Back to Journals » Drug Design, Development and Therapy » Volume 15
Chemistry, Biosynthesis, Physicochemical and Biological Properties of Rubiadin: A Promising Natural Anthraquinone for New Drug Discovery and Development
Authors Watroly MN , Sekar M , Fuloria S, Gan SH , Jeyabalan S , Wu YS , Subramaniyan V , Sathasivam KV, Ravi S, Mat Rani NNI, Lum PT, Vaijanathappa J, Meenakshi DU, Mani S, Fuloria NK
Received 9 September 2021
Accepted for publication 13 October 2021
Published 3 November 2021 Volume 2021:15 Pages 4527—4549
DOI https://doi.org/10.2147/DDDT.S338548
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Mohd Nasarudin Watroly,1 Mahendran Sekar,1 Shivkanya Fuloria,2 Siew Hua Gan,3 Srikanth Jeyabalan,4 Yuan Seng Wu,5,6 Vetriselvan Subramaniyan,7 Kathiresan V Sathasivam,8 Subban Ravi,9 Nur Najihah Izzati Mat Rani,10 Pei Teng Lum,1 Jaishree Vaijanathappa,11 Dhanalekshmi Unnikrishnan Meenakshi,12 Shankar Mani,13 Neeraj Kumar Fuloria2
1Department of Pharmaceutical Chemistry, Faculty of Pharmacy and Health Sciences, Universiti Kuala Lumpur Royal College of Medicine Perak, Ipoh, Perak, 30450, Malaysia; 2Faculty of Pharmacy & Centre of Excellence for Biomaterials Engineering, AIMST University, Kedah, 08100, Malaysia; 3School of Pharmacy, Monash University Malaysia, Bandar Sunway, Selangor Darul Ehsan, 47500, Malaysia; 4Department of Pharmacology, Sri Ramachandra Faculty of Pharmacy, Sri Ramachandra Institute of Higher Education and Research (DU), Chennai, Tamil Nadu, 600116, India; 5Centre for Virus and Vaccine Research, School of Medical and Life Sciences, Sunway University, Selangor, 47500, Malaysia; 6Department of Biological Sciences, School of Medical and Life Sciences, Sunway University, Selangor, 47500, Malaysia; 7Faculty of Medicine, Bioscience and Nursing, MAHSA University, Selangor, 42610, Malaysia; 8Faculty of Applied Science & Centre of Excellence for Biomaterials Engineering, AIMST University, Kedah, 08100, Malaysia; 9Department of Chemistry, Karpagam Academy of Higher Education, Coimbatore, Tamil Nadu, 640 021, India; 10Faculty of Pharmacy and Health Sciences, Universiti Kuala Lumpur Royal College of Medicine Perak, Ipoh, Perak, 30450, Malaysia; 11Department of Pharmaceutical Chemistry, School of Life Sciences, JSS Academy of Higher Education and Research Mauritius, Vacoas, Mauritius; 12College of Pharmacy, National University of Science and Technology, Muscat, 130, Oman; 13Department of Pharmaceutical Chemistry, Sri Adichunchanagiri College of Pharmacy, Adichunchanagiri University, Mandya, Karnataka, 571418, India
Correspondence: Neeraj Kumar Fuloria; Shivkanya Fuloria
Faculty of Pharmacy & Centre of Excellence for Biomaterials Engineering, AIMST University, Kedah, 08100, Malaysia
Tel +60 16 4037685
; +60 14 3034057
Email [email protected]; [email protected]
Abstract: Anthraquinones (AQs) are found in a variety of consumer products, including foods, nutritional supplements, drugs, and traditional medicines, and have a wide range of pharmacological actions. Rubiadin, a 1,3-dihydroxy-2-methyl anthraquinone, primarily originates from Rubia cordifolia Linn (Rubiaceae). It was first discovered in 1981 and has been reported for many biological activities. However, no review has been reported so far to create awareness about this molecule and its role in future drug discovery. Therefore, the present review aimed to provide comprehensive evidence of Rubiadin’s phytochemistry, biosynthesis, physicochemical properties, biological properties and therapeutic potential. Relevant literature was gathered from numerous scientific databases including PubMed, ScienceDirect, Scopus and Google Scholar between 1981 and up-to-date. The distribution of Rubiadin in numerous medicinal plants, as well as its method of isolation, synthesis, characterisation, physiochemical properties and possible biosynthesis pathways, was extensively covered in this review. Following a rigorous screening and tabulating, a thorough description of Rubiadin’s biological properties was gathered, which were based on scientific evidences. Rubiadin fits all five of Lipinski’s rule for drug-likeness properties. Then, the in depth physiochemical characteristics of Rubiadin were investigated. The simple technique for Rubiadin’s isolation from R. cordifolia and the procedure of synthesis was described. Rubiadin is also biosynthesized via the polyketide and chorismate/o-succinylbenzoic acid pathways. Rubiadin is a powerful molecule with anticancer, antiosteoporotic, hepatoprotective, neuroprotective, anti-inflammatory, antidiabetic, antioxidant, antibacterial, antimalarial, antifungal, and antiviral properties. The mechanism of action for the majority of the pharmacological actions reported, however, is unknown. In addition to this review, an in silico molecular docking study was performed against proteins with PDB IDs: 3AOX, 6OLX, 6OSP, and 6SDC to support the anticancer properties of Rubiadin. The toxicity profile, pharmacokinetics and possible structural modifications were also described. Rubiadin was also proven to have the highest binding affinity to the targeted proteins in an in silico study; thus, we believe it may be a potential anticancer molecule. In order to present Rubiadin as a novel candidate for future therapeutic development, advanced studies on preclinical, clinical trials, bioavailability, permeability and administration of safe doses are necessary.
Keywords: Rubiadin, Rubia cordifolia, biosynthesis, physicochemical properties, anticancer, pharmacology
Graphical Abstract:
Introduction
The most important translational research endeavor that contributes to human health and well-being is drug discovery. All efforts involved in changing a drug-like molecule to a final licensed drug product for marketing by the necessary regulatory authorities are referred to as drug development. Identifying and screening of small molecules for their therapeutic benefits and biological properties are critical phases for drug discovery and development. Most of the well-known compounds in the world has been documented in a library that was created over a long period of time. Among those compounds or small molecules, most of them are derived from natural resources in the past. Currently, the researchers have access to a wide range of chemical substances and hundreds of thousands of novel compounds have also been discovered through high-throughput and combinatorial chemical processes. It remains to be identified which of these millions of molecules have the features that will allow them to become drugs.
Anthraquinones (AQs) are the biggest group of compounds with natural colors, with over 700 chemicals identified thus far. Plants provided approximately 200 of these chemicals, with the remaining coming from lichens and fungus.1,2 AQs are found in all plant parts including the roots, rhizomes, fruits and flowers with the majority produced from 9,10-anthracenedione. These compounds are also found in peas, cabbage, lettuce and beans.3 Due to their wide range of applications, AQs and their derivatives are among the most extensively utilized phytomolecules in the food and pharmaceutical industries. Studies have shown that AQs have been their reported for antioxidant,4 antitumor,5–9 anti-inflammatory,9,10 diuretic,9 antiarthritic,11 antifungal,12 antibacterial,13 and antimalarial14 activities. AQ derivatives are a very useful category in the search for anticancer medicines. AQ-based drugs such as doxorubicin, valrubicin, mitoxantrone, idarubicin, and epirubicin have been used successfully to treat hematological and solid malignancies. Therefore, the AQs core continues to be a potential scaffold for developing novel therapeutic candidates.15
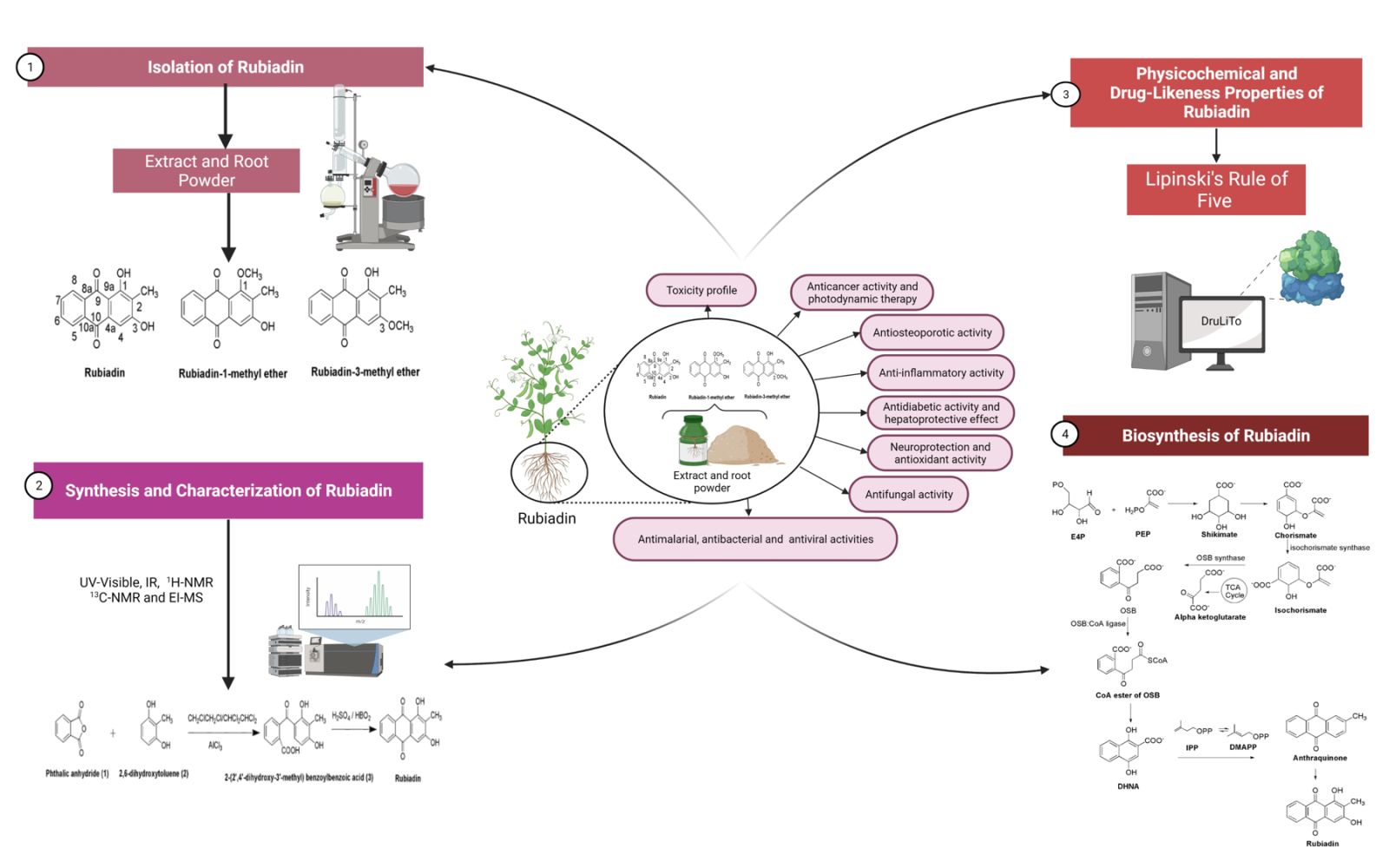
Rubiadin, a 1,3-dihydroxy-2-methyl anthraquinone (Figure 1), is primarily obtained from Rubia cordifolia Linn (R. cordifolia), which belongs to Rubiaceae family. It is an essential component in the Ayurvedic system of medicine in the treatment of various diseases.16 Rubiadin, which is ubiquitous in many natural products, has sparked a lot of interest in recent years due to its excellent pharmacological effects including anticancer, antiosteoporotic, hepatoprotective, neuroprotective, anti-inflammatory, antidiabetic, antioxidant, antibacterial, antimalarial, antifungal, and antiviral activities. Nevertheless, despite its various therapeutic effects, it lacks a thorough and comprehensive review, to date. Hence, in the present review, the phytochemistry, biosynthesis and physicochemical properties of Rubiadin is summarized, with a particular focus on its biological activity. In order to strengthen this review, we have carried out molecular docking studies with selected proteins to prove its anticancer mechanism. It is hoped that the scientific evidence from this review can serve as a solid foundation for further research and provide important information for developing Rubiadin as a therapeutic agent and health product.
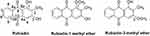 |
Figure 1 Chemical structures of Rubiadin and its analogues. Note: Created with ChemDraw Ultra 8.0. |
Methods
Relevant literature was collected from PubMed, ScienceDirect, Scopus and Google Scholar. The following keywords were used in the search: “Rubiadin” OR 1,3-dihydroxy-2-methyl anthraquinone OR 9,10-anthracenedione AND “Chemistry” OR “Biosynthesis” OR “in-vitro” OR “in-vivo” OR “Biological studies” OR “Pharmacological studies” OR “Toxicity” OR “Pharmacokinetics” OR “Pharmacodynamics” OR “Pharmacokinetics” OR “Pharmacodynamics”. An initial screening was performed on studies that were not written in English or did not have any abstracts. The review’s data was divided into two main categories: Rubiadin’s chemical and biological properties. The scientific evidence gathered is summarized and incorporated following a thorough screening.
Phytochemistry of Rubiadin
Origin and Distribution
Rubiadin is primarily isolated from the root of R. cordifolia, a Rubiaceae family. R. cordifolia is an important medicinal plant used in the Ayurvedic system of medicine in the treatment of a variety of diseases.16 Morinda officinalis (M. officinalis) which is another major source of Rubiadin17 is a commonly used traditional Chinese medicine which has been used in China for many years. Additionally, Rubiadin is also found in a traditional medicine named Manjisthadi churna, which consists of lesser cardamom, used in the treatment of hyperlipidemia in India’s Ayurvedic system of medicine and traditional medical practices.18 It is also found in some traditional Chinese medicines including Jia-Jian-Di-Huang-Yin-Zi decoction which is a seven-herb component consisting of Radix rehmanniae, Fructus corni, Radix morindae officinalis, Herba cistanches, Radix angelicae sinensis, Radix asparagi and Radix paeoniae alba19 as well as another decoction named Er Xian20 which is a six-herb component consisting of Herba epimedii, Radix morindae officinalis, Radix angelicae sinensis, Rhizoma anemarrhenae, Cortex phellodendri and Rhizoma curculiginis. Medicinal plants containing Rubiadin, Rubiadin-1-methyl ether (RBME) and Rubiadin-3-methyl ether are summarized in Table 1.
 |
Table 1 List of Medicinal Plants Containing Rubiadin |
Medicinal Uses of R. cordifolia
R. cordifolia, also known as Indian madder or Manjistha, is a medicinal plant that grows in the forests of Pakistan, India, China, Korea, Japan and Mongolia.21 Based on ethnobotanical reports, its roots are used in the treatment of jaundice while the stems are used to treat snake bites and scorpion stings. It is also useful against diabetic foot ulcers.22 Traditionally, R. cordifolia is used for chronic pyrexia and puerperal fever, as well as a common medicine to alleviate heat and itching in eczema, psoriasis, herpes and scabies. When combined with honey, it is also deemed as effective against vitiligo.23 Other therapeutic effects include as an immunomodulator, analgesic, diuretic, gastroprotective, hepatoprotective, antioxidant, wound healing, nephroprotective and antiviral properties.24–27
Isolation of Rubiadin
The powdered root parts of R. cordifolia are extracted using a cold percolation in 50% aqueous ethanol following by a drying step using a rotary evaporator under a reduced pressure and a controlled temperature. In a separating funnel, the solvent-free extract is portioned with a 90% aqueous methanol and n-hexane. The n-hexane layer is removed and the aqueous-methanolic layer is subsequently dried and suspended in water for a further extraction step using chloroform (fraction 2) and n-butanol extractions in successive manner. Fraction 2 is chromatographed on a silica gel followed by an elution step with hexane, toluene, ethyl acetate and butanol (in order of increasing polarity) using varied solvent ratios. The column is further eluted with a toluene–hexane (1:1) combination, yielding a yellow solid. The solid is purified by numerous recrystallization processes using toluene and is further validated by a thin layer chromatography using toluene:ethyl acetate (85:15) as the mobile phase (Rf = 0.58). The spots are finally observed under UV irradiation.16,28
Synthesis of Rubiadin
In the first step, the condensation of phthalic anhydride (1) and 2,6-dihydroxytoluene (2) with CH2ClCH2Cl/CHCl2CHCl2 in the presence of aluminium chloride yielded 2-(2ʹ,4ʹ-dihydroxy-3ʹ-methyl) benzoylbenzoic acid (3). Then, in the second step, cyclization via dehydration of the compound 3 is performed in the presence of fused boric acid with concentrated sulphuric acid at 100 °C for 25 min to yield Rubiadin29 (Figure 2).
 |
Figure 2 Synthesis of Rubiadin. Note: Created with ChemDraw Ultra 8.0. |
Structural Characterization of Rubiadin
A very detailed structural characterization of Rubiadin is given below based on the values obtained from spectroscopic methods including ultraviolet (UV), Fourier-transformed infrared (FTIR), 1H-nuclear magnetic resonance (NMR), 13C-NMR and EI-MS.30 The UV spectra of Rubiadin indicates a λmax at 408 nm for n-π* transition and at 279 and 251 for π-π* transitions. The IR spectra shows absorption bands at 3396 (OH), 2923, (C-H), 1661 (C=O non-chelated) and at 1623, 1589 (C=C< aromatic) representing the characteristic groups of Rubiadin. It also shows a molecular ion peak at m/z 254.12 in EI-MS and analysed for a molecular formula C15H10O4.
The 1H-NMR spectrum reveals the presence of a hydrogen-bonded hydroxyl group at δ 13.06 (1H, s] and another phenolic hydrogen at δ 11.22 (1H, br.s.). The strong singlet for three protons at δ 2.02 is due to the presence of a methyl group attached to the aromatic ring system while the singlet at δ 7.1 for one proton is due to an aromatic proton in H-4 position. On the other hand, the multiplet between δ 7.83 and 7.88 for two protons are attributed to aromatic protons at H-6 and H-7 positions. A pair of doublet of doublets at δ 8.08 (dd, 1H, J = 1.5, 7.5 Hz) and 8.12 (dd, 1H, J = 1.5, 7.5 Hz) are assigned to H-8 and H-5, respectively. In its 13C-NMR spectrum, there were 15 signals out of which two signals at δ 186.2 and 181.78 are due to conjugated carbonyl carbons and are characteristic signals of anthraquinoid nucleus. The signal present in the upfield region at δ 8.09 is due to the methyl carbon while the signal at δ 108.9 is assigned to C-2 carbon. The two phenolic carbons appeared at δ 162.82 and 162.46 where the carbon atoms adjacent to the phenolic carbons resonated at δ 107.35 (C-4) and 117.3 (C-9a). The remaining signals at δ 134.54 (C-8a), 134.44 (C-10a), 132.98 (C-4a), 132.87 (C-6), 131.70 (C-7), 126.70 (C-5) and 126.37 (C-8) are characteristic signals off the unsubstituted aromatic ring of the AQ system.30
Physicochemical and Drug-Likeness Properties of Rubiadin
The physicochemical properties of Rubiadin are mainly obtained from PubChem31 and other reliable databases such as DruLiTo. Certain physicochemical features allow a molecule to be converted into a drug-like molecule: 1) molecule should be small enough to be transported throughout the body, 2) hydrophilic enough to dissolve in the blood stream, 3) lipophilic enough to cross fat barriers within the body and 4) contain enough number of polar groups to bind to a receptor but not too many which leads elimination too quickly from the body via urine to exert the therapeutic effect (Table 2). The drug-like properties (molecular weight, H-bond donors, H-bond acceptors, log P value and rotatable bonds) as described in Lipinski’s rule of five32 are calculated using Biovia Discovery studio 19.0. Any drug-like compound should have 1) a molecular weight of 500, 2) a partition coefficient (log P)-value of 5, 3) H-bond donors of 5, 4) H-bond acceptors of 10 and 5) rotatable bonds of 10 according to Lipinski’s rule of five. Since compounds that do not break Lipinski’s rule of five may have improved folding, polarity and molecular size, the drug-like molecules are assumed to have the predicted therapeutic benefits.33 The objective of the suggested method is to deliver primary evidence defining the potential physical properties of the compounds, rather than serving as a rigid screening criteria in itself, since Lipinski’s rule of five is an effective and valid guide for forecasting the potential for oral exposure to enhanced chemical compounds. Overall, Rubiadin appears to match all five of Lipinski’s drug-likeness criteria (Table 2). According to the data acquired from DruLiTo software, Rubiadin also passed the Ghose filter, Veber’s rule, blood–brain barrier (BBB) likeness rule, Unweighted Quantitative Estimate of Drug-likeness (QED), and Weighted QED, but failed the CMC-50 like rule and MDDR like rule. All of the above findings indicate that it is a good potential therapeutic agent for a variety of disorders.
 |
Table 2 Computed Physicochemical Properties of Rubiadin |
Biosynthesis of Rubiadin
AQs are derived from a variety of precursors and pathways in nature. Two biosynthesis pathways have been established: 1) the polyketide and 2) the chorismate/o-succinylbenzoic acid pathways, despite the fact that their biosynthetic processes remain unknown. Rubiadin is made by combining phosphophenol pyruvate (PEP) with erythrose-4-phosphate (E4P) to produce chorismate and isochorismate, which are then transformed to o-succinylbenzoate (OSB) in the presence of -ketoglutarate (Figure 3). Rings A and B of Rubiadin are formed through the synthesis of 1,4-dihydroxy-2-naphthoic acid (DHNA) by a ring closure of OSB–CoA. Prenylation of the DHNA results in naphthoquinol or naphthoquinone, which is involved in the synthesis of ring C. Isopentenyl pyrophosphate (IPP), which can be generated from mevalonate (MVA) or 2-C-methyl-D-erythritol 4-phosphate (MEP), is required for the biosynthesis of AQs, including Rubiadin (Figure 4). In the late stages of biosynthesis, most AQs in the Rubiaceae undergo different alterations owing to hydroxylation or methylation, in which groups like as hydroxy and methyl groups are added to Rubiadin’s ring C.34
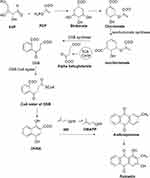 |
Figure 3 Biosynthesis of Rubiadin. Note: Created with ChemDraw Ultra 8.0. |
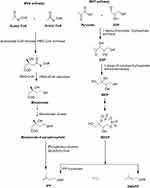 |
Figure 4 MVA and MEP pathways for the synthesis of IPP. Note: Created with ChemDraw Ultra 8.0. |
Biological Activities of Rubiadin
Rubiadin has been reported to possess many biological activities (Figure 5) as summarized below.
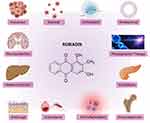 |
Figure 5 Biological properties and therapeutic potential of Rubiadin. Note: Created with BioRender.com. |
Toxicity Profile
One of the latest acute toxicity study reported that oral administration of Rubiadin (100, 200, 500 and 1000 mg/kg) to Swiss albino mice using staircase/up and down method showed no significant toxicity.35 Furthermore, mice given an aqueous extract of R. cordifolia and R. tinctorum (Rubiadin as one of the principal constituents) for 14 days indicated that the highest acceptable doses were 3500 mg/kg and 5000 mg/kg, respectively. In another study, both sexes of mice were administered with R. cordifolia root extract mixed in their diet (0–5%) for 90 days with no clinical signs of toxicity seen, although there were some changes in kidney and epidermal vaginal cyst in some animals based on histological analysis. Overall, the findings indicate that Rubiadin is safe at the investigated dose levels (Figure 6).
Anticancer Activity and Photodynamic Therapy
AQs have long been believed to have anticancer properties, acting primarily through DNA damage, cycle arrest and apoptosis.36 Very recently, Cogno et al37 evaluated the photoactivity of Rubiadin on monolayers and multicellular tumor spheroids following a photodynamic therapy. Rubiadin showed photosensitizing ability on monoculture of colon cancer cells (SW 480) at low concentration where necrosis has occurred. In addition, Comini et al38 investigated the potential role of Rubiadin and RBME as phototoxic agents against human breast cancer using MCF-7c3 cells. At 100 μM and a light exposure of 1 J/cm2, both Rubiadin and RBME showed considerable photocytotoxicity on cancer cells. The observed cellular death by the photoactivated Rubiadin and RBME were closely related to a singlet oxygen production, with decreased cell viability in relation to tumor cell uptake. Subsequently, another study by Vittar et al39 revealed that Rubiadin exhibited significant photocytotoxicity on human cancer cells (MCF-7c3) in a concentration-dependent manner. Additionally, biochemical analysis revealed the involvement of caspase-3, PARP cleavage and DNA fragmentation in Rubiadin-induced apoptosis (Figure 6).
In addition to the above studies, Rubiadin also exhibited significant cytotoxicity against HepG2 cells, with minimum inhibitory concentration (IC50) values of 3.6, 4.4 and 4.8 µM, respectively.40 It also conferred cytotoxicity towards the CEM-SS (T-lymphoblastic leukaemia), MCF-7 (breast carcinoma) and HeLa (cervical carcinoma) cell lines with IC50 of 3, 10 and >30 µg/mL, respectively.41 In another study, Rubiadin and RBME exhibited significant cytotoxicity against NCI-H187 cells, with IC50 values of 14.2 and 4.5 µg/mL, respectively,42 indicating its potential anticancer effects.
However, madder color, a food coloring made from Rubia tinctorum roots, has been shown to cause cancer in rats’ kidneys and liver. In F344 rats, treatment with Rubiadin (0.04%) for 23 weeks enhanced atypical renal tubules/hyperplasias and induced renal cell adenomas and carcinomas. Additionally, Rubiadin enhanced glutathione S-transferase placental form-positive liver cell foci and major intestinal dysplasias thus suggesting that Rubiadin enhances renal preneoplastic lesions, with a weaker effect on dysplasia. Additionally, Rubiadin may also target the liver and large intestine, implying that it plays a significant role in madder color-induced carcinogenicity.43 Based on another study by the same researcher, Rubiadin is a potent carcinogenic metabolite of madder color, targeting proximal tubule cells in the outer medulla,44 although oxidative stress increased by lucidin-3-O-primeveroside or alizarin may also be involved in renal carcinogenesis by madder color.
From another study, the excretion of lucidin and rubiadin was observed in rats following administration of lucidinprimeveroside (Lup). Lup was reduced to Rubiadin primeveroside which in turn, was hydrolyzed to Rubiadin, when treated with rat liver extract and nicotinamide adenine dinucleotide phosphate (NADPH). Rubiadin was more potent than lucidin but has similar effect to the positive control 7,12-di-methylbenz[α]anthracene in the unscheduled DNA synthesis experiment in primary rat hepatocytes. The uptake of the AQs glycosides alizarinprimeveroside (Alp) and Lup results in the production of a rodent carcinogen 1-hydroxyanthraquinone, as well as the highly genotoxic compounds lucidin and Rubiadin.45
The latest scientific evidence has demonstrated that novel AQs can inhibit cancer by paraptosis, autophagy, radiosensitization, thus overcoming chemoresistance.36 However, scientists are still far from having a full understanding on the anticancer properties of Rubiadin, since some report mentioned it is also carcinogenic.43,44 Thus, more in vivo and preclinical research are needed to fully understand its apparent potential in preventing and treating a variety of malignancies.
Although photoactivity is known for decades, only recently that it resurfaced as a potential therapy option for cancer and microbial diseases. Its key innovative component is light as the external factor since light can activate drugs locally, besides having a high level of selectivity and conferring minimal side effects.46 Animals that ingest the aerial parts of H. pustulata show a classic primary photoensitization reaction, which is clinically characterized as dermatitis and, in severe cases, blindness. H. pustulata grows mainly in the Andes region of northwest Argentina. Rubiadin, a major compound in H. pustulata, is characterized as either a Type I or II photosensitizer, depending on its physicochemical features. Oral administration of Rubiadin can reproduce natural in experimental animals.
The presence of Rubiadins in the serum was identified and quantified in the skin of experimental animals using high performance liquid chromatography.47 Since H. pustulata contains photosensitizing AQs such as Rubiadin and soranjidiol, the plant can cause dermal lesions through photosensitization.48 Another study reported that Rubiadin and soranjidiol should be administered between 24 and 72 hours after H. pustulata ingestion, in order to coincide with the time when clinical indications are more noticeable. The clinical findings were validated for the presence of Rubiadin and soranjidiol in sera, although Rubiadin and soranjidiol were absent in skin samples. Finally, toxicological investigations on both compounds are important, since several recent researches have suggested that they could be used in photodynamic therapy.49
Rubiadin confer a favorable response to photodynamic therapy and therefore should be further examined. Additionally, any interaction between its use in reversing drug-resistant phenotypes and its photosensitizing effect should be further investigated.
Antiosteoporotic Activity
Low bone mineral density (BMD) and micro-architectural deterioration of bone tissue are characteristic of osteoporosis, resulting in increased bone fragility and fracture risk.50 He et al51 investigated the in vitro effect of RBME on osteoclasts and the underlying mechanism. RBME inhibited the expression of osteoclast-related proteins such as nuclear factor of activated T-cells cytoplasmic 1 (NFATc1), cellular oncogene Fos (c-Fos), matrix metallopeptidase 9 (MMP-9) and cathepsin K (CtsK), as determined by Western blot analyses. RBM also reduced the nuclear translocation of p65 and inhibited the phosphorylation of nuclear factor kappa B (NF-κB) p65 and the degradation of nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor alpha (IκBα) indicating that RBME may be a promising agent for the prevention and treatment of bone disorders characterized by excessive bone resorption, due to its ability to inhibit osteoclastic bone resorption by blocking the NF-κB pathway (Figure 7).
Bao et al52 investigated the in vitro effects of Rubiadin on bone resorption activity and its mechanism on osteoclasts derived from rat bone marrow cells. In a co-culture system of osteoblasts and bone marrow cells, Rubiadin reduced the formation of bone resorption pits, the number of multinucleated osteoclasts and the activity of tartrate resistant acid phosphates (TRAP) and cathepsin K. Additionally, Rubiadin increased the death of osteoclasts caused by macrophage colony stimulation factor (M-CSF) and receptor activator of NF-κB ligand in bone marrow cells (RANKL). Rubiadin also 1) increased the ratio of osteoprotegerin (OPG), RANKL mRNA and protein expression in osteoblasts; 2) interfered the c-Jun N-terminal kinase (JNK) and NF-κB signaling pathways; and 3) reduced the expression of calcitonin receptor and carbonic anhydrase/II in osteoclasts induced from bone marrow cells with M-CSF and RANKL in osteoclasts. Overall, the findings suggest that Rubiadin may be a potential inhibitor for bone resorption.52 Since RBME can inhibit osteoclast TRAP activity and bone resorption, it may be useful against osteoporosis.53
Xia et al54 investigated the protective effect of M. officinalis on glucocorticoid-induced osteoporosis (GIOP)-modelled rats and osteoblasts. Eight weeks after dexamethasone (DEX) injection and M. officinalis treatment in female rats aged 12 weeks, the BMD, micro-architecture of the trabecular bone, serum level of bone metabolism markers and urine metabolomics were assayed in vivo. The cultured osteoblasts were injured with DEX before the effects of M. officinalis, RBME on osteoblastic proliferation, differentiation and mineralization were investigated in vitro. M. officinalis increased BMD, improved the micro-architecture and intervened with bone metabolism via regulating alkaline phosphatase (ALP), tartrate resistant acid phosphatase (TRAP) and c-terminal telopeptides of type I collagen (CTX-I) levels. The in vitro experiment showed that M. officinalis and RBME increased the cell proliferation, ALP activity and enhanced extracellular matrix mineralization in DEX-injured osteoblasts.54 Additionally, another study demonstrated that multiple ingredients in a traditional Chinese medicine decoction named Er-Xian that included Rubiadin has anti-osteoporotic activity.20 Although the data indicated that Rubiadin has action against bone tissue, more efficient and reliable bioassays should be urgently developed for thorough investigation of its antiosteoporotic mechanism before human clinical trials.
Anti-Inflammatory Activity
Inflammation is a set of activities that occur in response to tissue damage associated with oxidative stress or other factors that initiate repair processes including as extracellular matrix remodeling and fibrosis.55 Mohr et al56 investigated the anti-inflammatory efficacy of RBME using an in vitro model of RAW 264.7 macrophages induced by lipopolysaccharide (LPS). The results indicated that RBME decreased the levels of pro-inflammatory markers such as nitric oxide (NOx), interleukin (IL)-6 and IL-1 with increased in macrophage apoptotic rate seen (Figure 7). The anti-inflammatory effects of Rubiadin-1-methyl ether was also investigated using an in vivo acute lung injury (ALI) induced by LPS (5 mg/kg, P.O.) by a similar group of researchers. Administration of RBME (3, 10 and 30 mg/kg, P.O.) decreased leukocyte infiltration, fluid leakage, NOx, IL-6, IL-12p70, interferon gamma (IFN-γ), tumour necrosis factor α (TNF-α) and monocyte chemoattractant protein-1 (MCP-1) levels as well as MPO activity (Mohr et al, 2019). RBME also enhanced the levels of IL-10 in the bronchoalveolar lavage fluid (BALF). On the other hand, Rubiadin’s anti-inflammatory activity and potential mechanism of action, has yet to be confirmed and requires further research.
Antidiabetic Activity
Mujeeb et al57 evaluated the effectiveness of Rubiadin-loaded niosomes (RLN) in treating diabetic nephropathy (DN) that is induced by streptozotocin-nicotinamide (STZ-NA) in Wistar rats. In STZ-NA-induced DN rats, oral administration of RLN (100 and 200 mg/kg/week) markedly reduced blood glucose levels. Furthermore, RLN formulation significantly reduced urea, uric acid and creatinine levels while improving lipid, thiobarbituric acid reactive substances (TBARS), glutathione (GSH), superoxide dismutase (SOD) and catalase (CAT) levels in DN rats (Figure 8).57 Nevertheless, other than this study, no other evidence of Rubiadin’s anti-diabetic effectiveness has been found indicating that further studies are required to confirm Rubiadin’s anti-diabetic properties.
Hepatoprotective Activity
The hepatoprotective effect of Rubiadin was tested against carbon tetrachloride (CCl4)-induced liver injury in rats.16 In CCl4-induced rats (1 mL/kg, i.p), the co-treatment of Rubiadin (50, 100 and 200 mg/kg, P.O.) for 2 weeks restored serum glutamic oxaloacetic transaminase, glutamate pyruvate transaminase, alkaline phosphatase (ALP), γ-glutamyltransferase (γ-GT), glutathione S-transferase and glutathione reductase levels to normal. Rubiadin also inhibited the development of hepatic malondialdehyde and the depletion of reduced glutathione level in the liver of CCl4-intoxicated rats in a dose-dependent manner, thus strongly suggesting that Rubiadin has a hepatoprotective effect against CCl4-induced hepatic damage in rats (Figure 8).16 However, more research is needed in future to confirm Rubiadin’s hepatoprotective potential especially in models that greatly mimic humans liver disease.
Neuroprotection
The significant reduction in the onset of jerks and Straub tail along with a significant increase in onset of clonus and extensor were observed with pre-treatment of Rubiadin suspension (100 and 250 mg/kg, P.O.) for 3 days to the pentylene tetrazole (PTZ)-induced (80 mg/kg, i.p.) clonic-tonic convulsions in mice. Pre-treatment of Rubiadin suspension also significantly reduced the hind limb extension in maximal electro shock (MES)-induced seizures induced by delivering an electroshock at 50 mA using an electro-convulsometer through a pair of ear clip electrodes in Swiss albino mice. Rubiadin suspension (250 mg/kg) conferred some anticonvulsant effects in both MES- and PTZ-induced epileptic seizure models35 overall indicating that Rubiadin is a potential neuroprotectant (Figure 8). The above study is a novel platform for testing Rubiadin against neurodegenerative disorders including Alzheimer’s, Parkinson’s and Huntington’s disease.
Antioxidant Activity
The antioxidant effects of AQs including Rubiadin are well known.58–60 Rubiadin inhibits lipid peroxidation as induced by ferrous sulphate and t-butylhydroperoxide especially in Fe2+-induced lipid peroxidation (Figure 8). Rubiadin’s antioxidant properties were comparatively higher than ethylenediaminetetraacetic acid (EDTA), tris, mannitol, vitamin E and p-benzoquinone.28 Based on a follow-up study by Tripathi and Sharma,61 Rubiadin inhibits lipid peroxidation in a dose-dependent manner. Apart from the studies mentioned above, no in vitro or in vivo antioxidant investigations on Rubiadin have been reported making relating the findings reported in a few models to antioxidant potential a challenge. Further research using a variety of approaches, including animal models with established mechanisms of action, should be conducted in the future to confirm Rubiadin’s antioxidant potential.
Antifungal Activity
AQs and their derived compounds are well documented for their antifungal properties.62,63 Bacillus velezensis is a plant-growth-promoting rhizobacterium with enormous potential for agricultural development. Bacillus velezensis Lle-9 which is isolated from Lilium leucanthum bulbs has shown antifungal activities against plant pathogens such as Botryosphaeria dothidea, Fusarium oxysporum, Botrytis cinerea and Fusarium fujikuroi. The presence of Rubiadin and other antimicrobial compounds in the bacterial culture can assist in isolating Bacillus velezensis Lle-9’s antifungal activity.64
Marioni et al65 who investigated the antifungal effects of Rubiadin and RBME on Candida tropicalis confirmed that both compounds reduced biofilm formation and had an antifungal effect as mediated by oxidative and nitrosative stress under irradiation, with a significant increase in endogenous ROS and superoxide dismutase (SOD) activity. Rubiadin, in particular, altered the pro-oxidant-antioxidant balance. The most prominent effect of irradiation was oxidative stress, which altered the pro-oxidant-antioxidant balance and may contribute to an irreversible cell injury in the biofilm. Rubiadin is also an excellent synergistic combination with Amphotericin B indicating that the photosensitizing Rubiadin is a promising therapy option for Candida infections.65 A follow-up study suggested that the O2•− formed by an electron transfer quenches the Rubiadin and RBME excited states and is the main photosensitizing mechanism involved in the photo-induced antibiofilm activity.66 Additionally, RBME behaves exclusively as a photosensitizer.67
The antifungal activity of Rubiadin has been evaluated against three fungi such as Aspergillus ochraceus, Aspergillus niger and Candida lipolytica where it showed moderate antifungal activity against Aspergillus ochraceus [minimum inhibitory concentration (MIC) of >80 µg/disc] (Figure 9).41 Overall, the findings suggest that Rubiadin and RBME have antifungal activity against a variety of pathogenic Aspergillus and Candida species. Rubiadin has the potential to be employed as a lead compound in the investigation for the development of a potential antifungal agent.
Antimalarial, Antibacterial and Antiviral Activities
Rubiadin and RBME have been shown to have antimalarial activities with IC50 values of 13.00 and 1.56 µg/mL, respectively.68 The number of Plasmodium falciparum parasites (schizonts) decreased significantly in a dose-dependent manner with almost a 100% of inhibition by RBME (30 µg/mL)69 indicating that they have the potential to be developed as antiplasmodial agents. Rubiadin and RBME showed antibacterial activity against Staphylococcus aureus with MIC of 32–64 and >256 µg/mL, respectively. The mechanism of action seems to involve an increase in the levels of superoxide anion O2•− and/or singlet molecular oxygen 1O2.70 In another study, Rubiadin was investigated against hepatitis B virus (HBV) using HepG2.2.15 cells. Rubiadin inhibited HBV DNA replication and lowered hepatitis B e antigen (HBeAg) as well as hepatitis B core antigen (HBcAg) levels. It also decreased HBV x (HBx) protein expression and reduced intracellular free calcium suggesting that it is a promising anti-HBV drug candidate (Figure 9).30
Pharmacokinetics of Rubiadin
AQs which are absorbed mostly through the intestines and are primarily distributed in tissues and organs that receive good blood supply are transformed into another AQ, leading to potential pharmacological and/or toxicological effects.71 The main components of Morinda officinalis are Rubiadin and RBME. To investigate the pharmacokinetics and tissue distribution of these two compounds in rat plasma and tissues, Shi et al17 used an ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). When compared to the findings from Morinda officinalis without wood (MO), the salt-processed Morinda officinalis (SMO) groups had significant increase in the maximum concentration (Cmax) and area under the curve (AUC0-t) indicating that salt steaming can increase Rubiadin’s and RBME’s bioavailabilities. Rubiadin had a time to maximum concentration (Tmax) of 1.5 h, which was longer than RBME, although RBME had the highest Cmax, particularly in the SMO extract. Additionally, the small intestine had the highest concentrations of both Rubiadin and RBME. Nevertheless, due to the limited study focus on Rubiadin’s pharmacokinetic features, a comprehensive assessment of Rubiadin’s absorption, distribution, metabolism and excretion profiles remains lacking.
Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Properties of Rubiadin
The variable nearest neighbor (vNN)-ADMET webserver was used to predict ADMET properties72 of Rubiadin and to build new models based on vNN methodology. The parameters were classified as effects on the liver toxicity profile, metabolism, membrane transporters function, hERG (cardiotoxicity) activity, MMP (mitochondrial toxicity assay), mutagenicity (AMES test), and the maximum recommended therapeutic dose (MRTD). According to the unrestricted prediction model of the program, Rubiadin causes drug-induced liver injury (DILI) but not cytotoxicity, whereas the restricted model has no high confidence prediction. Rubiadin may produce positive findings for the human liver microsomal (HLM) stability assay and according to the unrestricted applicability domain, it may be rapidly metabolized. Rubiadin has the potential to inhibit the CYP 1A2, 2D6, 2C9, and 2C19 enzymes, but not the CYP 3A4 enzymes, as predicted by both models. Rubiadin has been found to have no effect on membrane transporters such as BBB and P-glycoprotein (P-gp). Rubiadin causes mutations and may cause mitochondrial malfunction, according to the results of the AMES and MMP assays. The MRTD for Rubiadin was found to be 665 mg/day, as predicted by the software.
Apart from the parameters listed above, the toxicity estimation software tool (TEST) was used to estimate Rubiadin toxicity using quantitative structure–activity relationships (QSARs) approaches.73 By employing TEST and QSAR techniques, the oral rat LD50 mg/kg (predicted value) of Rubiadin was found to be 1307.13, 3502.50, and 487.82 mg/kg, respectively, to consensus method, hierarchical clustering technique, and nearest neighbor technique. There is a possibility that the mutagenicity assay will be positive, as shown by the software when all three QSAR techniques are used. According to the consensus approach, hierarchical clustering technique, and nearest neighbor technique, the predicted value of the mutagenicity assay was 0.98, 0.97, and 1.00, respectively.
Molecular Docking of Rubiadin Against the Cancer Target Proteins
Molecular docking is a computational tool that can predict how a ligand will attach to a protein with a known three-dimensional structure. Docking may be used to do computer-generated screening on enormous collections of compounds, rate the outcomes, and offer structural models for in what manner the ligands inhibit the target, which is tremendously beneficial in the search for new inhibitors. There are no literature about molecular docking studies describing the interaction of Rubiadin with molecular targets involved in cancer development. To circumvent this constraint, we employed protein-ligand molecular docking to assess the Rubiadin’s binding mechanism and interaction energy with four important enzyme targets, which were identified in this research as being responsible for variety of cancers.
A number of cancer treatments have been evaluated in clinical trials to see if they can inhibit the cMET receptor tyrosine kinase, and resistance mutations in the cMET gene are beginning to be identified in a number of these medications. Molecular investigations are still required to further understand individual cMET modifications at the molecular level, particularly in terms of small molecule identification.74 In certain cancers, chromosomal translocation, amplification, or point mutations in the anaplastic lymphoma kinase (ALK) gene cause the tyrosine kinase to be constitutively activated. This gene has been found as a potential target for molecular docking studies, and it will be investigated extensively.75 PI5P4Ks have been shown to play a role in the growth of cancer cells as well as the development of other disorders. Due to a lack of effective and selective small drugs on the market, targeting these kinases for therapeutic purposes has gotten little attention.76 Hsp90 and Hsp90 have been linked to cancer and neurological disorders, although determining their precise role in these diseases has been difficult due to a lack of specific pharmacological studies.77 The protein with the PDB IDs 3AOX, 6OLX, 6OSP, and 6SDC were extracted from the protein data bank to commemorate the above four targets.
Molegro Virtual Docker (MVD) 6.0 was used to conduct the molecular docking study.78 The docking procedure includes the following steps. MVD was used to import the molecules, including protein and ligand (Rubiadin). In the protein molecule, potential binding sites and the configuration of the search space were determined. The Docking Wizard was used to run a docking simulation. The Pose Organizer and the ligand energy inspector tool were used to inspect the docking results, and the results were tabulated and the docked view was extracted (Table 3). The MolDock score with the lowest values was discovered to have the highest binding affinity to the target proteins. Rubiadin affinity for cancer targets was found to be 6SDC>6OLX>3AOX>6OSP, according to the report obtained. This result strengthens the anticancer potential of Rubiadin and that could help to enlighten this biologically active compound to the next level of drug discovery and development.
 |
Table 3 Docked Study Results of Rubiadin with the Cancer Target Proteins |
Cosmetic Formulation Containing Rubiadin
Rubiadin has been used as an active ingredient in a cream formulation conducted by a group of Korean researchers. The researchers reported that it had an excellent anti-allergy effect by inhibiting β-hexosaminidase secretion and the expression of caspase-1. The cream has been patented and relates to a cosmetic composition comprising ceramide, which can relieve atopic symptoms, additionally conferred by its moisturizing effect.79
Possible Structural Modifications in Rubiadin
Currently, it remains unclear whether the bioactivities of Rubiadin can be improved via structural modifications. Some analogues of Rubiadin have been isolated or synthesized using methylation and acylation reactions, which are possible due to their phenolic hydroxyl groups. The side-chain modifications is an impetus for further efforts to increase the therapeutic potential of this class of compounds (Figure 10). For example, Rubiadin analogues 1–3 have been synthesised and their cytotoxic activities against MCF-7 and K-562 cancer cell lines and the structure–activity relationship have been described.80 The structure–activity relationship suggested that methoxy and hydroxyl groups are important for the cytotoxicity and selectivity of the substituted AQs. Further RBME 4 and Rubiadin-3-methyl ether 5 were isolated from different medicinal plants and some of their biological activities were investigated (Table 1). Additionally, Rubiadin-1-methyl ether-3-O-β-primeveroside 6 has been isolated, characterised and reported from Pentas lanceolata.81
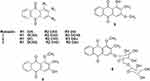 |
Figure 10 Possible structural modifications of Rubiadin. Note: Created with ChemDraw Ultra 8.0. |
The in silico design of more potent Rubiadin derivatives can be used as a way forward for novel drug discovery and development. Nevertheless, in vitro and in vivo studies should be conducted in future to confirm the safety and efficacy of all semi-synthetic derivatives of Rubiadin. Additionally, more studies relating to its structure–activity relationship (SAR) are warranted in the future to obtain several other novel compounds derived from Rubiadin.
Conclusion and Future Perspectives
In this review, the presence of Rubiadin in medicinal plants and its isolation, synthesis, structural characterization, physicochemical properties, along with its biosynthesis are described in detail. Additionally, the scientific updates on its biological and therapeutic potentials have been provided. Accumulating evidence provided by various preclinical studies has shown that Rubiadin can be a promising anticancer, anti-osteoporotic, hepatoprotective, neuroprotective, anti-inflammatory, antidiabetic, antioxidant, antibacterial, antimalarial, antifungal and antiviral drug candidate for further development. Rubiadin was proven to have the highest binding affinity to the cancer targeted proteins in an in silico study, thus we believe it may be a potential anticancer molecule. The in silico findings indicate that Rubiadin has a high ligand-potentiality for a wide range of macromolecules, leading us to believe that it may interact with a variety of other enzymes or proteins that were not included in this simulation. It is hoped that this review will stimulate further investigations on Rubiadin in relation to its pharmacokinetics, pharmacodynamics, clinical and SAR studies, which can help accelerate the development and utilization of Rubiadin as a promising drug candidate in the near future.
Consent for Publication
The final version of the manuscript was reviewed by all the authors who consented to its submission.
Acknowledgments
The authors acknowledge AIMST University, Kedah, Malaysia, and Universiti Kuala Lumpur Royal College of Medicine Perak, Ipoh, Perak, Malaysia, for providing the facilities and services required to complete the study. The figures and graphical abstract in this manuscript were created with BioRender.com and the support of https://biorender.com under a paid subscription.
Author Contributions
All the authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors have no conflict of interest associated with the publication and report no conflicts of interest for this work. There was also no significant financial support for this work.
References
1. Duval J, Pecher V, Poujol M, Lesellier E. Research advances for the extraction, analysis and uses of anthraquinones: a review. Ind Crops Prod. 2016;94:812–833. doi:10.1016/j.indcrop.2016.09.056
2. Seigler DS. Benzoquinones, naphthoquinones, and anthraquinones. In: Plant Secondary Metabolism. Springer; 1998.
3. Dave H, Ledwani L. A review on anthraquinones isolated from Cassia species and their applications. Indian J Natl Prod Resources. 2012;3(2012):291–319.
4. Malik EM, Müller CE. Anthraquinones as pharmacological tools and drugs. Med Res Rev. 2016;36(4):705–748. doi:10.1002/med.21391
5. Huang Q, Lu G, Shen HM, Chung MC, Ong CN. Anti‐cancer properties of anthraquinones from rhubarb. Med Res Rev. 2007;27(5):609–630. doi:10.1002/med.20094
6. Murdock K, Child R, Fabio P, et al. Antitumor agents. 1. 1, 4-Bis [(aminoalkyl) amino]-9, 10-anthracenediones. J Med Chem. 1979;22(9):1024–1030. doi:10.1021/jm00195a002
7. Shrestha JP, Fosso MY, Bearss J, Chang C-WT. Synthesis and anticancer structure activity relationship investigation of cationic anthraquinone analogs. Eur J Med Chem. 2014;77:96–102. doi:10.1016/j.ejmech.2014.02.060
8. Shrestha JP, Subedi YP, Chen L, Chang C-WT. A mode of action study of cationic anthraquinone analogs: a new class of highly potent anticancer agents. MedChemComm. 2015;6(11):2012–2022. doi:10.1039/C5MD00314H
9. Chien S-C, Wu Y-C, Chen Z-W, Yang W-C. Naturally occurring anthraquinones: chemistry and therapeutic potential in autoimmune diabetes. Evid Based Complement Altern Med. 2015;2015:1–13. doi:10.1155/2015/357357
10. Khan K, Karodi R, Siddiqui A, Thube S, Rub R. Development of anti-acne gel formulation of anthraquinones rich fraction from Rubia cordifolia (Rubiaceae). Int J Appl Res Nat Prod. 2011;4(4):28–36.
11. Davis RH, Agnew PS, Shapiro E. Antiarthritic activity of anthraquinones found in aloe vera for podiatric medicine. J Am Podiatr Med Assoc. 1986;76(2):1–8.
12. Wuthi-udomlert M, Kupittayanant P, Gritsanapan W. In vitro evaluation of antifungal activity of anthraquinone derivatives of Senna alata. J Health Res. 2010;24(3):117–122.
13. Fosso MY, Chan KY, Gregory R, Chang C-WT. Library synthesis and antibacterial investigation of cationic anthraquinone analogs. ACS Comb Sci. 2012;14(3):231–235. doi:10.1021/co2002075
14. Winter R, Cornell KA, Johnson LL, Ignatushchenko M, Hinrichs DJ, Riscoe MK. Potentiation of the antimalarial agent rufigallol. Antimicrob Agents Chemother. 1996;40(6):1408–1411. doi:10.1128/AAC.40.6.1408
15. Tikhomirov AS, Shtil AA, Shchekotikhin AE. Advances in the discovery of anthraquinone-based anticancer agents. Recent Pat Anticancer Drug Discov. 2018;13(2):159–183. doi:10.2174/1574892813666171206123114
16. Rao GMM, Rao CV, Pushpangadan P, Shirwaikar A. Hepatoprotective effects of rubiadin, a major constituent of Rubia cordifolia Linn. J Ethnopharmacol. 2006;103(3):484–490. doi:10.1016/j.jep.2005.08.073
17. Shi J, Ren X, Wang J, Wei X, Liu B, Jia T. Effects of the salt-processing method on the pharmacokinetics and tissue distribution of orally administered Morinda officinalis how. Extract J Analyt Methods Chem. 2020;2020:1–11.
18. Patel V, Patel R. Simultaneous analysis and quantification of markers of manjisthadi churna using high performance thin layer chromatography. Indian J Pharm Sci. 2013;75(1):106. doi:10.4103/0250-474X.113541
19. Zhang J, Zhang Z, Bao J, et al. Jia-Jian-Di-Huang-Yin-Zi decoction reduces apoptosis induced by both mitochondrial and endoplasmic reticulum caspase12 pathways in the mouse model of Parkinson’s disease. J Ethnopharmacol. 2017;203:69–79. doi:10.1016/j.jep.2016.12.053
20. Qin L, Han T, Zhang Q, et al. Antiosteoporotic chemical constituents from Er-Xian Decoction, a traditional Chinese herbal formula. J Ethnopharmacol. 2008;118(2):271–279. doi:10.1016/j.jep.2008.04.009
21. Bhatt P, Kushwah A. Rubia cordifolia overview: a new approach to treat cardiac disorders. Int J Drug Dev Res. 2013;5(2):47–54.
22. Ojha JK, Dwivedi KN, Chaurasiya AK. Effect of Rubia cordifolia on non healing diabetic foot ulcer. Nat Sem Trad Med Plants Skin Care. 1994:17.
23. Gogaṭe VU. Ramkrishnan S (editor). Ayurvedic Pharmacology and Therapeutic Uses of Medicinal Plants (Dravya-gunavignyan). Mumbai: Swami Prakashananda Ayurveda Research Centre; 2000.
24. Lodia S, Kansala L. Antioxidant activity of Rubia cordifolia against lead toxicity. Int J Pharma Sci Res. 2012;3(7):2224.
25. Tripathi YB, Singh AV. Role of Rubia cordifolia Linn. in radiation protection. Future Med Chem. 2007;12(7):627–644.
26. Karodi R, Jadhav M, Rub R, Bafna A. Evaluation of the wound healing activity of a crude extract of Rubia cordifolia L. (Indian madder) in mice. Int J Appl Res Nat Prod. 2009;2(2):12–18.
27. Prajapati SN, Parmar KA. Anti-viral and in-vitro free radical scavenging activity of leaves of Rubia cordifolia. Int J Phytomed. 2011;3(1):98.
28. Tripathi Y, Sharma M, Manickam M. Rubiadin, a new antioxidant from Rubia cordifolia. Indian J Biochem Biophys. 1997;34(3):302–306.
29. Takano T, Kondo T, Nakatsubo F. Facile synthesis of rubiadin by microwave heating. J Wood Sci. 2006;52(1):90–92. doi:10.1007/s10086-005-0727-6
30. Peng Z, Fang G, Peng F, et al. Effects of Rubiadin isolated from Prismatomeris connata on anti‐hepatitis B virus activity in vitro. Phytother Res. 2017;31(12):1962–1970. doi:10.1002/ptr.5945
31. National Center for Biotechnology Information. PubChem compound summary for CID 124062, Rubiadin. Avaiable from: https://pubchem.ncbi.nlm.nih.gov/compound/Rubiadin.
32. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi:10.1016/S0169-409X(96)00423-1
33. Zhang M-Q, Wilkinson B. Drug discovery beyond the ‘rule-of-five’. Curr Opin Biotechnol. 2007;18(6):478–488. doi:10.1016/j.copbio.2007.10.005
34. Shukla V, Asthana S, Gupta P, Dwivedi PD, Tripathi A, Das M. Toxicity of naturally occurring anthraquinones. In: Advances in Molecular Toxicology. Vol. 11. Elsevier; 2017.
35. Verma A, Mahalwal V, Kumar B. Antiepileptic activity of rubiadin isolated from the roots of Rubia cordifolia in mice. Int J Pharma Sci Res. 2019;10:3022–3028.
36. Tian W, Wang C, Li D, Hou H. Novel anthraquinone compounds as anticancer agents and their potential mechanism. Future Med Chem. 2020;12(7):627–644.
37. Cogno IS, Gilardi P, Comini L, Núñez-Montoya SC, Cabrera JL, Rivarola VA. Natural photosensitizers in photodynamic therapy: in vitro activity against monolayers and spheroids of human colorectal adenocarcinoma SW480 cells. Photodiagnosis Photodyn Ther. 2020;31:101852. doi:10.1016/j.pdpdt.2020.101852
38. Comini L, Fernandez I, Vittar NR, Montoya SN, Cabrera J, Rivarola V. Photodynamic activity of anthraquinones isolated from Heterophyllaea pustulata Hook f. (Rubiaceae) on MCF-7c3 breast cancer cells. Phytomedicine. 2011;18(12):1093–1095. doi:10.1016/j.phymed.2011.05.008
39. Vittar NBR, Comini L, Fernadez IM, et al. Photochemotherapy using natural anthraquinones: Rubiadin and Soranjidiol sensitize human cancer cell to die by apoptosis. Photodiagnosis Photodyn Ther. 2014;11(2):182–192. doi:10.1016/j.pdpdt.2014.02.002
40. Chiou C-T, Hsu R-Y, Lin L-C. Isolation and cytotoxic effect of anthraquinones from Morinda umbellata. Planta Med. 2014;80(13):1113–1117. doi:10.1055/s-0034-1382956
41. Ali A, Ismail N, Mackeen M, et al. Antiviral, cyototoxic and antimicrobial activities of anthraquinones isolated from the roots of Morinda elliptica. Pharm Biol. 2000;38(4):298–301. doi:10.1076/1388-0209(200009)38:4;1-A;FT298
42. Kanokmedhakul K, Kanokmedhakul S, Phatchana R. Biological activity of anthraquinones and triterpenoids from prismatomeris fragrans. J Ethnopharmacol. 2005;100(3):284–288. doi:10.1016/j.jep.2005.03.018
43. Inoue K, Yoshida M, Takahashi M, et al. Possible contribution of rubiadin, a metabolite of madder color, to renal carcinogenesis in rats. Food Chem Toxicol. 2009;47(4):752–759. doi:10.1016/j.fct.2009.01.003
44. Inoue K, Yoshida M, Takahashi M, et al. Carcinogenic potential of alizarin and rubiadin, components of madder color, in a rat medium‐term multi‐organ bioassay. Cancer Sci. 2009;100(12):2261–2267. doi:10.1111/j.1349-7006.2009.01342.x
45. Blömeke B, Poginsky B, Schmutte C, Marquardt H, Westendorf J. Formation of genotoxic metabolites from anthraquinone glycosides, present in Rubia tinctorum L. Mutat Res/Fundament Mol Mechan Mutagen. 1992;265(2):263–272. doi:10.1016/0027-5107(92)90055-7
46. Siewert B, Stuppner H. The photoactivity of natural products–an overlooked potential of phytomedicines? Phytomedicine. 2019;60:152985. doi:10.1016/j.phymed.2019.152985
47. Montoya SCN, Comini LR, Vittar BR, Fernández IM, Rivarola VA, Cabrera JL. Phototoxic effects of Heterophyllaea pustulata (Rubiaceae). Toxicon. 2008;51(8):1409–1415. doi:10.1016/j.toxicon.2008.03.011
48. Micheloud JF, Colque-Caro LA, Comini LR, et al. Spontaneous photosensitization by Heterophyllaea pustulata Hook. f. (Rubiaceae), in sheep from Northwestern Argentina. Trop Anim Health Prod. 2017;49(7):1553–1556. doi:10.1007/s11250-017-1354-0
49. Micheloud JF, Aguirre LS, Marioni J, et al. Experimental poisoning by Heterophyllaea pustulata Hook. f. (Rubiaceae) in goats. Clinical, biochemical and toxicological aspects. Toxicon. 2019;165:56–61. doi:10.1016/j.toxicon.2019.04.015
50. Sambrook P, Cooper C. Osteoporosis. Lancet. 2006;367(9527):2010–2018. doi:10.1016/S0140-6736(06)68891-0
51. He Y-Q, Zhang Q, Shen Y, et al. Rubiadin-1-methyl ether from Morinda officinalis How. Inhibits osteoclastogenesis through blocking RANKL-induced NF-κB pathway. Biochem Biophys Res Commun. 2018;506(4):927–931. doi:10.1016/j.bbrc.2018.10.100
52. Bao L, Qin L, Liu L, et al. Anthraquinone compounds from Morinda officinalis inhibit osteoclastic bone resorption in vitro. Chem Biol Interact. 2011;194(2–3):97–105. doi:10.1016/j.cbi.2011.08.013
53. Wu Y-B, Zheng C-J, Qin L-P, et al. Antiosteoporotic activity of anthraquinones from Morinda officinalis on osteoblasts and osteoclasts. Molecules. 2009;14(1):573–583. doi:10.3390/molecules14010573
54. Xia T, Dong X, Lin L, et al. Metabolomics profiling provides valuable insights into the underlying mechanisms of Morinda officinalis on protecting glucocorticoid-induced osteoporosis. J Pharm Biomed Anal. 2019;166:336–346. doi:10.1016/j.jpba.2019.01.019
55. Libby P. Inflammatory mechanisms: the molecular basis of inflammation and disease. Nutr Rev. 2007;65(suppl_3):S140–S146. doi:10.1301/nr.2007.dec.S140-S146
56. Mohr ETB, Dos Santos Nascimento MVP, da Rosa JS, et al. Evidence that the anti-inflammatory effect of Rubiadin-1-methyl ether has an immunomodulatory context. Mediators Inflamm. 2019;2019:1–12. doi:10.1155/2019/6474168
57. Mujeeb M, Ahad A, Aqil M, et al. Ameliorative effect of rubiadin-loaded nanocarriers in STZ-NA-induced diabetic nephropathy in rats: formulation optimization, molecular docking, and in vivo biological evaluation. Drug Deliv Transl Res. 2021;1–14. doi:10.1007/s13346-021-00971-0
58. Yen G-C, Duh P-D, Chuang D-Y. Antioxidant activity of anthraquinones and anthrone. Food Chem. 2000;70(4):437–441. doi:10.1016/S0308-8146(00)00108-4
59. Malterud KE, Farbrot TL, Huse AE, Sund RB. Antioxidant and radical scavenging effects of anthraquinones and anthrones. Pharmacology. 1993;47(Suppl. 1):77–85. doi:10.1159/000139846
60. Baghiani A, Charef N, Djarmouni M, Saadeh A, Arrar L, Mubarak S. Free radical scanvenging and antioxidant effects of some anthraquinone derivatives. Med Chem (Los Angeles). 2011;7(6):639–644. doi:10.2174/157340611797928424
61. Tripathi Y, Sharma M. Comparison of the antioxidant action of the alcoholic extract of Rubia cordifolia with rubiadin. Indian J Biochem Biophys. 1998;35(5):313–316.
62. Mishra BB, Kishore N, Tiwari VK, Singh DD, Tripathi V. A novel antifungal anthraquinone from seeds of Aegle marmelos Correa (family Rutaceae). Fitoterapia. 2010;81(2):104–107. doi:10.1016/j.fitote.2009.08.009
63. Mohamadzadeh M, Zarei M, Vessal M. Synthesis, in vitro biological evaluation and in silico molecular docking studies of novel β-lactam-anthraquinone hybrids. Bioorg Chem. 2020;95:103515. doi:10.1016/j.bioorg.2019.103515
64. Khan MS, Gao J, Chen X, et al. The endophytic bacteria Bacillus velezensis Lle-9, isolated from Lilium leucanthum, harbors antifungal activity and plant growth-promoting effects. J Microbiol Biotechnol. 2020;30(5):668–680. doi:10.4014/jmb.1910.10021
65. Marioni J, Da Silva MA, Cabrera JL, Montoya SCN, Paraje MG. The anthraquinones rubiadin and its 1-methyl ether isolated from Heterophyllaea pustulata reduces Candida tropicalis biofilms formation. Phytomedicine. 2016;23(12):1321–1328. doi:10.1016/j.phymed.2016.07.008
66. Marioni J, Bresolí-Obach R, Agut M, et al. On the mechanism of Candida tropicalis biofilm reduction by the combined action of naturally-occurring anthraquinones and blue light. PLoS One. 2017;12(7):e0181517. doi:10.1371/journal.pone.0181517
67. Montoya SCN, Comini LR, Sarmiento M, et al. Natural anthraquinones probed as type I and type II photosensitizers: singlet oxygen and superoxide anion production. J Photochem Photobiol B. 2005;78(1):77–83. doi:10.1016/j.jphotobiol.2004.09.009
68. Likhitwitayawuid K, Dej-adisai S, Jongbunprasert V, Krungkrai J. Antimalarials from Stephania venosa, Prismatomeris sessiliflora, Diospyros Montana and Murraya siamensis1. Planta Med. 1999;65(08):754–756. doi:10.1055/s-2006-960858
69. Koumaglo K, Gbeassor M, Nikabu O, De Souza C, Werner W. Effects of three compounds extracted from Morinda lucida on Plasmodium falciparum. Planta Med. 1992;58(06):533–534. doi:10.1055/s-2006-961543
70. Comini L, Montoya SN, Páez P, Argüello GA, Albesa I, Cabrera J. Antibacterial activity of anthraquinone derivatives from Heterophyllaea pustulata (Rubiaceae). J Photochem Photobiol B. 2011;102(2):108–114. doi:10.1016/j.jphotobiol.2010.09.009
71. Wang D, Wang X-H, Yu X, et al. Pharmacokinetics of anthraquinones from medicinal plants. Front Pharmacol. 2021;12:306.
72. Schyman P, Liu R, Desai V, Wallqvist A. vNN web server for ADMET predictions. Front Pharmacol. 2017;8:889. doi:10.3389/fphar.2017.00889
73. EPA. User’s guide for T.E.S.T. (version 5.1) (toxicity estimation software tool): a program to estimate toxicity from molecular structure. Avaiable from: https://www.epa.gov/chemical-research/toxicity-estimation-software-tool-test.
74. Collie GW, Koh CM, O’Neill DJ, et al. Structural and molecular insight into resistance mechanisms of first generation cMET inhibitors. ACS Med Chem Lett. 2019;10(9):1322–1327. doi:10.1021/acsmedchemlett.9b00276
75. Sakamoto H, Tsukaguchi T, Hiroshima S, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19(5):679–690. doi:10.1016/j.ccr.2011.04.004
76. Sivakumaren SC, Shim H, Zhang T, et al. Targeting the PI5P4K lipid kinase family in cancer using covalent inhibitors. Cell Chem Biol. 2020;27(5):525–537. doi:10.1016/j.chembiol.2020.02.003
77. Huck JD, Que NL, Sharma S, Taldone T, Chiosis G, Gewirth DT. Structures of Hsp90α and Hsp90β bound to a purine‐scaffold inhibitor reveal an exploitable residue for drug selectivity. Proteins Struct Func Bioinform. 2019;87(10):869–877. doi:10.1002/prot.25750
78. Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi:10.1021/jm051197e
79. Cosmetic components comprised of the rubiadin having anti-allergy activity. Patent number KR101337564B1. Korea Kolmar Co., Ltd.; 2010. Available from: https://patents.google.com/patent/KR101337564B1/en. Accessed October 28, 2021.
80. Akhtar MN, Zareen S, Yeap SK, et al. Total synthesis, cytotoxic effects of damnacanthal, nordamnacanthal and related anthraquinone analogues. Molecules. 2013;18(8):10042–10055. doi:10.3390/molecules180810042
81. Kumar P, Sharma AK, Shukla P. Physiochemical screening of marker anthraquinones and derivatives from the Pentas lanceolata (Forss. k.) deflers leaves. Eur J Pharm Med Res. 2017;4(5):439–446.
82. Dosseh C, Tessier A, Delaveau P. Nouvelles Quinones des Racines de Rubia cordifolia L., III. Planta Med. 1981;43(12):360–366. doi:10.1055/s-2007-971524
83. Shen C-H, Liu C-T, Song X-J, et al. Evaluation of analgesic and anti-inflammatory activities of Rubia cordifolia L. by spectrum-effect relationships. J Chromatogr B. 2018;1090:73–80. doi:10.1016/j.jchromb.2018.05.021
84. Liu Q, Kim SB, Ahn JH, Hwang BY, Kim SY, Lee MK. Anthraquinones from Morinda officinalis roots enhance adipocyte differentiation in 3T3-L1 cells. Nat Prod Res. 2012;26(18):1750–1754. doi:10.1080/14786419.2011.608676
85. Zhang J-H, Xin H-L, Xu Y-M, et al. Morinda officinalis How.–a comprehensive review of traditional uses, phytochemistry and pharmacology. J Ethnopharmacol. 2018;213:230–255. doi:10.1016/j.jep.2017.10.028
86. Zhao X, Wei J, Yang M. Simultaneous analysis of iridoid glycosides and anthraquinones in Morinda officinalis using UPLC-QqQ-MS/MS and UPLC-Q/TOF-MSE. Molecules. 2018;23(5):1070. doi:10.3390/molecules23051070
87. Mugas ML, Marioni J, Martinez F, et al. Inactivation of herpes simplex virus by photosensitizing anthraquinones isolated from heterophyllaea pustulata. Planta Med. 2021;80:716–723.
88. Schunck HE. III. On rubian and its products of decomposition. Philos Transact Royal Soc London. 1853;(143):67–107. doi:10.1098/rstl.1853.0003
89. Kawasaki Y, Goda Y, Yoshihira K. The mutagenic constituents of Rubia tinctorum. Chem Pharm Bull (Tokyo). 1992;40(6):1504–1509. doi:10.1248/cpb.40.1504
90. Cuoco G, Mathe C, Archier P, Chemat F, Vieillescazes C. A multivariate study of the performance of an ultrasound-assisted madder dyes extraction and characterization by liquid chromatography-photodiode array detection. Ultrason Sonochem. 2009;16(1):75–82. doi:10.1016/j.ultsonch.2008.05.014
91. Cooksey C. Quirks of dye nomenclature. 14. Madder: queen of red dyes. Biotech Histochem. 2020;95(6):474–482. doi:10.1080/10520295.2020.1714079
92. Tuntiwachwuttikul P, Butsuri Y, Sukkoet P, Prawat U, Taylor WC. Anthraquinones from the roots of Prismatomeris malayana. Nat Prod Res. 2008;22(11):962–968. doi:10.1080/14786410701650261
93. Rahman MM. Evaluation of Hymenodictyon excelsum phytochemical’s therapeutic value against prostate cancer by molecular Docking study. Jundishapur j Nat Pharma Prod. 2015;10(1). doi:10.17795/jjnpp-18216
94. Ahmad R, Shaari K, Lajis NH, Hamzah AS, Ismail NH, Kitajima M. Anthraquinones from Hedyotis capitellata. Phytochemistry. 2005;66(10):1141–1147. doi:10.1016/j.phytochem.2005.02.023
95. Usai M, Marchetti M. Anthraquinone distribution in the hypogeal apparatus of Rubia peregrina L. growing wild in Sardinia. Nat Prod Res. 2010;24(7):626–632. doi:10.1080/14786410902884842
96. Bussmann RW, Hennig L, Giannis A, Ortwein J, Kutchan TM, Feng X. Anthraquinone content in Noni (Morinda citrifolia L.). Evid Based Complement Altern Med. 2013;2013:1–5. doi:10.1155/2013/208378
97. Rajan R, Venkataraman R, Baby S. A new lupane-type triterpenoid fatty acid ester and other isolates from Ophiorrhiza shendurunii. Nat Prod Res. 2016;30(19):2197–2203. doi:10.1080/14786419.2016.1160232
98. Osman CP, Ismail NH, Ahmad R, Ahmat N, Awang K, Jaafar FM. Anthraquinones with antiplasmodial activity from the roots of Rennellia elliptica Korth. (Rubiaceae). Molecules. 2010;15(10):7218–7226. doi:10.3390/molecules15107218
99. Yuan S, Zhao Y. Chemical constituents of Knoxia valerianoides. Acta Pharmaceutica Sinica. 2006;41(8):735–737.
100. Yoo NH, Jang DS, Lee YM, et al. Anthraquinones from the roots of Knoxia valerianoides inhibit the formation of advanced glycation end products and rat lens aldose reductase in vitro. Arch Pharm Res. 2010;33(2):209–214. doi:10.1007/s12272-010-0204-7
101. Zhao F, Wang S-J, Lin S, et al. Anthraquinones from the roots of Knoxia valerianoides. J Asian Nat Prod Res. 2011;13(11):1023–1029. doi:10.1080/10286020.2011.606813
102. Lan M, Luo C, Tan C, Chen L, Wei S, Zhu D. Study on chemical constituents of the ethyl acetate extract from Blumea aromatica. J Chin Med Mater. 2012;35(2):229–231.
103. Jiang J-S, Feng Z-M, Zhang P-C. Chemical constituents from root of Prismatomeris tetrandra. China J Chin Materia Medica. 2005;30(22):1751–1753.
104. Huang W, Li Y, Jiang J. Chemical constituents from Hedyotis diffusa. China J Chin Materia Medica. 2009;34(6):712–714.
105. Liu G, Chen Z, Yao T, Ding W. Studies on the chemical constituents of Rhynchotechum vestitum Hook. F. et Thoms. Acta Pharmaceutica Sinica. 1990;25(9):699–704.
106. Haque MA, Khan G, Razzaque S, Khatun K, Chakraborty AK, Alam MS. Extraction of rubiadin dye from Swietenia mahagoni and its dyeing characteristics onto silk fabric using metallic mordants. Indian J Fibre Text Res. 2013;38:280–284.
107. Li S, Ouyang Q, Tan X, Shi S, Yao Z. Chemical constituents of Morinda officinalis how. China J Chin Materia Medica. 1991;16(11):675–676, 703.
108. Chokchaisiri S, Siriwattanasathien Y, Thongbamrer C, Suksamrarn A, Rukachaisirikul T. Morindaquinone, a new bianthraquinone from Morinda coreia roots. Nat Prod Res. 2019;1–7. doi:10.1080/14786419.2019.1705820
109. Zou X, Liang J, Ding L-S, Peng S-L. Studies on chemical constituents of Paederia scandense. China J Chin Materia Medica. 2006;31(17):1436–1441.
110. Li Y, Qi S, Chen X, Hu Z. Separation and determination of the anthraquinones in Xanthophytum attopvensis pierre by nonaqueous capillary electrophoresis. Talanta. 2005;65(1):15–20.
111. Tosa H, Iinuma M, Asai F, et al. Anthraquinones from Neonauclea calycina and their inhibitory activity against DNA topoisomerase II. Biol Pharm Bull. 1998;21(6):641–642. doi:10.1248/bpb.21.641
112. Khanh PN, Huong TT, Spiga O, et al. In silico screening of anthraquinones from Prismatomeris memecyloides as novel phosphodiesterase type-5 inhibitors (PDE-5Is). Revista Internacional De Andrologia. 2018;16(4):147–158. doi:10.1016/j.androl.2017.07.001
113. Chen -R-R, Liu J, Chen Z, Cai W-J, Li X-F, Lu C-L. Anthraquinones extract from morinda angustifolia roxb. Root alleviates hepatic injury induced by carbon tetrachloride through inhibition of hepatic oxidative stress. Evid Based Complement Altern Med. 2020;2020. doi:10.1155/2020/9861571
114. Lv Z, Zhang Q, Chen R, Yu D. Alkaloids and anthraquinones from branches and leaves of Uvaria kurzii. China J Chin Materia Medica. 2011;36(9):1190–1192.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.