Back to Journals » OncoTargets and Therapy » Volume 14
Circ_0006404 Accelerates Prostate Cancer Progression Through Regulating miR-1299/CFL2 Signaling
Authors Li P, Wang Z, Li S, Wang L
Received 19 August 2020
Accepted for publication 11 November 2020
Published 6 January 2021 Volume 2021:14 Pages 83—95
DOI https://doi.org/10.2147/OTT.S277831
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Peihuan Li,1 Zhijie Wang,2 Shuai Li,2 Liuxing Wang1
1Department of Oncology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, People’s Republic of China
Correspondence: Liuxing Wang
Department of Oncology, The First Affiliated Hospital of Zhengzhou University, No. 1 Jianshe East Road, Zhengzhou City, Henan, People’s Republic of China
Tel +86-13403712504
Email [email protected]
Background: Circular RNAs (circRNAs) have been proven to function as pivotal regulators in cancer occurrence and progression. However, the function of circ_0006404 (circRNA Forkhead box O3 (circFOXO3)in prostate cancer (PCa) is poorly understood.
Methods: The enrichment of circ_0006404, FOXO3, microRNA-1299 (miR-1299) and cofilin 2 (CFL2) was measured by quantitative real-time polymerase chain reaction (qRT-PCR). The viability, metastasis and proliferation were determined by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, transwell and colony formation assays, respectively. Flow cytometry was used to assess cell cycle progression and apoptosis. Circ_0006404/miRNAs interactions were explored using Circular RNA Interactome database, while TargetScan software was used for seeking the targets of miR-1299. Dual-luciferase reporter assay, RNA-pull down and RNA immunoprecipitation (RIP) assays were conducted to verify the target interaction between miR-1299 and circ_0006404 or CFL2. CFL2 protein level was analyzed by Western blot assay. Animal experiments were performed to test the role of circ_0006404 in PCa tumor growth in vivo.
Results: Circ_0006404 level was notably elevated in PCa. Circ_0006404 contributed to the viability, metastasis and proliferation and impaired the apoptosis of PCa cells. Circ_0006404 directly targeted miR-1299, and miR-1299 silencing largely reversed circ_0006404 interference-induced influences in PCa cells. CFL2 directly bound to miR-1299, and miR-1299-induced effects in PCa cells were largely attenuated by CFL2 overexpression. CFL2 was regulated by circ_0006404/miR-1299 axis in PCa cells. Circ_0006404 promoted PCa progression via miR-1299/CFL2 axis in vivo.
Conclusion: Circ_0006404 accelerated the survival, motility and proliferation while impeded the apoptosis of PCa cells via miR-1299/CFL2 axis. Circ_0006404 might be a stable potential bio-marker for PCa diagnosis and treatment.
Keywords: prostate cancer, circ_0006404, miR-1299, CFL2, proliferation, metastasis
Introduction
Prostate cancer (PCa) is a common malignancy that threatens the health of men. A continuous increase in the incidence of PCa in China has been noted in the past few decades,1 which is closely associated with the aged tendency of the population, the changes in dietary pattern and the advancement in diagnostic technology.2,3 Accumulating studies have reported the vital functions of the androgen receptor (AR) and several other pivotal molecules in PCa development.4,5 After endocrine therapy, most of the PCa patients eventually develop into castration-resistant prostate cancer (CRPC), which is resistant to chemo- or radio-therapy.6 Although great improvement has been made in the diagnosis and therapy of PCa, the prognosis of PCa patients with advanced tumor remains unsatisfactory owing to the reoccurrence and metastasis.7,8 PCa is a typical heterogeneous disease, and uncovering the precise molecular mechanism is essential for searching for effective bio-markers and treatment strategies.
Circular RNAs (circRNAs) are single-stranded non-coding RNAs (ncRNAs) that originate from matching linear genes.9 It is generally known that circRNAs could act as oncogenes or tumor suppressors in cancers via competitively binding to microRNAs (miRNAs).10 Circ_0006404 (circRNA Forkhead box O3 (circFOXO3)derives from the exon 2 of FOXO3 gene. The biological function of circ_0006404 in PCa was controversial in previous studies. Kong et al claimed that circ_0006404 promoted the cell cycle and proliferation and inhibited the apoptosis of PCa cells through its miR-29a-3p sponge activity.11 However, Shen et al found that circ_0006404 impeded cell viability, metastasis and drug resistance in PCa cells to docetaxel through regulating FOXO3 and EMT.12 Here, we explored the expression profile and working mechanism of circ_0006404 in PCa progression.
MiRNAs are a category of small ncRNAs with about 18–24 nucleotides.13 The dysregulation of miRNAs was widely observed in multiple cancers, and the abnormal regulation of genes related to proliferation, apoptosis and metastasis was also found to be associated with the dysregulation of miRNAs.14–16 MiR-1299 exerted an anti-tumor role in a variety of cancers. Lin et al demonstrated that circ-EGLN3 facilitated the growth and metastasis of renal cell carcinoma cells through targeting the miR-1299/IRF7 axis.17 MiR-1299 suppressed cell proliferation and motility through targeting NEK2 in PCa cells.18 Nevertheless, the molecular mechanism behind miR-1299-mediated suppression on PCa progression remains to be elucidated.
Cofilin 2 (CFL2) has been reported to be up-regulated in breast cancer and gastric cancer, and the abnormal up-regulation of CFL2 was related to the pathogenesis of these cancers.19,20 As for PCa, CFL2 was verified as a downstream gene of SOX2-OT/miR-369-3p axis in PCa cells, and CFL2 promoted the proliferation and migration of PCa cells.21 Nevertheless, the knowledge concerning the modulatory network of CFL2 in PCa remains limited.
We conducted functional experiments to investigate the role of circ_0006404 in the viability, metastasis, proliferation and apoptosis of PCa cells. Through predicting the downstream targets of circ_0006404 with bioinformatic software, the molecular mechanism behind circ_0006404 in PCa was further illustrated. These findings suggested that circ_0006404/miR-1299/CFL2 axis might be a promising therapeutic target for PCa.
Materials and Methods
Clinical Samples
A total of 30 pairs of PCa tissues and adjacent normal tissues were resected from 30 PCa patients who underwent radical prostatectomy in the First Affiliated Hospital of Zhengzhou University. No subject underwent chemotherapy or radiotherapy before the radical resection. The protocol was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. Written informed consent was provided by each participant before the operation.
Cell Lines
Human normal prostate epithelial cell line (WPMY-1) and two PCa cell lines (LNCaP-AI and DU145) were obtained from Bena Culture Collection (Beijing, China). Dulbecco’s Modified Eagle medium (DMEM; Gibco, Carlsbad, CA, USA) and Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco) were used for the cultivation of WPMY-1 and PCa cell lines, respectively. The base medium was supplemented with 10% fetal bovine serum (FBS; Biowest, Loire Valley, France) and 10% penicillin (100 U/mL)/streptomycin (100 μg/mL) mixture before use. Cell culture plates were maintained in a humidified 37°C incubator with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA samples were isolated using TRIzol reagent (Takara, Beijing, China). Reverse transcription was performed using reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and TaqMan reverse transcription kit (Applied Biosystems, Rotkreuz, Switzerland). qRT-PCR was carried out in a 25 μL reaction system with SYBR® Premix Ex TaqTM II (Takara). The abundance of relevant molecules was calculated using 2−ΔΔCt method. The abundance of circ_0006404, FOXO3 messenger RNA (mRNA) and CFL2 mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), while U6 small nuclear RNA (snRNA) served as the internal reference for miR-1299. The PCR condition was listed as below: 95°C for 10 min followed by repetition for 40 times of 95°C for 15 s and 63°C for 32 s. The specific primer sequences are displayed in Table 1.
 |
Table 1 Primer Sequences for qRT-PCR |
RNase R and Actinomycin D Treatment
The stability of circ_0006404 and FOXO3 mRNA was assessed by using RNase R and Actinomycin D treatment. PCa cells were lysed, and 40 U RNase R was incubated with 10 μg RNA for 15 min. qRT-PCR was carried out to detect the expression of circ_0006404 and FOXO3 mRNA. Actinomycin D was incubated with PCa cells, and qRT-PCR was applied to detect the expression of circ_0006404 and FOXO3 mRNA after treatment for 0 h, 6 h, 12 h, 18 h or 24 h.
Cell Transfection
To knockdown circ_0006404, circ_0006404-specific small interfering RNA (si-circ_0006404), negative control (si-NC), short hairpin RNA targeting circ_0006404 (sh-circ_0006404) and its control (sh-NC) were purchased from Genepharma (Shanghai, China). For miR-1299 overexpression and silencing, miR-1299 mimic and miR-1299 inhibitor and matching negative controls (miRNA NC and inhibitor NC) were also synthesized from Genepharma. For the overexpression of CFL2, pcDNA-CFL2 (pc-CFL2) was constructed from Genepharma, and matching empty vector (pc-NC) was used as the control.
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
After transfection for 72 h, 10 µL MTT reagent (Invitrogen) was added to the wells to incubate with viable PCa cells for 4 h. Dimethylsulfoxide (DMSO) was used to dissolve the formazan product, and the absorbance value at 570 nm was detected by a microplate reader.
Transwell Assays
The migration and invasion abilities of PCa cells were assessed by transwell migration and invasion assays, respectively. In transwell migration assay, after transfection for 24 h, the upper compartments were added with transfected PCa cells (1 × 104 cells/well) in serum-free medium, while culture medium added with 10% FBS was used to fill the lower compartments. After incubation for 24 h, migratory PCa cells were stained with crystal violet (Sigma, St. Louis, MO, USA) and counted.
In transwell invasion assay, extracellular matrix was simulated through adding Matrigel (Millipore, Billerica, MA, USA) into the upper compartments, and then transfected PCa cells (5 × 104 cells/well) were plated into the pre-coated upper compartments. The remaining protocol was similar to the above.
Colony Formation Assay
Transfected DU145 and LNCaP-AI cells were suspended in phosphate-buffered saline (PBS) at a low density to form single-cell suspension. The single-cell suspension was seeded into 6-well plates at the density of 200 cells/well. Cell culture plates were maintained in a humidified 37°C incubator with 5% CO2 for 2 weeks. The culture media was replaced every 4 d. Colonies were dyed with crystal violet.
Flow Cytometry
Flow cytometry was used to analyze the cell cycle and apoptosis of PCa cells.
For cell cycle detection, PCa cells were immobilized using 80% ice-cold ethanol. RNase (Sigma) was used to remove RNA at a concentration of 2 mg/mL for 30 min at 37°C. 20 mg/mL propidium iodide (PI; Sigma) was added to mark the DNA content for 20 min at 37°C. The percentages of PCa cells in different phases of cell cycle were analyzed by the flow cytometer.
For cell apoptosis detection, after 72-h transfection, PCa cells were simultaneously stained using Annexin V labeled fluorescein isothiocyanate (FITC; Solarbio, Beijing, China) and PI (Solarbio) in a dark room. Apoptotic PCa cells were identified by the flow cytometer.
Bioinformatic Prediction
The molecular targets of circ_0006404 and miR-1299 were predicted by Circular RNA Interactome database and TargetScan database.
Dual-Luciferase Reporter Assay
The partial sequences of circ_0006404 and CFL2 mRNA, containing the complementary binding sites with miR-1299, were amplified by PCR and cloned into pmirGLO vector (Promega, Madison, WI, USA), and these wild-type reporter plasmids were termed as WT-circ_0006404 and WT-CFL2-3ʹ untranslated region (3ʹ UTR). Also, the mutant sequences in circ_0006404 and CFL2 mRNA were also constructed into pmirGLO vector (Promega) to generate MUT-circ_0006404 and MUT-CFL2-3ʹ UTR. These luciferase re-constructed plasmids were co-transfected with miR-1299 mimic or miRNA NC into DU145 and LNCaP-AI cells. The Firefly luciferase activity was detected using Dual-luciferase reporter assay system kit (Promega) and normalized to Renilla luciferase activity.
RNA-Pull Down Assay
Wild-type or mutant type miR-1299 was biotinylated to generate Biotin-miR-1299-WT or Biotin-miR-1299-MUT. 2 μg cell lysate was incubated with 100 pmol Biotin-miR-1299-WT or Biotin-miR-1299-MUT. Streptavidin agarose beads were added to the reaction system to incubate for 1 h at room temperature. The beads were boiled using sodium dodecyl sulfate (SDS), and qRT-PCR was applied to detect the levels of circ_0006404 and GAPDH.
RNA Immunoprecipitation (RIP) Assay
Sepharose beads (Bio-Rad, Hercules, CA, USA) pre-coated with Argonaute-2 antibody (Anti-Ago2) or Immunoglobulin G antibody (Anti-IgG) were incubated with the cell lysate. After washing the beads three times, RNA was isolated and qRT-PCR was implemented to measure the expression of circ_0006404 and miR-1299.
Western Blot Assay
Protein extraction was performed using Radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China). The protein samples were subjected to 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and electro-transferred into the polyvinylidene fluoride (PVDF) membrane (Millipore). After blocking with 5% non-fat milk, primary antibodies were used to incubate with the membrane as follows: anti-CFL2 (ab96678; Abcam, Cambridge, MA, USA) and anti-GAPDH (ab37168; Abcam). The next day, secondary antibody was used to label the primary antibody for 2 h at room temperature. Finally, the abundance of protein was measured using the enhanced chemiluminescent (ECL) system (Beyotime). GAPDH was used as the control in this study.
Animal Experiments
BALB/c-nude mice (5-week old) from the Laboratory Animal Center of Nanjing Medical University (Nanjing, China) were routinely maintained under pathogen-free condition. DU145 cells stably expressing sh-NC or sh-circ_0006404 were subcutaneously injected into the right side of mice back (7 mice in each group). Bidimensional measure of tumor volume was carried out using the vernier caliper every 7 d after 7 d-inoculation with the formula of (width2 × length)/2. These nude mice were sacrificed after 28-d inoculation, and the resected tumors were weighed, and qRT-PCR and Western blot assay were used to detect the abundance of circ_0006404, miR-1299 and CFL2 protein. The protocol in animal experiments was authorized by the Animal Research Committee of the First Affiliated Hospital of Zhengzhou University. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
All data were displayed as mean and standard deviation (SD). Differences between two groups or multiple groups were assessed using Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test. Linear correlation was analyzed using Spearman correlation coefficient. P < 0.05 was considered as statistical significance.
Results
Circ_0006404 is Aberrantly Up-Regulated in PCa
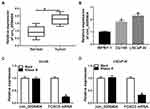
To test the expression pattern of circ_0006404 in PCa, we performed qRT-PCR to examine the abundance of circ_0006404 in 30 pairs of PCa tumor tissues and adjacent non-tumor tissues. A significant up-regulation in circ_0006404 level was observed in PCa tumor tissues compared with that in adjacent prostate normal tissues (Figure 1A). Furthermore, circ_0006404 was also up-regulated in two PCa cell lines in comparison with that in human normal prostate epithelial cell line WPMY-1 (Figure 1B). The stability of circ_0006404 in PCa cells was also tested through RNase R digestion and Actinomycin D (transcription inhibitor) exposure. RNase R treatment notably decreased the level of FOXO3 mRNA, which was the linear form of circ_0006404 (Figure 1C and D). However, little changes were found in circ_0006404 expression in RNase R group and Mock group (Figure 1C and D). After Actinomycin D treatment for 0 h, 6 h, 12 h, 18 h or 24 h, the abundance of circ_0006404 and FOXO3 mRNA was examined by qRT-PCR. Compared with FOXO3 mRNA, circ_0006404 was much more stable in PCa cells (Supplementary Figure 1A and 1B). Overall, circ_0006404 was a stable and up-regulated circRNA in PCa.
Circ_0006404 Interference Inhibits the Viability, Metastasis and Proliferation While Induces the Apoptosis of PCa Cells
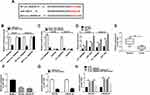
Si-circ_0006404 targeting the back-splicing sites of circ_0006404 was synthesized to specifically knockdown circ_0006404 expression rather than FOXO3. As mentioned in Figure 2A and B, circ_0006404 level was markedly reduced while the expression of FOXO3 mRNA remained almost unchanged in si-circ_0006404 transfected group compared with si-NC group, suggesting the specificity of circ_0006404 small interfering RNA. Loss-of-function experiments were used to analyze the influences of circ_0006404 silencing on the viability, migration, invasion, proliferation and apoptosis of PCa cells. Cell viability of PCa cells was markedly decreased with the silencing of circ_0006404 (Figure 2C). Circ_0006404 silencing restrained the migration and invasion abilities of PCa cells (Figure 2D). The proliferation ability of PCa cells was assessed by colony formation assay. The number of colonies was reduced when circ_0006404 was silenced compared with si-NC group (Figure 2E), demonstrating that circ_0006404 interference suppressed the proliferation of PCa cells. The percentages of cells in different phases of cell cycle were analyzed by flow cytometry. Circ_0006404 silencing elevated the percentage of PCa cells in G0/G1 phase, while the percentage of cells in S phase was reduced in si-circ_0006404 group (Figure 2F), suggesting that circ_0006404 interference halted cell cycle in G1/S transition. The early and late apoptotic PCa cells were measured by flow cytometry. The apoptosis rate was significantly enhanced in si-circ_0006404 group compared with si-NC group (Figure 2G), demonstrating that circ_0006404 silencing triggered the apoptosis of PCa cells. Taken together, circ_0006404 promoted the viability, metastasis and proliferation and impaired the apoptosis of PCa cells.
Circ_0006404 Directly Targets miR-1299 in PCa Cells
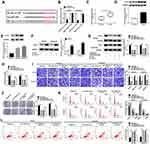
CeRNA (competing endogenous RNA) network is an important mechanism by which circRNAs function.10 We intended to explore if circ_0006404 functioned as a ceRNA of miRNA to regulate protein-coding gene. MiR-1299 was predicted as one of the candidate targets of circ_0006404 by Circular RNA Interactome database (Figure 3A). MiR-1299 played a tumor suppressor role in multiple cancers, including PCa.17,18,22 Furthermore, miR-1299 has been reported to be down-regulated in PCa.18 Hence, we focused on the interaction and functional association between miR-1299 and circ_0006404 in PCa cells. Dual-luciferase reporter assay was conducted to verify the direct target relationship between miR-1299 and circ_0006404 in PCa cells. The fragment of circ_0006404, harboring the wild-type or mutant type of miR-1299 binding sites, was cloned into luciferase reporter vector to generate WT-circ_0006404 or MUT-circ_0006404. MiR-1299 mimic or miRNA NC and these luciferase reporter plasmids were co-transfected into PCa cells. As shown in Figure 3B, miR-1299 mimic transfection conspicuously down-regulated the luciferase activity in wild-type circ_0006404 reporter plasmid group, while miR-1299 accumulation had no significant influence on the luciferase activity in mutant type circ_0006404 reporter plasmid group. Subsequently, RNA-pull down assay and RIP assay were conducted to further confirm the interaction between circ_0006404 and miR-1299 in PCa cells. Biotin-labeled miR-1299 and matching mutant counterpart were designed to pull down circ_0006404 in PCa cells. As mentioned in Figure 3C, Biotin-miR-1299-WT captured more circ_0006404 in comparison with that in Biotin-miR-1299-MUT group. The endogenous interaction between miR-1299 and circ_0006404 was tested by RIP assay. Anti-Ago2 was used to pull down RNA-induced silencing complex (RISC) that contains Ago2 protein. Circ_0006404 and miR-1299 were substantially enriched in Ago2 immunoprecipitation pellet (Figure 3D), suggesting the interaction between miR-1299 and circ_0006404 in PCa cells. MiR-1299 was abnormally down-regulated in PCa tissues and cell lines in contrast with that in normal tissues and WPMY-1 cell line (Figure 3E and F). The interference efficiency of miR-1299 inhibitor was high in PCa cells (Figure 3G). Circ_0006404 silencing up-regulated the expression of miR-1299, while the addition of miR-1299 inhibitor reduced miR-1299 level again in PCa cells (Figure 3H). The linear correlation between the levels of circ_0006404 and miR-1299 was analyzed using Spearman correlation coefficient. MiR-1299 expression was negatively correlated with the level of circ_0006404 (Supplementary Figure 2A), which further confirmed that miR-1299 was a target of circ_0006404. Collectively, circ_0006404 negatively regulated miR-1299 level through direct interaction in PCa cells.
Si-Circ_0006404-Mediated Influences in PCa Cells are Partly Weakened by the Addition of miR-1299 Inhibitor
To explore if circ_0006404 exerted an oncogenic role in PCa cells through acting as a sponge of miR-1299, we transfected si-circ_0006404 alone or together with miR-1299 inhibitor into PCa cells. Circ_0006404 silencing suppressed the viability of PCa cells, and viability of PCa cells was recovered in si-circ_0006404 + miR-1299 inhibitor co-transfected group (Figure 4A). Meanwhile, the results of transwell assays revealed that the metastasis of PCa cells was restrained with the interference of circ_0006404, and the addition of miR-1299 inhibitor recovered the migration and invasion abilities of PCa cells (Figure 4B). The proliferation ability was up-regulated in si-circ_0006404 + miR-1299 inhibitor group which was impaired by the interference of circ_0006404 (Figure 4C). Si-circ_0006404-mediated cell cycle suppression and apoptosis promotion in PCa cells were partly reversed by the introduction of miR-1299 inhibitor (Figure 4D and E). Overall, circ_0006404 acted as an oncogene to promote the proliferation, viability and metastasis and impede the apoptosis of PCa cells through sponging miR-1299.
CFL2 mRNA Directly Interacts with miR-1299 in PCa Cells
According to the data of TargetScan database, there might be a direct binding relationship between miR-1299 and the 3ʹ UTR of CFL2 mRNA via the predicted binding sites shown in Figure 5A. Meanwhile, we mutated the binding sequence in CFL2 mRNA by “AAGGUCU” to explore if this sequence was essential for the target relationship between miR-1299 and CFL2 mRNA (Figure 5A). DU145 and LNCaP-AI cells were co-transfected with miRNA NC or miR-1299 mimic and WT-CFL2-3ʹ UTR or MUT-CFL2-3ʹ UTR. As shown in Figure 5B, compared with miRNA NC and WT-CFL2-3ʹ UTR co-transfected group, luciferase activity was dramatically reduced in miR-1299 mimic and WT-CFL2-3ʹ UTR co-transfected group, suggesting the direct binding between miR-1299 and CFL2 mRNA. The expression feature of CFL2 in PCa was also explored. There was a notable up-regulation in CFL2 mRNA and protein levels in PCa tumor tissues compared with corresponding normal tissues (Figure 5C and D). CFL2 protein level was also up-regulated in PCa cell lines compared with WPMY-1 cell line (Figure 5E). The ectopic expression efficiency of pc-CFL2 was high in PCa cells (Figure 5F). To explore the regulatory relationship between these two genes, we co-transfected miR-1299 mimic and pc-CFL2 into PCa cells. As mentioned in Figure 5G, miR-1299 accumulation decreased the protein level of CFL2, and the introduction of pc-CFL2 ectopic expression plasmid recovered the protein expression of CFL2 in PCa cells, suggesting the negative regulatory relationship between miR-1299 and CFL2 in PCa cells. The expression of CFL2 was reversely correlated with the level of miR-1299 (Supplementary Figure 2B), which further verified that CFL2 was a target of miR-1299 in PCa cells. Similar transfection was also used to perform rescue experiments to test whether miR-1299 functioned through interacting with and down-regulating CFL2. MiR-1299 overexpression impeded the viability, metastasis and proliferation and promoted the apoptosis of PCa cells (Figure 5H–L). The addition of CFL2 ectopic expression plasmid partly recovered the malignant behaviors of PCa cells (Figure 5H–L). Overall, CFL2 was a direct target of miR-1299 in PCa cells, and miR-1299 suppressed the malignant phenotypes of PCa cells through down-regulating CFL2.
Circ_0006404 Acts as a Molecular Sponge of miR-1299 to Enhance the Level of CFL2 in PCa Cells
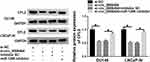
To explore the modulatory interaction among circ_0006404, miR-1299 and CFL2 in PCa cells, DU145 and LNCaP-AI cells were transfected with si-NC, si-circ_0006404, si-circ_0006404 + inhibitor NC or si-circ_0006404 + miR-1299 inhibitor. Circ_0006404 silencing down-regulated the protein expression of CFL2, and CFL2 level was recovered in si-circ_0006404 + miR-1299 inhibitor co-transfected group (Figure 6), suggesting that circ_0006404 functioned as a sponge of miR-1299 to up-regulate CFL2 expression in PCa cells.
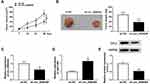
Circ_0006404 Silencing Suppresses the PCa Tumor Growth in vivo
Given our data that circ_0006404 promoted the progression of PCa in vitro, we further tested if circ_0006404 exerted a similar role in PCa tumor growth in vivo. DU145 cell line stably expressing sh-circ_0006404 and matching control cell line stably expressing sh-NC were built to conduct tumor xenograft assay. We generated a tumor growth curve by monitoring tumor volume every week. As mentioned in Figure 7A, tumors were smaller in sh-circ_0006404 group compared with sh-NC group. Besides, the tumor weight after 28-d injection was lighter in sh-circ_0006404 group in comparison with sh-NC group (Figure 7B). The key molecules in tumor tissues from sh-NC group and sh-circ_0006404 group were also detected by qRT-PCR and Western blot assay. As mentioned in Figure 7C–E, circ_0006404 level and CFL2 protein level were decreased in sh-circ_0006404 group than that in sh-NC group, while miR-1299 expression was up-regulated in sh-circ_0006404 group compared with sh-NC group. Overall, circ_0006404 promoted the tumor growth of PCa through miR-1299/CFL2 axis in vivo.
Discussion
Circular RNAs (circRNAs) are closed circular RNAs that derive from their parental linear RNAs by alternative splicing.23,24 Due to their high stability and resistance to RNase, circRNAs are ideal bio-markers for the early diagnosis and treatment of diverse cancers.25,26 Ji et al claimed that circ_0001649 level was reduced in colorectal cancer, and its level was closely related to pathological differentiation.27 Yin et al demonstrated that serum circ_0001785 was a stable bio-marker for breast cancer diagnosis.28 Circ_0006404 has been found to accelerate the proliferation and invasion of glioblastoma cells through up-regulating NFAT5 via its miRNA sponge activities for miR-138-5p and miR-432-5p.29 However, the role of circ_0006404 in PCa was debatable. Kong et al found that circ_0006404 promoted the development of PCa through targeting miR-29a-3p.11 Shen et al demonstrated that circ_0006404 silencing accelerated the progression and docetaxel resistance of PCa.12 Herein, an obvious up-regulation in circ_0006404 level was observed in PCa tissues and cell lines. Through silencing circ_0006404 using its specific siRNA si-circ_0006404, circ_0006404 was found to be an oncogene to contribute to the survival, metastasis and proliferation and impair the apoptosis of PCa cells, which was consistent with the report of Kong et al.
To investigate the potential working mechanism of circ_0006404-mediated development of PCa, circ_0006404-mediated ceRNA network was explored through the bioinformatic database Circular RNA Interactome. MiR-1299 was one of the candidate miRNA targets of circ_0006404. MiR-1299 was verified as a tumor suppressor in osteosarcoma, hepatocellular carcinoma and acute myeloid leukemia.30,31 Furthermore, Zhang et al claimed that miR-1299 abundance was decreased in PCa and it suppressed the proliferation and motility of PCa cells through regulating the level of NEK2.18 Due to the opposite expression trend of miR-1299 to circ_0006404 in PCa and its tumor suppressor role in multiple cancers, miR-1299 was selected for further experiments. The direct target interaction between miR-1299 and circ_0006404 in PCa cells was confirmed by dual-luciferase reporter assay, RNA-pull down assay and RIP assay. Through the expression pattern analysis using qRT-PCR, miR-1299 was found to be aberrantly down-regulated in PCa tissues and cell lines compared with adjacent normal prostate tissues and WPMY-1 cell line. Furthermore, there was a negative correlation between the expression of miR-1299 and circ_0006404. The rescue assays revealed that circ_0006404 contributed to PCa progression through sponging and down-regulating miR-1299.
CFL2 was predicted as a target of miR-1299 by TargetScan database. Dual-luciferase reporter assay subsequently validated the direct interaction between miR-1299 and CFL2 in PCa cells. CFL2 expression was negatively correlated with the level of miR-1299. CFL2 was an up-regulated gene in PCa tissues and cell lines. The oncogenic role of CFL2 in PCa has been reported before. Wo et al reported that SOX2-OT promoted the proliferation and migration of PCa cells through elevating CFL2 level via sequestering miR-369a-3p.21 In our study, CFL2 overexpression accelerated the malignant potential of PCa cells, which was in agreement with the previous article.21 Furthermore, miR-1299 accumulation-mediated suppression on the progression of PCa was largely reversed by the introduction of CFL2 overexpression plasmid, demonstrating that miR-1299 exerted an anti-tumor role in PCa through targeting and down-regulating CFL2. Further study revealed that circ_0006404 enhanced the level of CFL2 through sponging miR-1299 in PCa cells.
Animal experiments demonstrated that circ_0006404 promoted PCa tumor growth via miR-1299/CFL2 axis in vivo. More in vivo experiments are needed to illustrate the role of circ_0006404 in the metastasis of PCa tumors. Furthermore, circ_0006404 level in serum of PCa patients and healthy volunteers should be tested to verify if circulating circ_0006404 is suitable for the diagnosis of PCa patients.
In conclusion, circ_0006404 promoted the viability, metastasis and proliferation and restrained the apoptosis of PCa cells through sequestering miR-1299 to up-regulate CFL2 in vitro. Besides, circ_0006404 also accelerated tumor growth of PCa via miR-1299/CFL2 axis in vivo. Due to the dysregulated expression and stable circular structure of circ_0006404, circ_0006404 might be a potential bio-marker for PCa diagnosis and treatment. Furthermore, miR-1299 and CFL2 might also be potential therapeutic targets for PCa. Silencing of circ_0006404 and CFL2 and restoration of miR-1299 expression provide novel attractive therapeutic strategies for PCa.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Viktorov AA, Zharinov GM, Neklasova NJ, et al. Prediction of life expectancy for prostate cancer patients based on the kinetic theory of aging of living systems. Adv Gerontol. 2017;30(3):356–362.
3. Kalra S, Basourakos S, Abouassi A, et al. The implications of ageing and life expectancy in prostate cancer treatment. Nat Rev Urol. 2016;13(5):289–295. doi:10.1038/nrurol.2016.52
4. Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–256. doi:10.1016/j.cell.2009.04.056
5. Misawa A, Takayama K, Urano T, et al. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem. 2016;291(34):17861–17880. doi:10.1074/jbc.M116.718536
6. Grozescu T, Popa F. Prostate cancer between prognosis and adequate/proper therapy. J Med Life. 2017;10(1):5–12.
7. Fong MK, Hare R, Jarkowski A. A new era for castrate resistant prostate cancer: a treatment review and update. J Oncol Pharm Pract. 2012;18(3):343–354. doi:10.1177/1078155212437599
8. Rodrigues DN, Butler LM, Estelles DL, et al. Molecular pathology and prostate cancer therapeutics: from biology to bedside. J Pathol. 2014;232(2):178–184. doi:10.1002/path.4272
9. Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. doi:10.1093/nar/gkv045
10. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
11. Kong Z, Wan X, Lu Y, et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J Cell Mol Med. 2020;24(1):799–813. doi:10.1111/jcmm.14791
12. Shen Z, Zhou L, Zhang C, et al. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi:10.1016/j.canlet.2019.10.006
13. Wu CL, Ho JY, Chou SC, et al. MiR-429 reverses epithelial-mesenchymal transition by restoring E-cadherin expression in bladder cancer. Oncotarget. 2016;7(18):26593–26603. doi:10.18632/oncotarget.8557
14. Zhang X, Xu Y, Yamaguchi K, et al. Circular RNA circVAPA knockdown suppresses colorectal cancer cell growth process by regulating miR-125a/CREB5 axis. Cancer Cell Int. 2020;20:103. doi:10.1186/s12935-020-01178-y
15. Zhou G, Shen M, Zhang Z. ZW10 binding factor (ZWINT), a direct target of Mir-204, predicts poor survival and promotes proliferation in breast cancer. Med Sci Monit. 2020;26:e921659. doi:10.12659/MSM.921659
16. Antognelli C, Cecchetti R, Riuzzi F, et al. Glyoxalase 1 sustains the metastatic phenotype of prostate cancer cells via EMT control. J Cell Mol Med. 2018;22(5):2865–2883. doi:10.1111/jcmm.13581
17. Lin L, Cai J. Circular RNA circ-EGLN3 promotes renal cell carcinoma proliferation and aggressiveness via miR-1299-mediated IRF7 activation. J Cell Biochem. 2020;121(11):4377–4385. doi:10.1002/jcb.29620
18. Zhang FB, Du Y, Tian Y, et al. MiR-1299 functions as a tumor suppressor to inhibit the proliferation and metastasis of prostate cancer by targeting NEK2. Eur Rev Med Pharmacol Sci. 2019;23(2):530–538. doi:10.26355/eurrev_201901_16865
19. Luo D, Wilson JM, Harvel N, et al. A systematic evaluation of miRNA: mRNAinteractions involved in the migration and invasion of breast cancer cells. J Transl Med. 2013;11:57. doi:10.1186/1479-5876-11-57
20. Bian Y, Guo J, Qiao L, et al. miR-3189-3p mimics enhance the effects of S100A4 siRNA on the inhibition of proliferation and migration of gastric cancer cells by targeting CFL2. Int J Mol Sci. 2018;19(1):236. doi:10.3390/ijms19010236
21. Wo Q, Zhang D, Hu L, et al. Long noncoding RNA SOX2-OT facilitates prostate cancer cell proliferation and migration via miR-369-3p/CFL2 axis. Biochem Biophys Res Commun. 2019;520(3):586–593. doi:10.1016/j.bbrc.2019.09.108
22. Zhu H, Wang G, Zhou X, et al. miR-1299 suppresses cell proliferation of hepatocellular carcinoma (HCC) by targeting CDK6. Biomed Pharmacother. 2016;83:792–797. doi:10.1016/j.biopha.2016.07.037
23. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. doi:10.1080/15476286.2016.1271524
24. Eger N, Schoppe L, Schuster S, et al. Circular RNA splicing. Adv Exp Med Biol. 2018;1087:
25. Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067–2092. doi:10.2147/OTT.S233672
26. Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi:10.1016/j.trecan.2020.01.012
27. Ji W, Qiu C, Wang M, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497(1):122–126. doi:10.1016/j.bbrc.2018.02.036
28. Yin WB, Yan MG, Fang X, et al. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. doi:10.1016/j.cca.2017.10.011
29. Zhang S, Liao K, Miao Z, et al. CircFOXO3 promotes glioblastoma progression by acting as a competing endogenous RNA for NFAT5. Neuro Oncol. 2019;21(10):1284–1296. doi:10.1093/neuonc/noz128
30. Gao AM, Yuan C, Hu AX, et al. circ_ARF3 regulates the pathogenesis of osteosarcoma by sponging miR-1299 to maintain CDK6 expression. Cell Signal. 2020;72:109622. doi:10.1016/j.cellsig.2020.109622
31. Zhang L, Bu Z, Shen J, et al. A novel circular RNA (hsa_circ_0000370) increases cell viability and inhibits apoptosis of FLT3-ITD-positive acute myeloid leukemia cells by regulating miR-1299 and S100A7A. Biomed Pharmacother. 2020;122:
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.