to the ROC curve, circ_0040929 could be used to diagnose smokers and COPD patients, with an AUC of 0.9959 distinguishing smokers from non-smokers (Supplementary Figure 1A), 1.000 distinguishing COPD from non-smokers (Supplementary Figure 1B), and 0.6570 distinguishing COPD from smokers (Supplementary Figure 1C). Also, circ_0040929 expression was positively correlated with CRP level in COPD patients (Supplementary Figure 1D). In addition, circ_0040929 expression was dose-dependently increased in response to CSE treatment in 16HBE cells (Figure 1B). Likewise, circ_0040929 expression was gradually elevated by treatment with CSE in 16HBE cells in a time-dependent manner (Figure 1C). Generally, RNase R can digest linear RNA but not circRNA. RNase R digestion analysis suggested that circ_0040929 was more resistant to RNase R than linear ANKRD11 (Figure 1D). Actinomycin D assay exhibited that the half-life of circ_0040929 transcript exceeded 12 h, while the half-life of ANKRD11 transcript was about 8 h (Figure 1E), indicating that circ_0040929 transcript was more stable than ANKRD11 transcript. Next, we explored the localization of circ_0040929 in 16HBE cells. The data showed that circ_0040929 was mainly located in the cytoplasm (Figure 1F). Taken together, as a circular RNA, circ_0040929 might be involved in COPD development.
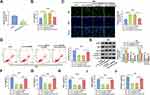
Knockdown of Circ_0040929 Attenuated CSE-Induced Apoptosis, Inflammation, and Airway Remodeling
Next, we explored the potential function of circ_0040929 in HBE cells exposed to 2% CSE. The expression of circ_0040929 was increased by exposure to CSE, which was weakened by transfection of si-circ_0040929 (Figure 2A). CCK-8 and EdU assay suggested that CSE treatment reduced cell viability and DNA synthesis, which could be reversed by knockdown of circ_0040929 (Figure 2B and C). The results of flow cytometry analysis depicted that CSE treatment induced apoptosis, which was attenuated by downregulating circ_0040929 (Figure 2D). Western blot assay was used to measure the protein levels of PCNA (essential for cellular DNA synthesis), Bax (pro-apoptotic molecule) and Bcl-2 (anti-apoptotic molecule), and Cleaved caspase 3 (a key executor in apoptotic process). The data showed that CSE treatment decreased PCNA and Bcl-2 protein levels and increased Bax and Cleaved caspase 3 protein levels, while these effects were overturned by circ_0040929 interference (Figure 2E). In addition, the secretion of inflammatory cytokines, including IL-1β, IL-6 and TNF-α, was detected in CSE-treated 16HBE cells after knockdown of circ_0040929. ELISA analysis showed that CSE treatment increased the concentrations of IL-1β, IL-6 and TNF-α, which was reversed by circ_0040929 downregulation (Figure 2F–H). The inflammation cytokines usually contribute to the airway remodeling process (a critical feature of COPD).23 Therefore, we further analyzed the effect of circ_0040929 on airway remodeling. Results of ELISA showed that both α-SMA and collagen I levels were increased by CSE treatment in 16HBE cells, while circ_0040929 inhibition counteracted airway remodeling caused by CSE (Figure 2I and J). The above findings manifested that circ_0040929 knockdown attenuated CSE-triggered injury in 16HBE cells.
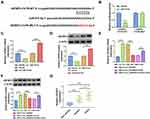
MiR-515-5p is a Direct Target of Circ_0040929
CircRNAs have been increasingly reported to regulate the expression of target genes via sponging certain miRNAs. Based on the combined prediction of circBANK, starbase, and circinteractome, only miR-515-5p was screened out as a candidate (Figure 3A). The complementary-binding sites of miR-515-5p and circ_0040929 are shown in Figure 3B. Transfection of miR-515-5p increased the expression of miR-515-5p in 16HBE cells (Figure 3C), indicating a high transfection efficiency. Dual-luciferase reporter, RNA pull-down and RIP assays were performed to further confirm the interaction between miR-515-5p and circ_0040929. Dual-luciferase reporter assay manifested that miR-515-5p enhancement remarkably decreased the luciferase activity of circ_0040929-WT but had no obvious impact on the luciferase activity of circ_0040929-MUT (Figure 3D). RNA pull-down assay showed that the enrichment of circ_0040929 was significantly increased in 16HBE cell transfection with Bio-miR-515-5p compared with Bio-NC group (Figure 3E). Moreover, circ_0040929 and miR-515-5p were enriched in Anti-Ago2 group compared to Anti-IgG group (Figure 3F). The results of qRT-PCR showed that miR-515-5p expression was lower in the serum of smokers than that in non-smokers, and its expression was the lowest in the serum of COPD patients (Figure 3G). Furthermore, the correlation between miR-515-5p and circ_0040929 expression was analyzed in the serum of COPD patients. As shown in Figure 3H, a negative correlation between miR-515-5p and circ_0040929 expression was observed. In addition, we found that miR-515-5p expression was reduced in 16HBE cells treated with CSE, which was restored by transfection of si-circ_0040929 (Figure 3I). Altogether, circ_0040929 sponged miR-515-5p in 16HBE cells.
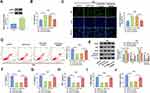
Circ_0040929 Silencing Alleviated CSE-Induced Injury via Sponging miR-515-5p in 16HBE Cells
Transfection of anti-miR-515-5p reduced the expression of miR-515-5p in 16HBE cells (Figure 4A). To illuminate whether circ_0040929 targeted miR-515-5p to affect CSE-induced cell injury, a range of rescue experiments were performed. The promoting effects of circ_0040929 knockdown on cell viability and DNA synthesis were reversed after the introduction of anti-miR-515-5p (Figure 4B and C). Furthermore, miR-515-5p inhibition restored the suppressive effect of si-circ_0040929 on cell apoptosis in CSE-exposed 16HBE cells (Figure 4D). Meanwhile, the increased PCNA and Bcl-2 protein levels and the decreased Bax and Cleaved caspase 3 protein levels caused by circ_0040929 knockdown were abrogated by miR-515-5p downregulation (Figure 4E). In addition, miR-515-5p suppression counteracted the inhibitory effects of circ_0040929 depletion on the levels of IL-1β, IL-6, TNF-α, α-SMA, and collagen I in CSE-treated 16HBE cells (Figure 4F–J). These data suggested that miR-515-5p inhibition reversed the roles of circ_0040929 knockdown in CSE-treated 16HBE cells.
IGFBP3 is a Direct Target Gene of miR-515-5p
Next, we explored the downstream target for miR-515-5p via starbase. As presented in Figure 5A, IGFBP3 possessed the possible binding sites for miR-515-5p, indicating that IGFBP3 was a potential target gene of miR-515-5p. To validate this assumption, dual-luciferase reporter assay was carried out. The results showed that miR-515-5p transfection markedly reduced the luciferase activity of IGFBP3-3’UTR-WT but did not affect the luciferase activity of IGFBP3-3’UTR-MUT in 16HBE cells (Figure 5B). Subsequently, we explored the impact of miR-515-5p on IGFBP3 expression. We observed that IGFBP3 mRNA and expression were reduced by overexpression of miR-515-5p and increased by inhibition of miR-515-5p in 16HBE cells (Figure 5C and D). Next, we explored whether circ_0040929 could regulate IGFBP3 expression via serving as a sponge of miR-515-5p. Results demonstrated that circ_0040929 interference reduced the mRNA and protein levels of IGFBP3, which was abolished by inhibiting miR-515-5p expression (Figure 5E and F), suggesting that circ_0040929 acted as a sponge of miR-515-5p to positively regulate IGFBP3 expression. In addition, IGFBP3 expression is increased in the serum of smokers compared to non-smokers, and IGFBP3 expression is further increased in the serum of COPD patients (Figure 5G). In a word, miR-515-5p directly targeted IGFBP3.
Overexpression of miR-515-5p Weakened CSE-Induced Injury by Targeting IGFBP3 in 16HBE Cells
Overexpression efficiency of IGFBP3 was determined by qRT-PCR. The data showed that transfection of IGFBP3 increased IGFBP3 protein expression in 16HBE cells (Figure 6A). Next, rescue experiments were performed to explore whether the function of miR-515-5p in CSE-treated 16HBE cells was regulated by IGFBP3. Overexpression of miR-515-5p increased cell viability and DNA synthesis and reduced cell apoptosis, while these effects were reversed by upregulating IGFBP3 (Figure 6B-D). Moreover, enforced expression of miR-515-5p resulted in a marked elevation in the protein levels of PCNA and Bcl-2, and a significant reduction in the protein levels of Bax and Cleaved caspase 3, which could be neutralized by increasing IGFBP3 expression (Figure 6E). Furthermore, the restoration of miR-515-5p reduced the levels of IL-1β, IL-6, TNF-α, α-SMA, and collagen I in CSE-treated 16HBE cells, whereas these effects were abolished by the overexpression of IGFBP3 (Figure 6F–J). These data indicated that miR-515-5p attenuated CSE-induced apoptosis, inflammation, and airway remodeling by regulating IGFBP3 in 16HBE cells.
Circ_0040929 is Upregulated in Exosomes Derived from COPD Patient Serum
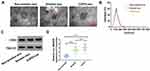
Exosomes were extracted from the serum of non-smokers, smokers, and COPD patients. As shown in Figure 7A, serum-derived exosomes were observed under a TEM, and the diameter of vesicles was about 40–120 nm, which was consistent with the morphological characteristics of exosomes. The particle size distribution was detected by NTA, and the result suggested that the size distribution of exosomes was approximately 100 nm in diameter (Figure 7B). The levels of exosome protein markers (CD63 and TSG101) were detected by Western blot. Results showed that CD63 and TSG101 were all highly enriched in the isolated exosomes from non-smokers, smokers, and COPD patient serum (Figure 7C). In addition, the results of qRT-PCR showed that circ_0040929 expression was increased in the serum exosomes from smokers compared with non-smokers, and its expression was further enhanced in the exosomes from COPD patient serum (Figure 7D). These data suggested that exosomal circ_0040929 might be involved in COPD development.
Discussion
Currently, it is still a great challenge to treat COPD. Recently, some circRNAs have been suggested to participate in the pathogenesis of COPD. For instance, circANKRD11 is elevated in the lung samples of COPD patients, and knockdown of circANKRD11 ameliorated CSE-triggered cell apoptosis, oxidative stress, and inflammation in human pulmonary microvascular endothelial cells.24 In addition, circ_0006892 is downregulated in lung tissue samples of COPD patients and CSE-treated bronchial epithelial cells, and upregulation of circ_0006892 weakened CSE-triggered apoptosis and inflammatory response25. However, the biological roles of most circRNAs in COPD have not been studied. Hence, the purpose of our research was to clarify the exact role and possible mechanism of circ_0040929 in COPD.
In this paper, circ_0040929 is strikingly enhanced in COPD patient serum samples and CSE-treated 16HBE cells, which is in accordance with a previous report.15 Besides, circ_0040929 knockdown weakened CSE-triggered cell injury by promoting cell proliferation and inhibiting apoptosis, inflammation, and airway remodeling in 16HBE cells. These data revealed that circ_0040929 might promote COPD progression.
Accumulating evidence has indicated that circRNAs in the cytoplasm can serve as competing endogenous RNAs (ceRNAs) or miRNA sponges, thereby regulating various biological processes.26 In this research, circ_0040929 was demonstrated to be primarily located in the cytoplasm. Hence, we hypothesized that circ_0040929 might serve as a sponge for miRNA. By analyzing bioinformatics tools and a series of experiments, we proved that circ_0040929 was a miR-515-5p sponge. Many researches manifested that miR-515-5p functioned as a tumor-suppressive miRNA in many cancers, including prostate cancer,27 hepatocellular carcinoma,28 and bladder cancer.29 Moreover, miR-515-5p has been revealed to be closely associated with inflammatory response.30,31 More importantly, Ma et al showed that CSE treatment could reduce miR-515-5p level in human bronchial epithelial cells, and miR-515-5p was implicated in CSE-mediated EMT and regulated FoxC1 and Snail expression in human bronchial epithelial cells.20 However, the impact of miR-515-5p on the proliferation, inflammation, apoptosis, and airway remodeling in CSE-induced human bronchial epithelial cells is still unclear. Herein, a decrease of miR-515-5p expression was detected in COPD patient serum samples and CSE-stimulated 16HBE cells. Additionally, miR-515-5p inhibition abated the roles of circ_0040929 interference in CSE-treated 16HBE cells, indicating that circ_0040929 silencing weakened CSE-triggered cell injury via sponging miR-515-5p.
Next, we continued to search for the target gene of miR-515-5p. Our research verified that IGFBP3 was a target for miR-515-5p according to bioinformatics tool and experimental verification. IGFBP3 is widely involved in regulating cell proliferation, apoptosis, airway inflammation, and airway remodeling.32,33 Moreover, Jiao et al disclosed that CSE treatment resulted in a marked increase of IGFBP3 expression in alveolar epithelial cells, CSE stimulation inhibited proliferation of alveolar epithelial cells and even induced apoptosis with a high expression of IGFBP3, indicating that IGFBP3 might play an important role in COPD.34 Besides, Jia et al declared that IGFBP3 was also increased after the treatment with CSE in NCI-H292 cells, and miR-212-5p exerted a protective role in COPD by downregulating IGFBP3 expression, implying that inhibition of COPD might be an effective strategy for the treatment of COPD.21 In our study, IGFBP3 was demonstrated to be highly expressed in COPD patient serum and CSE-exposed 16HBE cells. Overexpression of miR-515-5p facilitated cell proliferation and suppressed apoptosis, airway remodeling, and inflammation in CES-stimulated 16HBE cells, disclosing that miR-515-5p exerted a protective role in COPD by targeting IGFBP3. According to our results, we confirmed that IGFBP3 could promote the release of IL-1β, IL-6 and TNF-α. Therefore, we confirmed that IGFBP3 could aggravate inflammation response.
Exosomes are small vesicles with diameter range between 40 and 140 nm that originate from endosomal multivesicular bodies.35 The transfer of exosomes between cells allows the delivery of cellular contents, such as proteins, circRNAs, mRNAs, miRNAs, or other bioactive substances.36,37 CircRNAs are evolutionarily conserved, highly stable, and cell-/developmental stage-specific and have longer half-lives compared with linear RNAs, and circRNAs are plentiful in various body fluids including blood, saliva, serum, and even in exosomes. Moreover, exosomes can protect RNAs from degradation in the circulation, so exosomal circRNAs are thought to be novel therapeutic targets and disease biomarkers.38 The levels of exosomal circRNAs derived from COPD patients are still unknown. In this research, circ_0040929 was found to be upregulated in serum exosomes from COPD patients, implying that exosomal circ_0040929 might be a promising diagnostic marker for COPD patients.
In conclusion, we demonstrated that circ_0040929 knockdown accelerated cell proliferation and repressed cell apoptosis, inflammation, and airway remodeling in CES-exposed 16HBE cells via sponging miR-515-5p and mediating IGFBP3 expression. In addition, serum exosomal circ_0040929 might be a potential diagnostic indicator for COPD patients. Overall, our data might provide a promising therapeutic strategy for COPD therapy. However, the role of circ_0040929/miR-515-5p/IGFBP3 in COPD is still required to be further confirmed in animal models in the future study.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflict of interest.
References
1. Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (COPD). Lancet. 2004;364(9434):613–620. doi:10.1016/s0140-6736(04
2. Agustí A, Hogg JC. Update on the pathogenesis of chronic obstructive pulmonary disease. N Engl J Med. 2019;381(13):1248–1256. doi:10.1056/NEJMra1900475
3. Rahman I, Morrison D, Donaldson K, MacNee W. Systemic oxidative stress in asthma, COPD, and smokers. Am J Respir Crit Care Med. 1996;154(4 Pt 1):1055–1060. doi:10.1164/ajrccm.154.4.8887607
4. Rab A, Rowe SM, Raju SV, Bebok Z, Matalon S, Collawn JF. Cigarette smoke and CFTR: implications in the pathogenesis of COPD. Am J Physiol Lung Cell Mol Physiol. 2013;305(8):L530–541. doi:10.1152/ajplung.00039.2013
5. Arunachalam G, Yao H, Sundar IK, Caito S, Rahman I. SIRT1 regulates oxidant- and cigarette smoke-induced eNOS acetylation in endothelial cells: role of resveratrol. Biochem Biophys Res Commun. 2010;393(1):66–72. doi:10.1016/j.bbrc.2010.01.080
6. Chen Y, Luo H, Kang N, et al. Beraprost sodium attenuates cigarette smoke extract-induced apoptosis in vascular endothelial cells. Mol Biol Rep. 2012;39(12):10447–10457. doi:10.1007/s11033-012-1924-1
7. Sun Y, An N, Li J, et al. miRNA-206 regulates human pulmonary microvascular endothelial cell apoptosis via targeting in chronic obstructive pulmonary disease. J Cell Biochem. 2019;120(4):6223–6236. doi:10.1002/jcb.27910
8. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333.
9. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
10. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-6
11. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi:10.1016/j.ebiom.2018.07.036
12. Chen S, Yao Y, Lu S, et al. CircRNA0001859, a new diagnostic and prognostic biomarkers for COPD and AECOPD. BMC Pulm Med. 2020;20(1):311. doi:10.1186/s12890-020-01333-1
13. Qiao D, Hu C, Li Q, Fan J. Circ-RBMS1 knockdown alleviates CSE-induced apoptosis, inflammation and oxidative stress via up-regulating FBXO11 through miR-197-3p in 16HBE cells. Int J Chron Obstruct Pulmon Dis. 2021;16:2105–2118. doi:10.2147/COPD.S311222
14. Zhou F, Cao C, Chai H, Hong J, Zhu M. Circ-HACE1 aggravates cigarette smoke extract-induced injury in human bronchial epithelial cells via regulating toll-like receptor 4 by sponging miR-485-3p. Int J Chron Obstruct Pulmon Dis. 2021;16:1535–1547. doi:10.2147/COPD.S304859
15. Zeng N, Wang T, Chen M, et al. Cigarette smoke extract alters genome-wide profiles of circular RNAs and mRNAs in primary human small airway epithelial cells. J Cell Mol Med. 2019;23(8):5532–5541. doi:10.1111/jcmm.14436
16. Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2(4):161.
17. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
18. Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10. doi:10.1007/978-1-4939-6524-3_1
19. Salimian J, Mirzaei H, Moridikia A, Harchegani AB, Sahebkar A, Salehi H. Chronic obstructive pulmonary disease: microRNAs and exosomes as new diagnostic and therapeutic biomarkers. J Res Med Sci. 2018;23:27. doi:10.4103/jrms.JRMS_1054_17
20. Ma H, Lu L, Xia H, et al. Circ0061052 regulation of FoxC1/Snail pathway via miR-515-5p is involved in the epithelial-mesenchymal transition of epithelial cells during cigarette smoke-induced airway remodeling. Sci Total Environ. 2020;746:141181. doi:10.1016/j.scitotenv.2020.141181
21. Jia Q, Chang J, Hong Q, Zhang JJ, Zhou H, Chen FH. MiR-212-5p exerts a protective effect in chronic obstructive pulmonary disease. Discov Med. 2018;26(144):173–183.
22. Li D, Hu J, Wang T, et al. Silymarin attenuates cigarette smoke extract-induced inflammation via simultaneous inhibition of autophagy and ERK/p38 MAPK pathway in human bronchial epithelial cells. Sci Rep. 2016;6:37751. doi:10.1038/srep37751
23. Tam A, Churg A, Wright JL, et al. Sex differences in airway remodeling in a mouse model of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;193(8):825–834. doi:10.1164/rccm.201503-0487OC
24. Wang Z, Zuo Y, Gao Z. CircANKRD11 knockdown protects HPMECs from cigarette smoke extract-induced injury by regulating miR-145-5p/BRD4 axis. Int J Chron Obstruct Pulmon Dis. 2021;16:887–899. doi:10.2147/copd.S300332
25. Zhang C, Gu S, Kang X. CircRNA circ_0006892 regulates miR-24/PHLPP2 axis to mitigate cigarette smoke extract-induced bronchial epithelial cell injury. Biotechnol Appl Biochem. 2021. doi:10.1002/bab.2148
26. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi:10.1093/bib/bbw053
27. Zhang X, Zhou J, Xue D, Li Z, Liu Y, Dong L. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. doi:10.1016/j.ijbiomac.2019.01.127
28. Ni JS, Zheng H, Ou YL, et al. miR-515-5p suppresses HCC migration and invasion via targeting IL6/JAK/STAT3 pathway. Surg Oncol. 2020;34:113–120. doi:10.1016/j.suronc.2020.03.003
29. Cao G, Zhang C, Tian X, Jing G, Zhou X, Yan T. circCEP128 knockdown suppresses bladder cancer progression via regulating microRNA-515-5p/SDC1 axis. Cancer Manag Res. 2021;13:2885–2896. doi:10.2147/cmar.S288229
30. Wu R, Zhang F, Cai Y, et al. Circ_0134111 knockdown relieves IL-1β-induced apoptosis, inflammation and extracellular matrix degradation in human chondrocytes through the circ_0134111-miR-515-5p-SOCS1 network. Int Immunopharmacol. 2021;95:107495. doi:10.1016/j.intimp.2021.107495
31. Cai D, Hong S, Yang J, San P. The effects of microRNA-515-5p on the toll-like receptor 4 (TLR4)/JNK signaling pathway and WNT1-inducible-signaling pathway protein 1 (WISP-1) expression in rheumatoid arthritis fibroblast-like synovial (RAFLS) cells following treatment with receptor activator of nuclear factor-kappa-B ligand (RANKL). Med Sci Monit. 2020;26:e920611. doi:10.12659/msm.920611
32. Ingermann AR, Yang YF, Han J, et al. Identification of a novel cell death receptor mediating IGFBP-3-induced anti-tumor effects in breast and prostate cancer. J Biol Chem. 2010;285(39):30233–30246. doi:10.1074/jbc.M110.122226
33. Lee H, Kim SR, Oh Y, Cho SH, Schleimer RP, Lee YC. Targeting insulin-like growth factor-I and insulin-like growth factor-binding protein-3 signaling pathways. A novel therapeutic approach for asthma. Am J Respir Cell Mol Biol. 2014;50(4):667–677. doi:10.1165/rcmb.2013-0397TR
34. Jiao Z, Zhang Q, Chang J, et al. A protective role of sulforaphane on alveolar epithelial cells exposed to cigarette smoke extract. Exp Lung Res. 2013;39(9):379–386. doi:10.3109/01902148.2013.830162
35. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372.
36. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. doi:10.1016/j.bbagen.2012.03.017
37. Ge R, Tan E, Sharghi-Namini S, Asada HH. Exosomes in cancer microenvironment and beyond: have we overlooked these extracellular messengers? Cancer Microenviron. 2012;5(3):323–332. doi:10.1007/s12307-012-0110-2
38. Li Y, Feng W, Kong M, et al. Exosomal circRNAs: a new star in cancer. Life Sci. 2021;269:119039. doi:10.1016/j.lfs.2021.119039
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.