Back to Journals » Infection and Drug Resistance » Volume 17
Characterization of an NDM-1-Producing Citrobacter koseri Isolate from China
Authors Zi P, Fang M, Yang H, Zheng J, Ma N, Liu Q
Received 16 August 2023
Accepted for publication 21 December 2023
Published 6 January 2024 Volume 2024:17 Pages 61—67
DOI https://doi.org/10.2147/IDR.S435771
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Zhi Ruan
Panpan Zi,1,* Ming Fang,2,* Hongfu Yang,1 Jiahao Zheng,3 Ning Ma,1 Qilong Liu1
1Department of Critical Care Medicine, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, People’s Republic of China; 2Institute for Infection Disease Control, Shandong Center for Disease Control and Prevention, Jinan, People’s Republic of China; 3Institute of Animal Quarantine, Chinese Academy of Inspection and Quarantine, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Panpan Zi, Department of Critical Care Medicine, the First Affiliated Hospital of Zhengzhou University, No. 1 Longhu Zhonghuan Road, Jinshui District, Zhengzhou, Henan, People’s Republic of China, Tel +86-0371-66271356, Email [email protected]
Purpose: The continuous rise in carbapenemase-producing Enterobacteriaceae infections is a major public health concern. However, there is limited information available on New Delhi metallo-β-lactamase-1 (NDM-1) producing Citrobacter koseri. In this study, we isolated a blaNDM-1-carrying C. koseri from a stool sample of an inpatient. Our aim was to investigate the phenotypic and genomic features of this clinically derived carbapenem-resistant C. koseri isolate and to characterize the transmission pattern of the IncFII/IncN plasmid that carries the blaNDM-1 gene.
Methods and Results: S1-PFGE, Southern blot and conjugation assay confirmed the presence of blaNDM-1 gene in a conjugative plasmid. C. koseri L2395 and transconjugant L2395-EC600 strains showed similar resistance spectrum. Whole-genome analysis revealed that pL2395_NDM is an IncFII/IncN plasmid with a length of 67,839 bp. Moreover, blaNDM-1 gene was found encoded in the ISKpn19-blaNDM-1-ble-tnpF-dsbD-cutA-ISKpn19 cassette array. Phylogenetic analysis revealed that strain L2395 was close to an IMP-4-bearing C. koseri from Australia.
Conclusion: Ongoing surveillance will be essential to control and prevent the spread of carbapenem-resistant Citrobacter spp. in the future.
Keywords: Citrobacter koseri, carbapenem-resistant, NDM-1, whole-genome sequencing
Introduction
Citrobacter spp. are facultative anaerobic Gram-negative bacteria within the Enterobacteriaceae family.1 They are frequently found in food, water, soil and intestines of humans and are considered as environmental contaminants.2 Among the 11 species of Citrobacter spp., the most dominant are Citrobacter freundii and Citrobacter koseri, and regarded as commensal in humans.3 Citrobacter spp. are now gaining importance as a clinical pathogen causing healthcare-associated infections such as urinary tract, liver, and biliary tract infections.4 Moreover, C. koseri could cause central nervous system (CNS) infections in immunocompromised individuals.5 C. koseri is intrinsically resistant to aminopenicillins and carboxypenicillins.6 Infections caused by C. koseri are often treated with aminoglycosides, cephalosporins, chloramphenicol, quinolones and carbapenems.6,7 However, the unceasing increase in infections with carbapenemase-producing Enterobacteriaceae is of significant public health concern.8 Carbapenem-resistant C. koseri strains have been associated with a higher rate of in-hospital mortality compared to susceptible strains.9 To date, several studies have reported the occurrence of infections involving C. koseri producing KPC, VIM and OXA.3,7 However, reports of C. koseri carrying blaNDM are rare compared to other carbapenemase genes, and there is a paucity of genomic analysis of NDM-producing C. koseri.10,11 Yao et al reported that the detection rate of carbapenem-Resistant Citrobacter spp. has gradually increased.2 Carbapenem-resistant Citrobacter spp. is fast gaining importance as a clinical multidrug-resistant pathogen.
In this study, we identified a clinical isolate of C. koseri L2395 carrying blaNDM, and described the complete sequence of a conjugative IncFII/IncN plasmid encoding NDM-1. S1-PFGE and southern blotting indicated the location of blaNDM-1 gene, and conjugation assay was performed to reveal the potential transmission mechanisms of blaNDM-1. Whole-genome sequencing (WGS) revealed the genetic characterization of the isolate. Our study aims to evaluate the phenotypic characteristics of NDM-harboring C. koseri and emphasizes the importance of further monitoring of carbapenem-resistant C. koseri in the clinic.
Methods and Materials
Sample Collection
In the routine screening of the intestinal colonization carbapenems-resistant bacteria in a teaching hospital in Zhejiang Province since 2016, fecal samples were collected from patients with diarrhea. A female patient of 56-year-old was admitted to hospital for postoperative recurrence of cholangiocarcinoma in July 2020. Patients with pulmonary infections were treated with levofloxacin and piperacillin-tazobactam on admission. The patient developed diarrhea 6 days after admission. A fecal sample was collected from an inpatient on July 27, 2020. The sample was cultured in Brain Heart Infusion Broth (BD, Sparks, USA) and spread on MacConkey agar plate (OXOID, Hampshire, United Kingdom) with 2 mg/L meropenem (Meilunbio, Dalian, China) for preliminary screening. The isolate L2395 was detected and identified as C. koseri using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker, Bremen, Germany). The carbapenemase genes were identified using PCR as previously reported.12
Antibiotic Susceptibility Testing
Antimicrobial Susceptibility Testing (AST) was determined using VITEK 2 system with AST-GN16 panel.13 Results were interpreted based on both the Clinical and Laboratory Standards Institute (CLSI) guidelines (https://clsi.org) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/clinical_breakpoints/).
S1-PFGE, Southern Blotting and Conjugation Assay
The plasmid characterization of C. koseri L2395 was determined by S1 Nuclease-Pulsed Field Gel Electrophoresis (S1-PFGE). Briefly, DNA plugs were digested using the S1 Nase restriction enzyme (Takara Bio Inc., Japan). Then, plugs undertaken on a CHEF-DR III (Bio-Rad, Hercules, CA, USA). Salmonella enterica serotype Braenderup H9812 was used as a size marker. The location of NDM was determined with Southern blotting and hybridization by a digoxigenin-labelled blaNDM-1 probe. Conjugation assays were conducted using Escherichia coli J53/EC600 as the recipient strains. Transconjugants were selected on plates contained 2 mg/L meropenem and 200 mg/L rifampicin/sodium azide (Meilunbio, Dalian, China) and verified with PCR and MALDI-TOF/MS identification.
Whole-Genome Sequencing (WGS) and in silico Analyses
Whole-genomic DNA of L2395 was extracted using Gentra Puregene Yeast/Bact. Kit (QIAGEN, Hilden, Germany) and subsequently sequenced on the Nanopore PromethION platform (Oxford Nanopore Technologies, Oxford, United Kingdom) and Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States) by Novo Gene Co., Ltd. (Beijing, China). Assembly was performed using Unicycler v0.4.2. The acquired antimicrobial resistance genes (ARGs) and replicon types of plasmids were identified through the ResFinder (http://genepi.food.dtu.dk/resfinder) and PlasmidFinder (https://cge.food.dtu.dk/services/PlasmidFinder/).14,15 Genomic sequences were annotated using Prokka v1.14.0, and the comparative maps of plasmids were drawn under the BLAST Ring Image Generator (BRIG) v0.95.16,17
Phylogenetic Analysis
A total of 220 publicly available C. koseri sequences were downloaded from the National Center for Biotechnology Information (NCBI) GenBank database. Genomes encoding carbapenemase genes were selected for phylogenetic analysis. The identification of ARGs was performed using ResFinder 4.0 database.14 Finally, 43 genomes were identified carrying carbapenemase genes and selected for subsequent analysis (Table S1). The alignment file was generated by Roary v3.11.2, a rapid large-scale prokaryote pan genome analysis tool.18 The multi-alignment of genomes was conducted with MAFFT.19 A total of 3,507 core genes were identified. The substitution model was General Time Reversible (GTR) model, and the bootstrap replications was 100. Finally, the maximum likelihood tree was constructed and visualized by MEGA 11 and iTOL v6.8, respectively.20,21
Results and Discussion
A female patient of 56-year-old was admitted to hospital for postoperative recurrence of cholangiocarcinoma in July 2020. Patients with pulmonary infection were treated with levofloxacin and piperacillin-tazobactam on admission. The patient developed diarrhea 6 days after admission. Fecal sample was collected and spread on the surface of selected plates. C. koseri L2395 was recovered from the medium. NDM-1 was detected using PCR.
As shown in Table 1, C. koseri L2395 exhibited resistance to multiple antibiotics, including amoxicillin/clavulanate (MIC ≥ 32 μg/mL), piperacillin/tazobactam (MIC ≥ 128 μg/mL), cefuroxime (MIC ≥ 64 μg/mL), cefuroxime axetil (MIC ≥ 64 μg/mL), cefoxitin (MIC ≥ 64 μg/mL), ceftazidime (MIC ≥ 64 μg/mL), ceftriaxone (MIC ≥ 64 μg/mL), cefoperazone/sulbactam (MIC ≥ 64 μg/mL), cefepime (MIC = 16 μg/mL), ertapenem (MIC ≥ 8 μg/mL), imipenem (MIC ≥ 16 μg/mL), levofloxacin (MIC = 4 μg/mL), with the exception of amikacin (MIC ≤ 2 μg/mL), tigecycline (MIC = 2 μg/mL) and trimethoprim/sulfamethoxazole (MIC = 40 μg/mL).
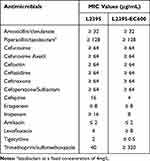 |
Table 1 Antimicrobial Susceptibilities of Strain C. koseri L2395 and Transconjugant L2395-EC600 |
According to the results of S1-PFGE and Southern-blot, the blaNDM-1 gene was located on a ~67 kb plasmid (designated as pL2395_NDM) (Figure 1). Whole-genome sequencing confirmed strain L2395 consist of a circular chromosome and two plasmids. And plasmid pL2395_NDM belonged to the IncFII/IncN type plasmid, with 67,839 bp in length and an average GC content of 51.7%. Conjugation assays revealed that the transmission of blaNDM-1 from C. koseri L2395 into E. coli 600 was successful. PCR and MALDI-TOF/MS identification confirmed pL2395_NDM was successfully transferred into recipient E. coli 600. In addition, the transconjugant L2395-EC600 showed similar resistance to antibiotics with L2395 (Table 1). This result indicated that the antimicrobial resistance phenotypes of transconjugant L2395-EC600 were acquired from pL2395_NDM. The transferability of plasmids increases the risk of transmission of drug-resistant bacteria and poses a great challenge to clinical treatment. In silico analysis identified strain L2395 also carry other antibiotic resistance genes (ARGs) including β-lactams (blaMAL-1), trimethoprim (dfrA14) and quinolone (qnrS1). Multiple ARGs confer isolate multidrug-resistance, which increases the difficulty of clinical treatment.
A search of the nr/nt database found plasmid pL2395_NDM shared high similarity with plasmids unnamed2 (ON111450.1), pECC33-57 (CP098488.1) and pHKN-2 (MW191859.1) which identified from Klebsiella pneumoniae, Enterobacter hormaechei and E. coli, respectively (Figure 2).22–24 Of note, plasmids unnamed2 and pECC33-57 were identified from clinic-originated isolates in China. The genetic environment of blaNDM-1 gene in L2395 was ISKpn19-blaNDM-1-ble-tnpF-dsbD-cutA-ISKpn19. This genetic structure was also found in plasmids unnamed2 and pHKN-2 but was different with pECC33-57. The latter contained an incomplete dsbD and lacked of cutA. Restriction modification systems encoded ecoRIIR gene were found upstream of the blaNDM-1. This restriction modification system could assist in the defense against phage infection and conduce to the spread and maintenance of plasmids.25 Such a system can also be found in blaIMP-4-carrying C. freundii.26
The genomes of 43 C. koseri isolates harboring the carbapenemase encoding genes were downloaded from the NCBI database and performed phylogenetic analysis together with C. koseri L2395. As shown in Figure 3 and Table S1, C. koseri isolates were detected across various countries, including Austria, China, France, Germany, Singapore, Spain, the United States, and the United Kingdom, suggesting the wide distribution. Strains isolated from different countries were not significantly clustered together, suggesting that carbapenemase-carrying C. koseri may be polyclonal at present. Multiple carbapenem resistance genes have been detected in C. koseri isolates, with blaKPC genes being most popular. As one of the most common carbapenemase genes, blaKPC has been found to be prevalent in the Enterobacteriales. However, reports of Citrobacter spp. carrying KPC are still limited. C. koseri L2395 is closely related to (GCA 023687185.1) from Australia carrying blaIMP-4, but is far from the other three isolates from China (GCA 023156325.3, GCA 902387665.1, and GCA 003184045.1). These three isolates carrying different KPC variants, two of which carried KPC-2 (GCA 902387665.1, and GCA 003184045.1) showed close genetic relationships, and another isolate carries KPC-34 (GCA 023156325.3).
In conclusion, we isolated a multidrug resistant C. koseri strain L2395 carrying blaNDM-1 gene from a clinic in China. The genetic environment of NDM was elucidated, and the transferability of IncFII/IncN plasmid was confirmed. The emergence of carbapenem-resistant C. koseri highlights the importance of continuously monitored carbapenem-resistant Enterobacteriaceae in clinic.
Ethics Approval and Consent to Participate
This study was conducted following the Declaration of Helsinki and obtained approval from the clinical research ethics committee of The First Affiliated Hospital of Zhengzhou University [no. 2023-KY-0264]. The patient provided written informed consent to allow the case details to be published.
Nucleotide Sequence Accession Numbers
The whole-genome sequence of the C. koseri L2395 was submitted to GenBank under the following BioSample number: SAMN33442942.
Acknowledgments
This work is supported by research grants from Henan Provincial Medical Science and Technology Research Project (LHGJ20190213).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Canario DG, Remé P, Cunha BA. Citrobacter koseri infection and abscess associated with Harrington rods. Am J Infect Control. 2004;32(6):372–374. doi:10.1016/j.ajic.2003.07.009
2. Yao Y, Falgenhauer L, Falgenhauer J, et al. Carbapenem-Resistant Citrobacter spp. as an Emerging Concern in the Hospital-Setting: results From a Genome-Based Regional Surveillance Study. Front Cell Infect Microbiol. 2021;11:744431. doi:10.3389/fcimb.2021.744431
3. Mavroidi A, Neonakis I, Liakopoulos A, et al. Detection of Citrobacter koseri carrying beta-lactamase KPC-2 in a hospitalised patient, Greece, July 2011. Euro Surveill. 2011;16(41):19990.
4. Arana DM, Ortega A, González-Barberá E, et al. Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J Antimicrob Chemother. 2017;72(12):3283–3287. doi:10.1093/jac/dkx325
5. Lechowicz M, Dąbek K, Majewska U, Bekesińska-Figatowska M, Borszewska-Kornacka MK, Bokiniec R. Multiple Brain Abscesses Caused by Citrobacter Koseri in a Preterm Neonate - Case Report. Pol J Radiol. 2017;82:837–841. doi:10.12659/PJR.903276
6. Deveci A, Coban AY. Optimum management of Citrobacter koseri infection. Expert Rev Anti Infect Ther. 2014;12(9):1137–1142. doi:10.1586/14787210.2014.944505
7. Ekwanzala MD, Dewar JB, Kamika I, Momba MNB. Genome sequence of carbapenem-resistant Citrobacter koseri carrying blaOXA-181 isolated from sewage sludge. J Glob Antimicrob Resist. 2020;20:94–97. doi:10.1016/j.jgar.2019.07.011
8. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62(2):e01882–17. doi:10.1128/AAC.01882-17
9. Mohamed NA, Said HM, Hussin H, Abdul Rahman N, Hashim R. Carbapenem-Resistant Enterobacteriaceae: clinicoEpidemiological Perspective. Trop Biomed. 2018;35(2):300–307.
10. Arana DM, Ortega A, et al. Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J Antimicrob Chemother. 2017;72(12). doi:10.1093/jac/dkx325
11. Kocsis E, Gužvinec M, Butić I, et al. blaNDM-1 Carriage on IncR Plasmid in Enterobacteriaceae Strains. Microb Drug Resist. 2016;22(2):123–128. doi:10.1089/mdr.2015.0083
12. Zheng B, Zhang J, Ji J, et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother. 2015;59(11):7086–7089. doi:10.1128/AAC.01363-15
13. Lellouche J, Schwartz D, Elmalech N, et al. Combining VITEK® 2 with colistin agar dilution screening assist timely reporting of colistin susceptibility. Clin Microbiol Infect. 2019;25(6):711–716. doi:10.1016/j.cmi.2018.09.014
14. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi:10.1093/jac/dkaa345
15. Carattoli A, Hasman H. PlasmidFinder and In Silico pMLST: identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol Biol. 2020;2075:285–294. doi:10.1007/978-1-4939-9877-7_20
16. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
17. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi:10.1186/1471-2164-12-402
18. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi:10.1093/bioinformatics/btv421
19. K K, Dm S. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evolution. 2013;30(4). doi:10.1093/molbev/mst010
20. Foster G, Evans J, Tryland M, et al. Use of citrate adonitol agar as a selective medium for the isolation of Escherichia fergusonii from a captive reindeer herd. Vet Microbiol. 2010;144(3–4):484–486. doi:10.1016/j.vetmic.2010.01.014
21. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi:10.1093/nar/gkab301
22. Huang Y, Li J, Wang Q, Tang K, Cai X, Li C. Detection of carbapenem-resistant hypervirulent Klebsiella pneumoniae ST11-K64 co-producing NDM-1 and KPC-2 in a tertiary hospital in Wuhan. J Hosp Infect. 2023;131:70–80. doi:10.1016/j.jhin.2022.09.014
23. Dong X, Zhu M, Li Y, et al. Whole-Genome Sequencing-Based Species Classification, Multilocus Sequence Typing, and Antimicrobial Resistance Mechanism Analysis of the Enterobacter cloacae Complex in Southern China. Microbiol Spectr. 2022;10(6):e0216022. doi:10.1128/spectrum.02160-22
24. Zhu L, Yuan L, Shuai XY, et al. Deciphering basic and key traits of antibiotic resistome in influent and effluent of hospital wastewater treatment systems. Water Res. 2023;231:119614. doi:10.1016/j.watres.2023.119614
25. Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29(18):3742–3756. doi:10.1093/nar/29.18.3742
26. Chi X, Guo J, Zhou Y, et al. Complete-Genome Sequencing and Comparative Genomic Characterization of an IMP-4 Producing Citrobacter freundii Isolate from Patient with Diarrhea. Infect Drug Resist. 2020;13:1057–1065. doi:10.2147/IDR.S244683
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



