Back to Journals » Infection and Drug Resistance » Volume 14
Characterization of Antigen Escape Mutations in Chronic HBV-Infected Patients in Upper Egypt
Authors El-Mokhtar MA , Hetta HF , Mekky MA , Abd El-Kareem DM , Ramadan M, Salah M, Mohamed NA, El-Masry EA, Adel S, Sayed IM
Received 11 April 2021
Accepted for publication 24 May 2021
Published 28 June 2021 Volume 2021:14 Pages 2419—2427
DOI https://doi.org/10.2147/IDR.S315299
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Mohamed A El-Mokhtar, 1,* Helal F Hetta, 1,* Mohamed A Mekky, 2 Doaa M Abd El-Kareem, 3 Mohammed Ramadan, 4 Mohammed Salah, 5 Nahed A Mohamed, 6 Eman A El-Masry, 7, 8 Sara Adel, 9 Ibrahim M Sayed 1, 10
1Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, 71515, Egypt; 2Department of Tropical Medicine and Gastroenterology, Assiut University Hospital, Assiut, Egypt; 3Department of Clinical Pathology, Faculty of Medicine Assiut University, Assiut, Egypt; 4Microbiology and Immunology Department, Faculty of Pharmacy Al-Azhar University-Assiut branch, Assiut, 71526, Egypt; 5Microbiology and Immunology Department, Faculty of Pharmacy Port Said University, Port Said, 42526, Egypt; 6Department of Medical Biochemistry, Faculty of Medicine, Assiut University, Assiut, Egypt; 7Microbiology and Immunology Unit, Department of Pathology, College of Medicine, Jouf University, Al-Jouf, Saudi Arabia; 8Department of Medical Microbiology and Immunology, College of Medicine, Menoufia University, Menoufia, Egypt; 9Department of Clinical Pathology, Faculty of Medicine, Al-Azhar University, Assiut, 71515, Egypt; 10Department of Pathology, School of Medicine, University of California, San Diego, CA, 92093, USA
*These authors contributed equally to this work.
Correspondence: Ibrahim M Sayed
Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, 71515, Egypt
Email [email protected]
Background: Mutations within the “a” determinant region (position 124– 147) that is present in the major hydrophilic region (MHR, position 99– 160) of the hepatitis B surface antigen (HBsAg) are associated with vaccine-escape, lack of diagnosis, and failure to hepatitis B immunoglobulin (HBIG) therapy. Data regarding the amino acid changes of “a” determinant region of HBsAg are limited in Egypt. The prevalence and mutations in this region among chronic HBV (CHB)-infected patients in Upper Egypt are not known.
Material and Methods: Blood samples were collected from HBsAg-positive CHB-infected patients (n=123) admitted to Assiut University Hospitals. Serum samples were screened for HBsAg, HBeAg, anti-HBs and anti-HBe antibodies using commercially available ELISA kits. Viral load was determined by qPCR. In addition, mutational analysis was carried out targeting the HBV surface gene to determine the HBV genotype and vaccine escape mutations.
Results: Sequencing analysis of HBV DNA revealed that genotype D is the major circulating type (81.3%), followed by genotype E (18.7%). Analysis of the HBV genome revealed that 103/123 (83.7%) patients showed wild-type sequences and 20/123 (16.3%) showed mutations in the HBsAg gene. Mutation in seventeen patients (17/20, 85%) showed only one mutation, and three patients showed two mutations (3/20, 15%) in the “a” determinant region. The observed mutations were T115S (3/20, 15%), P120T/S (3/20, 15%), T126S (1/20, 5%), Q129R (2/20, 10%), M133T (2/20, 10%), S143L (5/20, 25%), D144E/A (3/20, 15%), and G145R/A (4/20, 20%). Mutations in the “a” determinant region were detected in genotype D isolates only.
Conclusion: We described for the first time the prevalence and characterization of vaccine escape mutants in CHB patients in Upper Egypt. Mutational analysis of the “a” determinant region revealed the presence of a wide spectrum of mutants in the circulating HBV isolates that could be a potential threat to HBV diagnosis, therapy success, and HBV vaccination program in Upper Egypt.
Keywords: HBV, “a” determinant, mutational analysis, vaccine escape, Upper Egypt
Graphical Abstract:
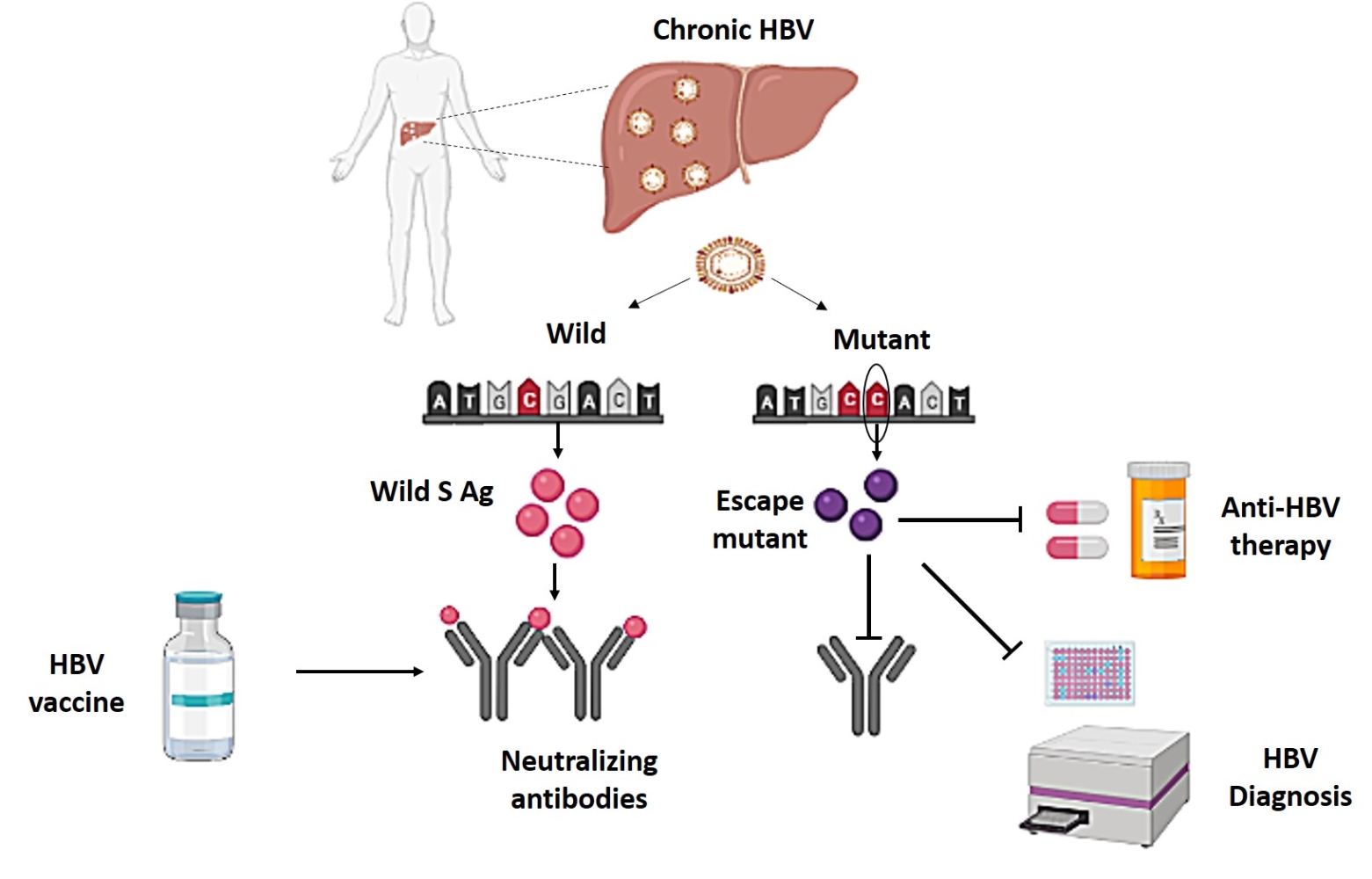
Introduction
Hepatitis B virus (HBV) is the smallest hepatotropic DNA virus (about 3200 bp) infecting humans. HBV belongs to the Hepadnaviridae and causes a potentially life-threatening disease.1 HBV causes acute and chronic infections; however, chronic infections can lead to a high risk of death due to HBV-associated complications such as cirrhosis and hepatocellular carcinoma (HCC).2 The WHO estimated that, in 2015, 257 million individuals were infected with HBV and 887,000 patients died of HBV-associated cirrhosis and liver cancer.3
Hepatitis B surface antigen (HBsAg) was the first discovered HBV protein. It is a glycoprotein located in the envelope of HBV, and is released from infected cells mainly as subviral parts in different particle forms (competent Dane particles, filaments, and spherical particles), representing empty viral envelopes rather than infectious virions.4,5 Detection of serum HBsAg is a key diagnostic indicator of HBV-infection, and quantitative estimation of the HBsAg has been suggested to be a biomarker for predicting the response to treatment of chronic HBV-infected patients.5,6 HBsAg is the first HBV marker that appears in serum and its persistence for >6 months indicates a chronic infection.7 Besides, HBsAg is highly antigenic, and anti-HBsAg antibodies can neutralize the virus and provide a protective humoral immune response against infection.8
Serum HBsAg is encoded by a single open reading frame and is translated into 3 HBsAg proteins of variable lengths. The small (S) protein is the highest expressed protein. It is a secreted protein that consists of 226 amino acids. It is the major protein in virions and the subviral parts.9 The (S) protein contains the “a” determinant region which is located in amino acid positions 124–147. This region codes for a neutralizing conformational epitope of high antigenicity and is formed by folding of the cysteine-rich major hydrophilic region (MHR), which extends from amino acids 99–160, resulting in the formation of loop‐like structures.10 Antibodies that bind regions 1–137 and 139–147 have been described in serum samples of vaccinated people.8,11
Importantly, in the past years, HBV mutants with variations in the amino acid composition of the “a” determinant region have been reported. Some of these HBV mutants have been identified even before the introduction of the vaccine program.12 These naturally occurring HBV variants can infect vaccinated individuals and are not neutralized by the vaccine-induced anti-HBsAg antibodies because mutations introduced in this epitope can decrease the affinity of antibodies to the HBsAg.13,14 The development of mutants that escape the neutralization by the vaccine-induced antibodies (vaccine-escape mutants) poses a serious threat to control HBV infections, including reactivation of HBV by such mutants, vaccination failure or underestimation of HBsAg levels.15 Furthermore, the HBV polymerase gene overlaps with the HBsAg gene. Therefore, mutations in the polymerase (pol) gene, which could be induced by antiviral therapy or immunological pressure, can lead to mutations in the HBsAg regions as well.16
The prevalence of HBV (based on the serological marker of HBsAg) among adults aged 15 −59 in Egypt is 1.4–1.7%.17,18 Spatial analysis revealed that HBV infection is mainly localized to urban areas of Upper Egypt.17 Data regarding the amino acid changes in the “a” determinant region of HBsAg among chronic HBV (CHB)-infected patients in Upper Egypt is limited. Therefore, this study aimed to characterize HBsAg MHR variants, especially in the “a” determinant region in cases with CHB infection from Upper Egypt to determine the vaccine escape mutations.
Materials and Methods
Ethical Statement
The study was approved by the ethical committee of the Faculty of Medicine, Assiut University (IRB no: 17300006). Informed written consent was given by all participants.
Patient Samples
One hundred twenty-three patients with HBV infection were enrolled in this study. HBV-infected patients were admitted to Assiut University Hospitals, Egypt, in the period from October 2018 to December 2020. Serum samples were screened for hepatitis viruses according to the protocol of Assiut University hospitals and as described previously.19,20 All patients were positive for HBsAg while those who had coinfection with HAV, HCV, HEV, or HIV were excluded from the study.
Detection of HBV Markers
Serum samples were screened for HBsAg, HBeAg, anti-HBs and anti-HBe antibodies using commercially available ELISA kits (Wkea Medical Supplies Corporation, China) following the manufacturer’s instructions. Plates were read using the Biotek microplate reader (Biotek version Elx508v, USA). For determination of HBV viral load, viral nucleic acid was extracted from 200 ul of serum samples using QIAamp MinElute Virus Spin kit (Qiagen, Germany) then analyzed by real-time PCR (qPCR). HBV DNA was quantified using the Artus-HBV RT PCR kit (Qiagen, Germany) on the Applied Biosystems 7500 Fast RT-PCR machine (Applied biosystems, USA).
Determination of HBV Genotype and S Gene Mutations
HBV genotyping was determined by amplification and sequencing of 332 bp of HBV S gene using nested PCR reaction as described previously.21,22 Briefly, the first PCR was carried out using the primer pairs HBV-022 (sense primer) and antisense primers HBV-065 and HBV-066. The nested reaction was performed primers HBV-024 (sense primer) and antisense primers HBV-041 and HBV-064. Nested PCR products of approximately 332 bp were purified by GeneJET Gel Extraction Kit (Thermo Scientific™, USA) and sequenced at Macrogen sequencing facility (Macrogen, Seoul, Korea) using primer HBV-024. PCR reactions were carried out using the HotStarTaq Master Mix (Qiagen, Germany) and included an initial denaturation at 95°C for 15 min, followed by 40 cycles of 94°C for 30 sec, 55°C for 30 sec (54°C in the second reaction), and 72°C for 45 sec, followed by a 5 min final extension at 72°C. HBV genotyping was determined by alignment and comparing the obtained sequences with sequences of HBV reference genotypes (genotypes A-J) downloaded from NCBI GenBank using the MEGA X software. Each sequence was blasted into NCBI BLASTn to determine the best match isolates according to the highest Max score. To confirm HBV genotyping, these sequences were compared with randomly selected HBV sequences isolated from Egypt and deposited in GenBank. Sequences were translated into amino acids to identify amino acid substitutions and determine the vaccine escape mutations. Sequences of the used primers are shown in Supplemental Table 1. The HBV sequences were deposited in the GenBank and assigned the following accession numbers (MZ197872-MZ197892).
Statistical Analyses
Statistical analyses were performed using the GraphPad Prism software 9 (GraphPad Software, La Jolla, USA). Results are expressed as median with interquartile range (IQ) unless otherwise specified. Comparison between groups was carried out using Student’s t-test. P < 0.05 was considered significant.
Results
Demographic and Laboratory Characterization of HBV-Infected Patients
This study included CHB-infected patients (n=123) admitted to Assiut University Hospitals from 2018 to 2020. The median age of patients was 45 years and 67/123 (54.47%) were male and 56/123 (45.53%) were female. All the enrolled patients were positive for HBs Ag. HBe Ag was recorded in 25/123 (20.3%) of patients, and anti-HBe Ag was detected in 88/123 (71.54%). While 10/123 (8.1%) of the patients were negative to HBe Ag and anti-HBe Ag (Table 1). Four out of 123 patients were positive for anti-HBs Ab (3.25%). The mean HBV plasma load was 1.29×106 IU/mL (median with IQR, 2.34×105 (2.4x104- 6.7×105) IU/mL). The median (interquartile range, IQR) of ALT in CHB patients was 240 U/l (Table 1). Sequencing analysis revealed that the prevalent circulating genotype belonged to group D (100/123, 81.3%), and genotype E (23/123, 18.7%). No mixed isolates were detected in this cohort. All patients received 0.5 mg/day entecavir.
 |
Table 1 Characteristics of CHB Patients Enrolled in the Study |
Assessment of HBV Antigen Escape Mutations in the CHB-Infected Patients
To assess the HBV vaccine escape mutations in this cohort, we did a mutational analysis on the CHB-infected patients in the major hydrophilic region (MHR) (aa 99–169), especially in “a” determinant region of the small surface protein of HBs Ag (aa 124–147). We focused mainly on the “a” determinant region since it is highly conserved and found in all genotypes and serotypes of HBV, also major vaccine escape mutants were recorded in this region. Analysis of the HBV surface gene revealed that 103/123 (83.7%) patients showed wild-type sequences, and 20/123 (16.3%) showed mutations in this region. First, we found two mutations in the region aa 115–123, where the mutations were recorded in the aa positions 115 and 120 (Table 2). In the “a” determinant region, which also is known as major B-cell antigenic epitopes, we recorded mutations in the aa positions 126, 129, 133, 144, and 145 (Table 2). No mutation was recorded in the aa position 148–160 (Table 2).
 |
Table 2 Mutations in the MHR in the Current Study |
Mutation analysis revealed that only one mutation was detected in seventeen patients (17/20, 85%), and three patients showed two mutations (3/20, 15%) (Figure 1 and Table 3). The following mutation pairs were observed: T115S (3/20, 15%), P120T/S (3/20, 15%), T126S (1/20, 5%), Q129R (2/20, 10%), M133T (2/20, 10%), S143L (5/20, 25%), D144E/A (3/20, 15%), and G145R/A (4/20, 20%) (Table 3). Ag escape mutations were detected in genotype D isolates, and no mutation was detected in HBV genotype E isolates.
 |
Table 3 Mutations in the “a” Determinant Domain of Egyptian HBV Isolates |
Distribution of HBV Antigen Escape Mutations Among the Egyptian CHB
Then, we assessed the distribution of wild-type (WT) HBV isolates and HBV mutants among the Egyptian CHB patients in terms of age, gender, the status of other HBV markers, HBV load, HBV genotypes, and liver enzyme ALT (Table 4). We did not find a significant difference between the distribution of WT and mutant HBV isolates regarding age, gender, the status of HBeAg and anti-HBe Ag, HBV load, ALT level, and the level of HBs Ag/ anti-HBs Ab (Table 4). Three patients (3/100, 3%) who tested positive for HBs Ag/anti-HBs Ab showed wild HBV sequence, and one patient (1/23, 4.3%) who tested positive for HBs Ag/anti-HBs Ab showed mutant HBV sequence. No mutant HBV isolates were recorded in patients tested negative for HBeAg and anti-HBe. Also, all mutant HBV isolates belonged to genotype D and none of them belonged to genotype E (Table 4).
 |
Table 4 Distribution of “a” Determinant Mutants in the Egyptian CHB |
Discussion
The emergence of HBV escape mutants raised concerns for HBsAg immunoassays, the development of vaccine-escape mutants, and failure to hepatitis B immunoglobulin (HBIG) therapy.23–25 HBV mutants can escape the host immune response leading to the development of complications such as fulminant hepatitis, cirrhosis, and hepatocellular carcinoma.26 The development of HBsAg mutations is caused by administration of HBV vaccine and/or HBIG, or natural selection by the host immune response, which changes the amino acid composition of the HBsAg.27,28 Escape mutations are mostly recorded in the major hydrophilic region (MHR) of HBsAg, which codes for aa 99–160, more specifically in the “a” determinant region (aa 124–147), suggesting that this region was more affected by immune selection or antiviral therapy than other regions.25 Ma and Wang reported that eight mutations (P120T, T126S, Q129H, G130N, S143L, D144A, and G145A/R) are associated with diagnostic failure and mutations in the positions 120, 126, 129, 130, 133, 134, 137, 140, 143, 144, and 145 are recorded in escape variants hat evade vaccine or immunoglobulin therapy.25 The well-characterized escape mutations in the HBsAg are P120T, D144E/A, and G145R, the latter mutant affects the polymerase gene.27,29,30 HBV variants with combinations of HBs Ag (G145R or P120T) and polymerase (L526M plus M550V) mutations showed increased HBV replication resulting in a severe clinical course in transplanted patients.30
In Egypt, characterization of HBs Ag mutants is limited among CHB patients and occult blood donors.31,32 HBs Ag mutation has not been previously described in Upper Egypt and this is the aim of our study.
In this study, we assessed HBs Ag mutations in the CHB patients admitted to Assiut University hospitals. First, we characterized the HBV genotypes circulating among these patients, and we found that genotype D (HBV-D) (81.3%) is the major circulating isolate among the CHB patients, followed by genotype E (17.3%). In a parallel line, other studies revealed that genotype D is the predominant isolate circulating among Egyptian CHB and blood donors.31–34 HBV-D is widely distributed throughout the Mediterranean region, from Europe to North Africa.33,35,36 No mixed isolates were recorded in our cohort. Contrary to our findings, Zekri et al reported that 15.7% of HBV infection among Egyptian pediatric cancer patients was caused by mixed A/D genotype infections.36 The difference in the patients’ age and criteria could explain the difference in the circulating HBV isolates among the two cohorts.
In this study, we assessed the mutations in the MHR, especially the aa positions 115–160. This region is highly conserved and found in all HBV genotypes and serotypes.37 Our results showed that 103/123 (83.7%) were WT sequences, while 20/123 (16.3%) had mutations in this region. Previous studies in Egypt revealed that the percentage of MHR variants was 14.8%, 37.8%, and 50% in CHB patients, blood donors with overt infection, and blood donors with occult infection, respectively.31,32 Other studies reported that the prevalence of HBsAg vaccine escape mutations ranged 0.7% to 28% depending on the age of patients, region, and endemicity of HBV.21,28,30
In the region aa115-123, we found two mutations, the first mutation at the position aa115 (T115S) in three patients and the second mutation at the position aa120 (P120T) in three patients. In the region aa124-147, we found 6 different mutations at the positions 126 (T126S), 129 (Q129R), 133 (M133T), 143 (S143L), 144 (D144E/A) and 145 (G145R/A). S143L, G145R/A, and D144E/A were the highest reported mutations among our cohort, which were detected in 25%, 20%, and 15% of the patients, respectively. While Q129R, M133T, and T126S were recorded in 10%, 10%, and 5% of the patients, respectively. Carman et al described the first vaccine escape mutation resulted from the substitution of a glycine (G) residue at position 145 by an arginine (A) residue (G145R). This widely reported mutation is stable and reduces the binding of HBsAg to anti-HBs monoclonal antibody.8 Several other studies reported other substitutions in the “a” determinant region and associated with vaccine escape, such as T116N, P120S/E, I/T126A/N/I/S, Q129H/R, M133L, K141E, and D144A/E.38,39 Similar to our findings, Zeid et al reported that S143L mutation is the predominant mutation (6.8%) among Egyptian CHB patients.31 T115S, P120T, T125M, P127T, Q129R, K141R, and S143L were HBV mutants recorded among Egyptian blood donors with occult infection.32,40 Up to our knowledge, our study is the first report that describes the vaccine escape mutants among CHB in Upper Egypt. In addition, this study shows the presence of different vaccine mutants circulating among Egyptian CHB patients, some mutants such as G145R/A, and D144E/A are recognized as the commonly known vaccine escape mutants. The presence of different vaccine escape mutants suggests that there could be a potential threat to the HBV vaccination program in Upper Egypt.
In this study, we correlated between the presence of WT/mutant HBsAg isolate and patients’ criteria. We did not find a significant difference between patients infected with WT HBV isolates and patients infected with vaccine escape mutants regarding age, sex, viral load, the status of HBe Ag and anti-HBe Ag, and liver enzyme level. Similarly, Al Baqlani et al reported that there was no correlation between WT HBV isolates and HBV mutants regarding the geographic distribution, gender, age, HBV genotypes, the status of HBeAg, and anti-HBe, and HBV load in Oman.21 However, there was a significant difference between WT HBV isolates and HBV mutants in patients who tested negative for HBe Ag and anti-HBe Ag, suggesting that CHB patients who tested negative for HBe Ag and anti-HBe Ag could be more likely to develop vaccine escape mutations. Further studies need to ascertain this point.
The emergence and prevalence of HBV vaccine mutants have been increasing since the application of the HBV vaccine program. The World Health Organization (WHO) recommended the implementation of HBV vaccination for children worldwide. In 1992, the HBV vaccination program was started in Egypt with a schedule of 2, 4, and 6 months of age.41 However, we do not have an actual percentage for the adults who vaccinated against HBV in Egypt. Different factors could affect the prevalence of vaccine mutation such as cohort analysis, sample size, time of vaccine implementation, viral genotype, etc.
In this study, four patients (4/123, 3.25%) tested positive for both HBs Ag and anti-HBs Ab. Similarly, other studies reported the prevalence of HBsAg and anti-HBs double-positive serological profiles among CHB patients ranged 2.8% to 3.6%.42,43 Three out of four patients showed a wild-type HBV sequence, and the fourth patient showed a mutant HBV sequence (Pt#14). Colson et al reported that HBsAg+/anti-HBsAb+ CHB patients exhibited significantly higher amino acid variability in the “a” determinant region than HBs Ag+ CHB.42 Also, they showed that mutation in “a” determinant region at positions 120, 126, 144 and 145 were detected in HBsAg+ patients without the presence of coexistence of anti-HBs Ab.42 However, the number of HBsAg+/anti-HBsAb+ patient in our cohort is small (n=4). Therefore, we assume that further studies including a relatively larger number of double-positive (HBsAg+/anti-HBsAb+) CHB patients could help to evaluate the impact of the presence of anti-HBsAg with HBs Ag on the prevalence of vaccine escape mutants.
There are some limitations to this study. First, we have not analyzed the mutation in the polymerase gene which is associated with antiviral resistance. Due to gene overlapping between HBs Ag gene and polymerase gene, the mutation in polymerase gene induced by anti-HBV therapies could affect HBs Ag detection and vaccine escape mutation. Also, we have not determined HBV sub-genotypes which could link to the vaccine escape mutation. A recent study showed that HBV mutants associated with vaccine escape, diagnosis failure, and drug resistance could be genotypic and sub-genotypic specific.44 Further studies should be done to assess the impact of anti-HBV therapies on the incidence and spread of HBV escape mutants.
In conclusion, this study describes for the first time the prevalence and characterization of vaccine escape mutants in CHB patients in Upper Egypt. We found that genotype D is the predominant isolates, and mutational analysis in the “a” determinant region revealed the presence of a wide spectrum of mutants that could be a potential threat to HBV diagnosis, therapy success, and HBV vaccination program in Upper Egypt.
Institutional Review Board Statement
The study was approved by the ethical committee of the Faculty of Medicine, Assiut University (IRB no: 17300006). Informed written consent was given by all participants. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board.
Acknowledgments
The authors are grateful to the Assiut Medical Research center for providing some instruments required for doing the experiments.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This paper is based upon work supported by Science, Technology & Innovation Funding Authority (STDF) under grant number 30208.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350(11):1118–1129. doi:10.1056/NEJMra031087
2. Chaturvedi VK, Singh A, Dubey SK, Hetta HF, John J, Singh MP. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb Pathog. 2019;128:184–194. doi:10.1016/j.micpath.2019.01.004
3. CfDAWA C. Estimates of the coverage of diagnosis and treatment for hepatitis B and C virus infection, by WHO region and income group. Global hepatitis report 2017. Geneva: World Health Organization; 2018. (WHO/CDS/HIV/1847) 2015.
4. Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64(1):51–68.
5. Nguyen DH, Ludgate L, Hu J. Hepatitis B virus-cell interactions and pathogenesis. J Cell Physiol. 2008;216(2):289–294. doi:10.1002/jcp.21416
6. Jaroszewicz J, Calle Serrano B, Wursthorn K, et al. Hepatitis B surface antigen (HBsAg) levels in the natural history of hepatitis B virus (HBV)-infection: a European perspective. J Hepatol. 2010;52(4):514–522. doi:10.1016/j.jhep.2010.01.014
7. Kao J-H. Diagnosis of hepatitis B virus infection through serological and virological markers. J Exp Rev Gastroenterol Hepatol. 2008;2(4):553–562. doi:10.1586/17474124.2.4.553
8. Carman WF, Zanetti AR, Karayiannis P, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336(8711):325–329. doi:10.1016/0140-6736(90)91874-A
9. Simon K, Lingappa VR, Ganem D. Secreted hepatitis B surface antigen polypeptides are derived from a transmembrane precursor. J Cell Biol. 1988;107(6 Pt 1):2163–2168. doi:10.1083/jcb.107.6.2163
10. Eble B, Lingappa V, Ganem D. Hepatitis B surface antigen: an unusual secreted protein initially synthesized as a transmembrane polypeptide. J Mol Cell Biol. 1986;6(5):1454–1463. doi:10.1128/MCB.6.5.1454
11. Brown SE, Howard CR, Zuckerman AJ, Steward MW. Affinity of antibody responses in man to hepatitis B vaccine determined with synthetic peptides. Lancet. 1984;2(8396):184–187. doi:10.1016/S0140-6736(84)90479-3
12. Okamoto H, Yano K, Nozaki Y, et al. Mutations within the S gene of hepatitis B virus transmitted from mothers to babies immunized with hepatitis B immune globulin and vaccine. J Pediatr Res. 1992;32(3):264–268. doi:10.1203/00006450-199209000-00002
13. Oon C. Evolution and transmission of hepatitis B virus mutants. J Hepatitis B Asia-Pacific Region London Royal College Physicians. 1997;177–190.
14. El Chaar M, Candotti D, Crowther RA, Allain JP. Impact of hepatitis B virus surface protein mutations on the diagnosis of occult hepatitis B virus infection. Hepatology. 2010;52(5):1600–1610. doi:10.1002/hep.23886
15. Sheldon J, Soriano V. Hepatitis B virus escape mutants induced by antiviral therapy. J Antimicrob Chemother. 2008;61(4):766–768. doi:10.1093/jac/dkn014
16. Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002;25(2):97–106. doi:10.1016/S1386-6532(02)00049-5
17. Ismail SA, Cuadros DF, Benova L. Hepatitis B in Egypt: a cross-sectional analysis of prevalence and risk factors for active infection from a nationwide survey. Liver Int. 2017;37(12):1814–1822. doi:10.1111/liv.13469
18. Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386(10003):1546–1555. doi:10.1016/S0140-6736(15)61412-X
19. El-Mokhtar MA, Seddik MI, Osman AOB, et al. No evidence of HEV genotype 1 infections harming the male reproductive system. Virology. 2021;554:37–41. doi:10.1016/j.virol.2020.12.002
20. Sayed IM, El-Mokhtar MA, Mahmoud MAR, et al. Clinical Outcomes and Prevalence of Hepatitis E Virus (HEV) Among Non-A-C Hepatitis Patients in Egypt. Infect Drug Resist. 2021;14:59–69. doi:10.2147/IDR.S289766
21. Al Baqlani SA, Sy BT, Ratsch BA, et al. Molecular epidemiology and genotyping of hepatitis B virus of HBsAg-positive patients in Oman. PLoS One. 2014;9(5):e97759. doi:10.1371/journal.pone.0097759
22. Hoan NX, Hoechel M, Tomazatos A, et al. Predominance of HBV Genotype B and HDV Genotype 1 in Vietnamese Patients with Chronic Hepatitis. Viruses. 2021;13:2. doi:10.3390/v13020346
23. Oon CJ, Lim GK, Ye Z, et al. Molecular epidemiology of hepatitis B virus vaccine variants in Singapore. Vaccine. 1995;13(8):699–702. doi:10.1016/0264-410X(94)00080-7
24. Weber B. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J Clin Virol. 2005;32(2):102–112. doi:10.1016/j.jcv.2004.10.008
25. Ma Q, Wang Y. Comprehensive analysis of the prevalence of hepatitis B virus escape mutations in the major hydrophilic region of surface antigen. J Med Virol. 2012;84(2):198–206. doi:10.1002/jmv.23183
26. Bartholomeusz A, Locarnini S. Hepatitis B virus mutants and fulminant hepatitis B: fitness plus phenotype. Hepatology. 2001;34(2):432–435. doi:10.1053/jhep.2001.26764
27. Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30(5):1312–1317. doi:10.1002/hep.510300511
28. Carman WF, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345(8962):1406–1407. doi:10.1016/S0140-6736(95)92599-6
29. Locarnini S, McMillan J, Bartholomeusz A. The hepatitis B virus and common mutants. Semin Liver Dis. 2003;23(1):5–20. doi:10.1055/s-2003-37587
30. Bock CT, Tillmann HL, Torresi J, et al. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology. 2002;122(2):264–273. doi:10.1053/gast.2002.31015
31. Abu Zeid WM, Ramadan DI, Shemis MA. Prevalence of mutations within major hydrophilic region of hepatitis B virus and their correlation with genotypes among chronically infected patients in Egypt. Arab J Gastroenterol. 2016;17(1):34–40. doi:10.1016/j.ajg.2016.03.001
32. Kishk R, Nemr N, Elkady A, et al. Hepatitis B surface gene variants isolated from blood donors with overt and occult HBV infection in north eastern Egypt. Virol J. 2015;12:153. doi:10.1186/s12985-015-0389-y
33. Zaky S, Farghaly AM, Rashed HA, et al. Clinicopathologic features and genotyping of patients with chronic HBV infection in the Upper Egypt. Cell Immunol. 2010;265(2):97–104. doi:10.1016/j.cellimm.2010.07.008
34. Elbahrawy A, Alaboudy A, El Moghazy W, Elwassief A, Alashker A, Abdallah AM. Occult hepatitis B virus infection in Egypt. World J Hepatol. 2015;7(12):1671–1678. doi:10.4254/wjh.v7.i12.1671
35. Schaefer S. Hepatitis B virus genotypes in Europe. Hepatol Res. 2007;37(s1):S20–26. doi:10.1111/j.1872-034X.2007.00099.x
36. Zekri AR, Hafez MM, Mohamed NI, et al. Hepatitis B virus (HBV) genotypes in Egyptian pediatric cancer patients with acute and chronic active HBV infection. Virol J. 2007;4:74. doi:10.1186/1743-422X-4-74
37. Lazarevic I, Banko A, Miljanovic D, Cupic M. Immune-Escape Hepatitis B Virus mutations associated with viral reactivation upon immunosuppression. Viruses. 2019;11(9):778. doi:10.3390/v11090778
38. Echevarría JM, Avellón A. Hepatitis B virus genetic diversity. J Med Virol. 2006;78(Suppl 1):S36–42. doi:10.1002/jmv.20605
39. Lazarevic I. Clinical implications of hepatitis B virus mutations: recent advances. World J Gastroenterol. 2014;20(24):7653–7664. doi:10.3748/wjg.v20.i24.7653
40. Elkady A, Iijima S, Aboulfotuh S, et al. Characteristics of escape mutations from occult hepatitis B virus infected patients with hematological malignancies in South Egypt. World J Hepatol. 2017;9(9):477–486. doi:10.4254/wjh.v9.i9.477
41. Mansour E, Abdul-Rahim S, Batouty G, Zaghloul I, Abdel-Hadi S. Integration of hepatitis B immunization in the expanded program on immunization of the child survival project. J Egypt Public Health Assoc. 1993;68(5–6):487–494.
42. Colson P, Borentain P, Motte A, et al. Clinical and virological significance of the co-existence of HBsAg and anti-HBs antibodies in hepatitis B chronic carriers. Virology. 2007;367(1):30–40. doi:10.1016/j.virol.2007.05.012
43. Kwak M-S, Chung G-E, Yang JI, Yim JY. Long-term outcomes of HBsAg/anti-HBs double-positive versus HBsAg single-positive patients with chronic hepatitis B. Sci Rep. 2019;9(1):19417. doi:10.1038/s41598-019-56015-8
44. Di Lello FA, Ridruejo E, Martínez AP, Pérez PS, Campos RH, Flichman DM. Molecular epidemiology of hepatitis B virus mutants associated with vaccine escape, drug resistance and diagnosis failure. J Viral Hepat. 2019;26(5):552–560. doi:10.1111/jvh.13052
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

