Back to Journals » Infection and Drug Resistance » Volume 15
Characterization of Silver Resistance and Coexistence of sil Operon with Antibiotic Resistance Genes Among Gram-Negative Pathogens Isolated from Wound Samples by Using Whole-Genome Sequencing
Authors Wang H, Li J, Min C, Xia F , Tang M, Li J, Hu Y, Zou M
Received 16 January 2022
Accepted for publication 12 March 2022
Published 31 March 2022 Volume 2022:15 Pages 1425—1437
DOI https://doi.org/10.2147/IDR.S358730
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Haichen Wang,1,2 Jia Li,2,3 Changhang Min,1,2 Fengjun Xia,1,2 Mengli Tang,1,2 Jun Li,1,2 Yongmei Hu,1,2 Mingxiang Zou1,2
1National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan Province, People’s Republic of China; 2Department of Clinical Laboratory, Xiangya Hospital, Central South University, Changsha, Hunan Province, People’s Republic of China; 3Shanghai Institute of Immunology, Department of Immunology and Microbiology, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China
Correspondence: Mingxiang Zou, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, 41008, People’s Republic of China, Tel/Fax +86 7384327440, Email [email protected]
Purpose: Due to the extensive consumption of silver-containing compound, silver resistance spreads among gram-negative pathogens and is regarded as a great public problem. In this study, we investigated silver resistance mechanisms and antibiotic resistance genes co-harbored with sil operon among gram-negative pathogens isolated from wound samples.
Methods: A total of 193 strains of gram-negative pathogens were collected from wound samples between 2018 and 2020 in Xiangya hospital. Silver resistance was obtained by broth microdilution method. The silver resistance mechanisms and the prevalence, genetic environments, and coexistence with antibiotic resistance genes of sil operon were investigated by polymerase chain reaction (PCR) and whole genome sequencing (WGS).
Results: Among 193 strains, nine strains (4.7%) were resistant to Ag+ and assigned to the following species: Klebsiella pneumoniae (n = 5) and Enterobacter hormaechei (n = 4). WGS confirmed that 24 strains carried the entire sil operon, including the four Ag+-resistant E. hormaechei and 20 Ag+-susceptible strains, while PCR failed to detect some sil genes, especially silE, due to sequence variations. In seven strains, Tn7 transposon was identified in the upstream of sil operon. Spontaneous mutants resistant to Ag+ were induced in 15 out of 20 Ag+-susceptible strains, including K. pneumoniae strains belonged to high-risk groups (ST11 and ST15). The sil-positive strains harbored various antibiotic resistance genes, including blaESBL and blaApmC. WGS revealed that a single mutation in cusS gene and loss of major porins conferred silver resistance in the five K. pneumoniae strains.
Conclusion: Our findings emphasize the cryptic silver resistance is prevalent among Enterobacteriaceae with sil operon or with the combination of cus operon and major porin loss and increase the understanding of the prevalence of sil operon with antibiotic resistance genes, especially blaESBL and blaApmC.
Keywords: silver resistance, gram negative pathogen, sil operon, antibiotic resistance gene, whole-genome sequencing
Introduction
The silver compounds exhibit excellent bactericidal effect on various pathogens and minimal toxicity towards human cells.1 Silver targets a variety of bacterial components, ranging from cell wall to nucleic acid. Silver ions damage the integrality of cell wall, increasing the membrane permeability, and bind to the protein and enzymes through thiol groups, inhibiting the respiratory chain reaction. Furthermore, the silver ions generate reactive oxygen species and interfere with the replication of DNA.2
Silver has been employed as antibacterial material for thousands of years.3 Nowadays, it has been intensively used in hospital and in daily life. However, the large and uncontrolled consumption of silver-containing materials raises the concern about the widely spread of silver resistance.
Both endogenous and exogenous mechanisms confer silver resistance. Endogenous mechanism involves a single mutation in cusS, which increases the expression of cus operon, and the loss of outer membrane porins. So far endogenous mechanism is only successfully induced in vitro.4 The sil operon is firstly identified in a plasmid pMG101 from a Salmonella enterica strain in 1975 and plays an important role in exogenous silver resistance due to horizontal gene transfer.5 The sil operon consists of nine genes, including silCFBA (ORF105aa) PRSE, and encodes efflux pumps (SilCBA and SilP), Ag+ chaperone or binding proteins (SilF and SilE) and is regulated by a two-component regulatory system (genes silRS).6,7 The sil operon and pco operon often locate on plasmid and compose the copper hemostasis and silver resistance island (CHASRI).8
Earlier studies indicate that silver resistance mainly distributes in Enterobacteriaceae. A research involved 752 strains isolated from bloodstream and demonstrated that 13% of Enterobacter spp. were phenotypic resistant to Ag+.9 Another study reported that 1006 strains of Staphylococcus aureus were susceptible to Ag+ with the minimum inhibition concentration (MIC) values all below or equal to 16 μg/mL and no resistant mutants were selected during a 42-day exposure experiment.10 But a recent study in 2019 at Egypt reported that both silver resistance and sil genes were firstly identified in S. aureus, Pseudomonas aeruginosa and Acinetobacter baumannii.11 Unfortunately, data on the occurrence of silver resistance and the prevalence of sil operon in China is lacking. In addition, previous studies usually employed polymerase chain reaction (PCR) method for the screening of sil and antibiotic resistance genes, whole-genome sequencing (WGS) analysis of the sil-positive strains is scarce.
In this study, we screened 193 strains of gram-negative pathogens isolated from wound samples and performed WGS on 26 strains with sil operon and/or silver resistance, in order to investigate silver resistant mechanisms and antibiotic resistance genes co-harbored with sil operon in Hunan province, China.
Materials and Methods
Bacteria Source
Non-duplicate gram-negative pathogens from wound samples were collected between 2018 and 2020 in Xiangya hospital, a university-affiliated tertiary teaching hospital with a 3000-bed capacity in Changsha, Hunan province, China. All samples were routinely tested in the microbiology laboratory. The strains were identified by MicroflexTM MALDI-TOF MS system (Bruker Daltonik, Bremen, Germany). All the strains were stored at −80 °C for further analysis.
Antimicrobial Susceptibility Tests to Silver Nitrate and Antibiotics
MIC to silver nitrate of all strains in our study was determined by broth microdilution method with Mueller-Hinton broth (MHB, Oxoid, unipath, UK) according to the Clinical and Laboratory Standards institute (CLSI) guidelines.12 Concentrations range between 2 and 512 μg/mL were tested in our study. Bacteria with MIC above or equal to 512 μg/mL were considered as strains resistant to Ag+.9,11
Antibiotic susceptibility data were extracted from the laboratory database. The routine antibiotic susceptibility test was performed by VITEK-2 Compact system (bioMérieux, Marcy L’Etoile, France), followed by the manufacturers’ instructions. Escherichia coli ATCC25922 and P. aeruginosa ATCC27853 were used as quality controls.
Detection of Silver Resistance Genes
Further detection experiments on silver resistance genes were completed by means of PCR. The genomic DNA were extracted by boiling method. The silver resistance genes, including silS, silR, silE, silA, silB, silCBA, silP and silF were analyzed in our study. The primers used in our study were listed in Supplemental Table S1. PCR products were electrophoresed with 1.2% agarose gel and visualized under a UV transilluminator.
In vitro Selection of Ag+ Resistance Mutants
All the silS-positive strains with MIC value below 512 μg/mL were subjected to in vitro select spontaneous mutants resistant to Ag+, according to previous work with modification.13 Briefly, 100 μL of an overnight culture (~ 108 cfu/mL) was placed on MH agar supplemented with 128 μg/mL silver nitrate. Each strain was tested on ten agars to detect the mutation at frequencies of one in 109 bacteria. The experiments were repeated twice. The full length of silS and silR genes of the Ag+ susceptible parent strains and corresponding resistant mutants were amplified, sequenced and compared with using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Relative change in the fitness cost of Ag+ resistance was determined by growth curves between the susceptible parent strains and the corresponding mutants.14
Whole-Genome Sequencing of Strains with sil Operon and/or Silver Resistance
A chosen subcollection of strains (n = 26) with sil operon and/or silver resistance were analyzed by whole-genome sequencing (WGS). One colony of each strain was cultured in 3 mL LB broth (Oxoid, unipath, UK) at 37 °C for 12 h. The bacterial genomic DNA were extracted by Ezup Column Bacteria Genomic DNA purification Kit (Sangon Biotech, Shanghai, China), according to the manufacturer's recommendation. The quality and quantity of DNA were examined by agarose gel electrophoresis and Qubit fluorometric instrument (Invitrogen, USA). Fragmented DNAs were end repaired, A-tailed, adapter ligated and amplified using a NEB Next® Ultra™ DNA Library Prep Kit for Illumina® (NEB, USA). The libraries were sequenced with an Illumina HiSeq platform. The read quality was assessed using FastQC software (v0.11.2; http://www.bioinformatics.babraham.ac.uk) according to the developers’ recommendations. Genome assembly was conducted by SPAdes (v3.5.0) (Bankevich et al, 2012). Draft genomes were annotated by use of the annotation software Prokka (v1.10) and RAST.15,16 The antibiotics resistance genes and types of plasmids were identified by ResFinder and PlasmidFinder from the Centre for Genomic Epidemiology, respectively.17,18 In silico analyses of ST type for Klebsiella pneumoniae strains was performed by MLST 2.0 (Multi-Locus Sequence Typing).19 Pan-genome dendrograms describing single nucleotide polymorphisms of K. pneumoniae and Enterobacter hormaechei strains were constructed by Roary: the Pan Genome Pipeline.20
The WGS of the strains was deposited at GenBank and the accession numbers were listed in Supplemental Table S2.
Statistical Analysis
Numbers were presented for categorical variables. Pearson chi-square test or Fisher’s exact test were employed to compare categorical variables, when appropriate. P values < 0.05 was considered as statistically significant. SPSS (version 22, IBM Corporation, USA) was used for all analysis.
Results
Bacterial Source
A total of 193 strains of gram-negative pathogens were collected during the study period. The strains belonged to Acinetobacter (n = 33), Klebsiella (n = 33), Escherichia (n = 30), Pseudomonas (n = 29), Enterobacter (n = 29), Proteus (n = 18), Serratia (n = 6), Morganella (n = 6), Stenotrophomonas (n = 4), Citrobacter (n = 3), Myroides (n = 1) and Alcaligenes (n = 1) genera.
Susceptibility Profile to Silver Nitrate
Among the 193 tested strains, the MIC range of silver nitrate for all strains was between 4 μg/mL and > 512 μg/mL. The MIC at which 50% and 90% strains were inhibited (MIC50 and MIC90) were 16 μg/mL and 32 μg/mL, respectively. In total, nine (4.7%) strains were resistant to silver nitrate with MIC values above 512 μg/mL and were classified to the following species: K. pneumoniae (n = 5) and E. hormaechei (n = 4).
Screening for Silver Resistance Genes
PCR was employed to screen the prevalences of sil genes among 193 strains. The 68 strains of Nonfermenters were negative for all the silver resistance genes tested in our study.
Among the 125 strains of Enterobacteriaceae, PCR revealed that 26 strains carried one or more sil genes. Two Enterobacter spp. were positive for one (silF) or two sil genes (silF and silA). In the rest 24 strains, only ten strains were positive for the eight primers while the other strains mostly lacked silE gene. The MIC values of the 24 sil positive strains ranged from 8 to > 512 μg/mL. The distribution of sil genes and MIC values for the 24 strains were listed in Supplemental Table S2. Notably, none of the five Ag+-resistant K. pneumoniae strains harbored any sil genes.
Whole-Genome Sequencing
In our study, a subcollection of strains (n = 26) were subjected to WGS, including two sil-negative Ag+-resistant K. pneumoniae strains to reveal the resistant mechanism, and 24 sil-positive strains.
Molecular Mechanism of Ag+ Resistance K. pneumoniae
Due to the same ERIC-PCR fingerprint (Supplemental Result Figure S1), two strains (WHC1 and WHC2) were analyzed by WGS to reveal the molecular mechanism of Ag+ resistance. After assembled by SPAdes (v3.5.0), the sum of the contig length were 5,375,743 and 5,377,294 bp, respectively, and the G + C content were 57.31% for both strains. The Prokka software revealed the genome of WHC1 and WHC2 harbored 5195 and 5200 protein-encoding sequences, respectively. The average gene lengths were 914 bp for both strains.
According to the genes annotated by Prokka software, several heavy metal transport-related genes were identified in K. pneumoniae strains WHC1 and WHC2, consisting of cation efflux systems. The cus operon, including cusRSCFBA genes, was identified in both strains, while the sil operon was confirmed absent in both strains. The sequences of cus operon were identical to the corresponding region of K. pneumoniae strains SMKP03 (GenBank no. AP023148.1), except for gene cusS, in which a single mutation was found, resulting in an amino acid change (Pro209Ser). The sequences of ompK35 and ompK36 genes were analyzed by WGS and further verified with sanger sequencing and compared with those of K. pneumoniae KCTC 2242 (GenBank no. CP002910) and K. pneumoniae NTUH-K 2044 (GenBank no. AP006725). For gene ompK36, a deletion of a 1 bp (at nucleotide position 46) created a premature stop codon at amino acid position 32, resulting in early termination of translation. For gene ompK35, the sequences of WHC1 and WHC2 were the same as those in K. pneumoniae KCTC 2242. However, three-point mutations were identified in the promoter region at nucleotide 121, 163 and 453 upstream of the start codon.
The antibiotics resistance genes for two strains included oqxAB, qnrS1, aac(6’)-Ib-cr, fosA, tet(A), aadA16, dfrA27, sul1, sul2, mph(A), ARR-3, aph(3’)-Ia, aph(6)-Id, aac(3)-IId, aph(3”)-Ib, floR, blaSHV-27, blaCTX-M-3, blaTEM-1B and blaNDM-1. The strains belonged to ST967 and harbored IncFIB, IncFII, IncQ1 and IncX3 type plasmids.
WGS Results for sil-Positive Strains
WGS was performed on 24 strains with at least five sil genes according to PCR results, including four Ag+-resistant strains and 20 Ag+-susceptible strains. The strains belonged to Klebsiella spp. (n = 13), Enterobacter spp. (n = 9), Escherichia spp. (n = 1) and Citrobacter spp. (n =1). WGS revealed that the 24 strains carried the entire sil operon, ie, silESRCFBAP. Furthermore, 17 strains carried the cus operon, while the pco operon was detected alongside with sil operon in 23 strains and together formed a copper homeostasis and silver resistance island (CHASRI). In strain WHC182, only pcoE was detected with WGS and PCR also failed to detect pcoD and pcoR (data not shown). The sequence of silS, silR and silE were compared with the corresponding region of pMG101 (GenBank no. AF067954) (Table 1). The silR genes showed sequence variations between 7.42% and 1.31% at the nucleotide level. For silE gene, six strains showed 100% overall identities (Ident) to the silE gene in pMG101 while the rest strains showed a variation up to 9.49% at the nucleotide level. For silS gene, in strain WHC21, an IS5-like element was inserted into the silS gene and the full length for of silS gene amplified by PCR was 2548 bp, with a query coverage (QC) of 58% with silS gene in plasmid pMG101. The silS genes in other strains also show a variation between 5.89% and 1.10%.
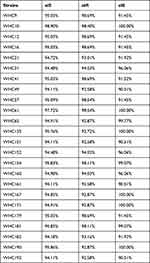 |
Table 1 Overall Identities of sil Genes Compared with pMG101 |
As for the antibiotic resistance genes, the most prevalent gene was fosA (n =22). Various β-lactamase encoding genes were detected, including blaSHV (n = 15), blaTEM-1B (n = 13), blaCTX-M (n = 10), blaACT (n = 7), blaOXA (n = 6), blaDHA (n =6), blaLAP (n = 4), blaSFO (n = 2), blaMIR (n = 1), blaCMY (n = 1), blaLEN (n = 1) and blaOKP (n = 1). Ten blaCTX-M genes were classified as blaCTX-M-15 (n = 5), blaCTX-M-3 (n = 2), blaCTX-M-65 (n = 1), blaCTX-M-27 (n = 1) and blaCTX-M-14 (n = 1). Two carbapenem-resistance genes were identified as blaKPC (n = 1) and blaNDM (n = 2). The rates for qnr-, sul-, dfr-, tet-, oqx-, aad-, mph-, ARR- and flo-genes were 66.7% (n = 16), 62.5% (n = 15), 58.3% (n = 14), 58.3% (n = 14), 50.0% (n = 12), 45.8% (n = 11), 37.5% (n = 9), 33.3% (n = 8) and 29.2% (n = 7) (Table 2).
 |
Table 2 Antibiotic Resistance Genes and Plasmid Inc Type Among Gram-Negative Pathogens Harboring sil Operon |
In four strains, no plasmid hit was found with PlasmidFinder. For other strains, various plasmid types were found, including IncFIB(K), IncFII(K), IncM2, IncQ1, IncHI2A, IncX3, IncR, IncI1-I, IncN, IncU, repB and Col. Only four strains carried IncHI type plasmids (Table 2).
According to the assembled sequences and annotations, the genetic environments of sil operon were also analyzed. Tn7 transposon was identified in the upstream of sil operon in seven strains, including the four strains in which no plasmid hit was found (ie, WHC61, 65, 167 and 171, Table 2). In five strains (ie, WHC9, 12, 16, 57 and 179), two mobile elements were identified in the upstream of sil operon and the assembled contigs showed an Iden of 100% with a QC of 100% with the corresponding region in K. pneumoniae strain C2972 plasmid pC2972-2 (GenBank no. CP039803), except for WHC179 (Iden 92% and QC 99.82%). In strain WHC181, a contig was assembled and carried tra and sil operon. The only similar region was found in E. hormaechei strain SH19PTE2 plasmid pYUSHP2-2 (GenBank no. CP073773, BLASTn, NR database, Iden 100% and QC 100%).
The MLST type of K. pneumoniae were also analyzed. The 11 strains were assigned to the following ST type: ST15 (n = 4), ST35 (n = 1), ST45 (n = 1), ST39 (n = 1), ST11 (n = 1), ST65 (n = 1), ST3393 (n = 1) and ST412 (n = 1) (Table S2).
Pan-Genome Dendrogram Based on WGS Results
Pan-genome dendrograms of K. pneumoniae and E. hormaechei strains were constructed by Roary according to the prevalence of different genes categories, including core (genes shared by 99–100% of the strains), soft core (genes shared by 95–99% of the strains), shell (genes shared by 15–95% of the strains) and cloud genes (genes shared by 0–15% of the strains).
In K. pneumoniae, 3763 core, 3201 shell and 45 cloud genes were identified. The pan-genome dendrogram revealed that the 13 strains of K. pneumoniae were clustered into four groups (Figure 1). Notably, the sil-negative Ag+-resistant strains (WHC1 and 2) were identified in one group with minor genetic variations. Strains WHC9, 12 and 16 also showed high genetic similarity.
 |
Figure 1 Phylogeny of K. pneumoniae isolated based on differences in SNPs. |
In E. hormaechei, 2928 core, 2075 shell and 51 cloud genes were identified. The pan-genome dendrogram revealed that the 7 strains of E. hormaechei were clustered into three groups (Figure 2). The three Ag+-susceptible strains (WHC181, 171 and 182) were identified into one group, while four resistant strains were clustered into two groups with different genetic similarities.
 |
Figure 2 Phylogeny of E. hormaechei isolated based on differences in SNPs. |
Co-Resistance Between Silver and Antibiotics
The antibiotic susceptibility profiles, silver resistance and the prevalence of silS gene among the Enterobacteriaceae were also analyzed. The results were listed in Table 3. For the nine Ag+ resistant Enterobacteriaceae, eight strains were carbapenem resistant and were highly resistant to the tested antibiotics compared with Ag+ susceptible strains, except for amikacin (0.0% vs 4.3%, P = 1.000). Considering the relationship between silS gene and antibiotics resistance, strains which harbored silS gene were more resistant to ceftriaxone (75.0% vs 45.5%, P = 0.012), ceftazidime (58.3% vs 25.7%, P = 0.003) and aztreonam (58.3% vs 28.7%, P = 0.009) than the silS-negative strains. No connection between the resistance of other antibiotics and the prevalence of silS was revealed in our study.
 |
Table 3 Antimicrobial Susceptibility Profiles in Enterobacteriaceae (n=125) |
Spontaneous Mutation Resistance to Silver Nitrate
The spontaneous mutation frequency was also determined among the 20 silS positive strains. Fifteen strains showed selected mutants which exhibited silver resistance (MIC > 512μg/mL) and belonged to Klebsiella spp. (n = 10), Enterobacter spp. (n = 4) and Escherichia spp. (n = 1). The frequencies were between 2.2×10−7 and 9.8×10−9. The silver resistance was induced in K. pneumoniae strains belonged to ST11 and ST15. The complete sequences of silS and silR genes were sequenced to reveal the amino acid substitution responsible for silver resistance. The results were listed in Table 4. Fourteen strains with selected mutants showed point mutations in different positions in silS gene. Only one strain showed a point mutation (Asp187Tyr) in silR gene. Growth curves were measured to determine the fitness cost of Ag+ resistance. The parent strains and the derivative mutants showed similar growth curves, except for strain WHC182 (Figure S2).
 |
Table 4 In vitro Selection of Ag+ Resistance Mutants of the silS-Positive Strains |
Discussion
In the current study, 193 strains of gram-negative pathogens isolated from wound samples were collected in Xiangya hospital in Hunan province, China. Among the pathogens, Acinetobacter spp. were predominant, followed by Klebsiella spp., Escherichia spp. and Pseudomonas spp. The distribution of species was in accordance with studies conducted in St. Louis and Mashhad, which both reported P. aeruginosa, Acinetobacter and Klebsiella were the leading causes among gram-negative pathogens from burn samples.21,22 However, another study demonstrated that K. pneumoniae, P. aeruginosa and E. coli were the leading causes for skin and skin structure infection among gram-negative pathogens in Greece.23 These differences could be explained by the antibiotics commonly used in the areas where the studies were conducted.
It is not surprising that only nine strains were resistant to Ag+, while all strains of Nonfermenters were susceptible to Ag+. Previous studies confirmed that the silver resistant strains were not prevalent, and all belonged to Enterobacteriaceae.24,25 However, the MIC results from ours and previous studies were not comparable because the methods chosen to detect silver resistance were variable. The differences include the culture medium (MHB, Luria-Bertani broth without salt or IsoSensitent broth) and the MIC cut-off value chosen.24,26,27 The culture medium significantly influences the bactericidal potency of Ag+ due to NaCl or thiol-containing components, which can precipitate or bind to Ag+.28 Moreover, no widely accepted cut-off value for Ag+ is available to date. It is urgent to standardize the procedure of antimicrobial susceptibility test for Ag+.
In our study, WGS confirmed that the 24 strains carried the entire sil operons. To the best of our knowledge, this is the first time to verify PCR results for sil genes with WGS. In earlier studies, the sil genes were only detected by PCR and the authors claimed in some strains one or more sil genes were missing, mostly silE and silRS genes.11,24 Our PCR results also showed the absent of several sil genes, mainly silE. We further verified primer silRS used in Finley et al and only ten silRS genes were detectable out of 24 strains (Supplemental Table S2).24 The low detection rates for PCR method is due to the variation of sil genes. The sequences of silE, silR and silS genes showed variations (0–10%) when compared with that of the corresponding regions in pMG101, which is in consistence with minor variation (up to 4%) found in previous work.29 Our results emphasize that the primers for sil genes should be designed specifically for the highly conserved regions.
Among the 24 stains with sil operon, only four E. hormaechei strains were phenotypically resistant to Ag+. Ag+ susceptible strains with sil operon were reported in earlier studies and it is presumed that the sil operon is not constitutively expressed due to the regulation mediated by silRS. But importantly, the phenotypic resistance was easily induced, as indicated in our study that 15 out of 24 sil positive strains showed spontaneous mutants resistant to Ag+. Previous study demonstrated that spontaneous mutants resistant to Ag+ is prevalent among Enterobacteriaceae and the phenotypic change is due to the single missense mutations in silS, while no differences were found in other sil genes.9,26 In our study, only a single mutation in silR gene of strain WHC171 was identified before and after the exposure of Ag+, indicating that silR also involved in the spontaneous development of silver resistance. MLST analysis showed that silver resistance can be induced within K. pneumoniae strains belonged to both high-risk group (ST11 and ST15) and minor clone groups, which is in line with previous study.14
The most important factor contributing to the dissemination of sil operon is Tn7 transposon, which presents in the upstream region of sil operon. Tn7 element, encoding tnsABCDE, recognizes the attachment site (attTn7) though tnsD, which locates downstream of glmS gene, and can be inserted into chromosomes of all bacteria.30 In four strains, no plasmid hit was found by PlasmidFinder and their sil operon were all flanking with Tn7 element, so the sil operon may be transposed into the chromosome.31 Another important pathway for Tn7 is mediated by tnsE and targets mobile plasmids and filamentous bacteriophage, which facilitates the spread of sil operon into new host.30 The sil operon is first discovered in plasmid belonged to the Incompatibility group H. The IncH plasmids are large plasmid (> 150 Kb) with various antibiotic resistance genes and are transmissible under the temperature between 22 to 30 °C, contributing to the dissemination of resistance genes in soil or water.32 In our study, WGS confirmed the plasmid types in isolates with sil operon were variable, including IncFIB, IncFII, IncM2, IncQ1, IncHI2A, IncX3, IncR, IncI1-I, IncN, IncU, repB and Col, which is in line with previous study.11 The transmission of sil cassette into different plasmids may be mediated by Tn7 transposon.
Another important route for the spread of sil operon is co-selection under the pressure of antibiotics, especially β-lactams.33,34 Previous works have reported the relationship of silE and CTX-M-15 in E. coli strains from human and avian.35 Another study showed higher prevalence of silC gene in blaNDM-1-positive Enterobacteriaceae than that in blaCTM-M-15-producing strains and in susceptible strains.36 In current study, we also disclosed the relationship between silS and resistance to ceftriaxone, ceftazidime and aztreonam and detected various β-lactamase encoding genes, including blaESBL and blaApmC, in sil-positive strains. Furthermore, various plasmids with sil-operon were reported. A conjugative plasmid pSTM6-275 was reported to harbor sil operon and various antibiotics resistance genes, including blaTEM, strA, strB, sul3, aadA2, cmlA, aphA2 and tet(A).37 Similarly, an IncHI2 plasmid with sil operon, blaCTX-M-14 and aac(6’)-Ib-cr was identified from a E. coli strain isolated among a pig in Guangzhou province, China.38 In our study, a contig carrying tra operon, a well-known conjugation component, and sil operon was assembled in strain WHC181, indicating a putative conjugative plasmid.39 In BLAST NR database, this contig in WHC181 showed identical to the corresponding region in E. hormaechei strain SH19PTE2 plasmid pYUSHP2-2, isolated from pig feces in Shanghai, China in 2021. The geographic distance and the different sources indicated the possible dissemination of sil operon through a conjugative plasmid.
As for sil-negative K. pneumoniae strains, WGS revealed that a single mutation of cusS in cus operon with loss of outer membrane porins conferred high-level resistance, which is identical to endogenous resistance mechanism elaborated in E. coli BW25113 by Randall et al.4 Silver resistance was also induced in vitro in K. pneumoniae K5024 strain, due to a single mutation in cusS and a nonsense mutation in ompK36 gene.14 However, the authors failed to investigate the sequence of ompK35. In our study, we identified three-point mutations in the promoter region of ompK35, which was linked to loss of OmpK35.40 Roary identified the genetic variations between two strains were negligible, which were consistent to ERIC-PCR result, indicating that clone dissemination was possible. To the best of our knowledge, this is the first time to identify silver resistance strains from clinical samples with endogenous resistance mechanism. This finding emphasized that endogenous resistance mechanism also contribute to cryptic silver resistance. Because cus operon distributes widely in Enterobacteriaceae, including E. coli, C. freundii, Shigella sonnei and K. pneumoniae.41 Major porin loss is also prevalent, especially in multi-drug resistant K. pneumoniae. Recently, a global analysis based on WGS results of 2076 K. pneumoniae strains reported 29% strains lacked ompK35 and mainly distributed in K. pneumoniae CC258 while 3.7% strains showed mutations in ompK36.42 Another global research among 487 strains of ertapenem-non-susceptible K. pneumoniae reported 83.0% strains showed mutations in either or both of ompK35 and ompK36 gene.43 A study in Taiwan revealed 46.4% strains lost OmpK35 and OmpK36 among 347 carbapenem non-susceptible K. pneumoniae strains.44 It is reasonable to believe that silver resistant strains with endogenous mechanism could be induced during Ag+-based therapeutics.
Taken together, although phenotypic silver resistance is not prevalent in current study, our findings indicate that gram-negative pathogens can develop silver resistance via two different routes: a single mutation in silS or silR in strains with sil operon or a single mutation in cusS in strains with cus operon and major porins loss. Due to cryptic silver resistance, it is not sufficient to detect phenotypic silver resistance only. The screening at genetic level is essential but the researchers must be aware of the variation of sil genes. Moreover, silver or silver-containing materials should be used more discreetly, especially against multi-drug resistant pathogens, to eliminate the possibility to develop silver resistance and to prevent further spread of silver resistance genes.
Conclusion
The silver resistant rate for gram negative pathogen isolated from wound samples in our hospital is low. However, our findings indicate that silver resistance is easily induced in pathogens with two genetic backgrounds: strains with sil operon or with the combination of cus operon and major porin loss, especially in Klebsiella spp., so the screening of silver resistance at genetic level is essential. Furthermore, strains with sil operon often harbor various antibiotic resistance genes, including blaESBL and blaApmC. Therefore, it is of great significance to restrict the uncontrolled use of silver to prevent the further spread of silver resistance.
Abbreviations
BLAST, the Basic Local Alignment Search Tool; CHASRI, a copper homeostasis and silver resistance island; CLSI, Clinical and Laboratory Standards institute; Iden, overall identity; MIC, minimum inhibition concentration; MLST, multilocus sequence typing; PCR, polymerase chain reaction; QC, query coverage; WGS, whole genome sequencing.
Ethics Statement and Informed Consent
This research was conducted according to the recommendations of the Ethics Committee of Central South University (Changsha, Hunan Province, China). Ethical review and approval were waived for this study since the human samples were routinely collected and patients’ data remained anonymous.
Consent for Publication
All authors approved the manuscript and gave their consent for submission and publication.
Acknowledgments
We thank all staff in the Microbiology Department of Xiangya hospital for their assistance with bacterial collection.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Funding
This study was supported by grants from the Hunan Provincial Natural Science Foundation (No. 2019JJ50958 and 2020JJ5901).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Chernousova S, Epple M. Silver as antibacterial agent: ion, nanoparticle, and metal. Angewandte Chemie. 2013;52(6):1636–1653. doi:10.1002/anie.201205923
2. Mijnendonckx K, Leys N, Mahillon J, Silver S, Van Houdt R. Antimicrobial silver: uses, toxicity and potential for resistance. Biometals. 2013;26(4):609–621. doi:10.1007/s10534-013-9645-z
3. Talapko J, Matijević T, Juzbašić M, Antolović-Požgain A, Škrlec I. Antibacterial activity of silver and its application in dentistry, cardiology and dermatology. Microorganisms. 2020;8(9):1400. doi:10.3390/microorganisms8091400
4. Randall CP, Gupta A, Jackson N, Busse D, O’Neill AJ. Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J Antimicrob Chemother. 2015;70(4):1037–1046. doi:10.1093/jac/dku523
5. McHugh GL, Moellering RC, Hopkins CC, Swartz MN. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet. 1975;1(7901):235–240. doi:10.1016/S0140-6736(75)91138-1
6. Gupta A, Matsui K, Lo JF, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5(2):183–188. doi:10.1038/5545
7. Blanco Massani M, Klumpp J, Widmer M, et al. Chromosomal Sil system contributes to silver resistance in E. coli ATCC 8739. Biometals. 2018;31(6):1101–1114. doi:10.1007/s10534-018-0143-1
8. Staehlin BM, Gibbons JG, Rokas A, O’Halloran TV, Slot JC. Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in Enterobacteria. Genome Biol Evol. 2016;8(3):811–826. doi:10.1093/gbe/evw031
9. Sütterlin S, Dahlö M, Tellgren-Roth C, Schaal W, Melhus Å. High frequency of silver resistance genes in invasive isolates of Enterobacter and Klebsiella species. J Hosp Infect. 2017;96(3):256–261. doi:10.1016/j.jhin.2017.04.017
10. Randall CP, Oyama LB, Bostock JM, Chopra I, O’Neill AJ. The silver cation (Ag+): antistaphylococcal activity, mode of action and resistance studies. J Antimicrob Chemother. 2013;68(1):131–138. doi:10.1093/jac/dks372
11. Hosny AEM, Rasmy SA, Aboul-Magd DS, Kashef MT, El-Bazza ZE. The increasing threat of silver-resistance in clinical isolates from wounds and burns. Infect Drug Resist. 2019;12:1985–2001. doi:10.2147/IDR.S209881
12. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
13. O’Neill AJ, Cove JH, Chopra I. Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus. J Antimicrob Chemother. 2001;47(5):647–650. doi:10.1093/jac/47.5.647
14. Hanczvikkel A, Fuzi M, Ungvari E, Toth A. Transmissible silver resistance readily evolves in high-risk clone isolates of Klebsiella pneumoniae. Acta Microbiol Immunol Hung. 2018;65(3):387–403. doi:10.1556/030.65.2018.031
15. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
16. Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi:10.1186/1471-2164-9-75
17. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi:10.1093/jac/dkaa345
18. Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi:10.1128/AAC.02412-14
19. Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi:10.1128/JCM.06094-11
20. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi:10.1093/bioinformatics/btv421
21. Zilberberg MD, Shorr AF, Micek ST, et al. Epidemiology and outcomes of hospitalizations with complicated skin and skin-structure infections: implications of healthcare-associated infection risk factors. Infect Control Hosp Epidemiol. 2009;30(12):1203–1210. doi:10.1086/648083
22. Rezaei E, Safari H, Naderinasab M, Aliakbarian H. Common pathogens in burn wound and changes in their drug sensitivity. Burns. 2011;37(5):805–807. doi:10.1016/j.burns.2011.01.019
23. Nodaras C, Kotsaki A, Tziolos N, et al. Microbiology of acute bacterial skin and skin-structure infections in Greece: a proposed clinical prediction score for the causative pathogen. Int J Antimicrob Agents. 2019;54(6):750–756. doi:10.1016/j.ijantimicag.2019.08.020
24. Finley PJ, Norton R, Austin C, Mitchell A, Zank S, Durham P. Unprecedented silver resistance in clinically isolated Enterobacteriaceae: major Implications for burn and wound management. Antimicrob Agents Chemother. 2015;59(8):4734–4741. doi:10.1128/AAC.00026-15
25. Percival SL, Salisbury AM, Chen R. Silver, biofilms and wounds: resistance revisited. Crit Rev Microbiol. 2019;45(2):223–237. doi:10.1080/1040841X.2019.1573803
26. Elkrewi E, Randall CP, Ooi N, Cottell JL, O’Neill AJ. Cryptic silver resistance is prevalent and readily activated in certain Gram-negative pathogens. J Antimicrob Chemother. 2017;72(11):3043–3046. doi:10.1093/jac/dkx258
27. Sutterlin S, Tellez-Castillo CJ, Anselem L, Yin H, Bray JE, Maiden MCJ. Heavy metal susceptibility of Escherichia coli isolated from urine samples from Sweden, Germany, and Spain. Antimicrob Agents Chemother. 2018;62(5). doi:10.1128/AAC.00209-18
28. Liau SY, Read DC, Pugh WJ, Furr JR, Russell AD. Interaction of silver nitrate with readily identifiable groups: relationship to the antibacterial action of silver ions. Lett Appl Microbiol. 1997;25(4):279–283. doi:10.1046/j.1472-765X.1997.00219.x
29. Gupta A, Phung LT, Taylor DE, Silver S. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology. 2001;147(Pt 12):3393–3402. doi:10.1099/00221287-147-12-3393
30. Peters JE. Tn7. Microbiol Spectr. 2014;2(5). doi:10.1128/microbiolspec.MDNA3-0010-2014
31. Hooton SPT, Pritchard ACW, Asiani K, et al. Laboratory stock variants of the archetype silver resistance plasmid pMG101 demonstrate plasmid fusion, loss of transmissibility, and transposition of Tn7/pco/sil into the host chromosome. Front Microbiol. 2021;12:723322. doi:10.3389/fmicb.2021.723322
32. Phan MD, Wain J. IncHI plasmids, a dynamic link between resistance and pathogenicity. J Infect Dev Ctries. 2008;2(4):272–278. doi:10.3855/jidc.221
33. Baker-Austin C, Wright MS, Stepanauskas R, McArthur JV. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14(4):176–182. doi:10.1016/j.tim.2006.02.006
34. Pal C, Asiani K, Arya S, et al. Metal resistance and its association with antibiotic resistance. Adv Microb Physiol. 2017;70:261–313.
35. Sütterlin S, Edquist P, Sandegren L, et al. Silver resistance genes are overrepresented among Escherichia coli isolates with CTX-M production. Appl Environ Microbiol. 2014;80(22):6863–6869. doi:10.1128/AEM.01803-14
36. Yang QE, Agouri SR, Tyrrell JM, Walsh TR. Heavy metal resistance genes are associated with bla(NDM-1)- and bla(CTX-M-15)-carrying Enterobacteriaceae. Antimicrob Agents Chemother. 2018;62(5). doi:10.1128/AAC.02642-17
37. Billman-Jacobe H, Liu Y, Haites R, et al. pSTM6-275, a conjugative IncHI2 plasmid of Salmonella enterica that confers antibiotic and heavy-metal resistance under changing physiological conditions. Antimicrob Agents Chemother. 2018;62(5). doi:10.1128/AAC.02357-17
38. Fang L, Li X, Li L, et al. Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli<isolates of food-producing animals. Sci Rep. 2016;6(1):25312. doi:10.1038/srep25312
39. Wong JJ, Lu J, Glover JN. Relaxosome function and conjugation regulation in F-like plasmids - a structural biology perspective. Mol Microbiol. 2012;85(4):602–617. doi:10.1111/j.1365-2958.2012.08131.x
40. Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M, et al. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2018;52(6):898–905. doi:10.1016/j.ijantimicag.2018.03.020
41. McNeilly O, Mann R, Hamidian M, Gunawan C. Emerging concern for silver nanoparticle resistance in Acinetobacter baumannii and other bacteria. Front Microbiol. 2021;12:652863. doi:10.3389/fmicb.2021.652863
42. Rocker A, Lacey JA, Belousoff MJ, et al. Global trends in proteome remodeling of the outer membrane modulate antimicrobial permeability in Klebsiella pneumoniae. MBio. 2020;11(2):e00603–e00620. doi:10.1128/mBio.00603-20
43. Wise MG, Horvath E, Young K, Sahm DF, Kazmierczak KM. Global survey of Klebsiella pneumoniae major porins from ertapenem non-susceptible isolates lacking carbapenemases. J Med Microbiol. 2018;67(3):289–295. doi:10.1099/jmm.0.000691
44. Chiu SK, Wu TL, Chuang YC, et al. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One. 2013;8(7):e69428. doi:10.1371/journal.pone.0069428
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
