Back to Journals » Infection and Drug Resistance » Volume 16
Characterization and Molecular Mechanism of Aminoglycoside-6-Adenyl Transferase Associated with Aminoglycoside Resistance from Elizabethkingia meningoseptica
Authors Zhang S, Zhang Y, Liu R, Yuan S, Chen Y, Li W, Lu X, Tong Y, Hou L, Chen L, Sun G
Received 21 June 2023
Accepted for publication 9 August 2023
Published 22 August 2023 Volume 2023:16 Pages 5523—5534
DOI https://doi.org/10.2147/IDR.S423418
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shaoxing Zhang,1 Yuxin Zhang,1 Ruijie Liu,1 Shuying Yuan,2 Yanwen Chen,1 Wenjie Li,1 Xinrong Lu,3 Yongliang Tong,3 Linlin Hou,4 Li Chen,3 Guiqin Sun1
1School of Medical Technology and Information Engineering, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, People’s Republic of China; 2Clinical Laboratory Department, Jiaxing Maternity and Child Health Care Hospital, Jiaxing, Zhejiang, People’s Republic of China; 3Key Laboratory of Medical Molecular Virology (MOE/NHC/CAMS), School of Basic Medical Sciences, Fudan University, Shanghai, People’s Republic of China; 4College of Veterinary Medicine, Qingdao Agricultural University, Qingdao, Shandong, People’s Republic of China
Correspondence: Guiqin Sun, School of Medical Technology and Information Engineering, Zhejiang Chinese Medical University, 548 Binwen Road, Binjiang District, Hangzhou, Zhejiang, People’s Republic of China, Tel +86 13868118601, Fax +86 571-86633307, Email [email protected] Li Chen, Key Laboratory of Medical Molecular Virology (MOE/NHC/CAMS), School of Basic Medical Sciences, Fudan University, 131 Dongan Road, Xuhui District, Shanghai, People’s Republic of China, Tel +86 15821138980, Fax +86 21-54237381, Email [email protected]
Purpose: Elizabethkingia meningoseptica (EM) is a multi-drug-resistant bacterium of global concern for its role in nosocomial infection and is generally resistant to aminoglycoside antibiotics. In the whole genome of an EM strain (FMS-007), an aminoglycoside-6-adenyl transferase gene (ant(6)FMS-007) was predicted. This study aimed to characterize the biochemical function of ANT(6)FMS-007 and analyze the relationship between genotype and phenotype of ant(6) in clinical EM isolates, so as to provide evidence for clinical precision drug use. This study could establish a method for the verification of known or unknown functionally resistant genes.
Methods: A total of 42 EM clinical isolates were collected from clinical departments during 2015– 2023. The phenotype of aminoglycoside antibiotics was analyzed by broth microdilution (BMD) and Kirby-Bauer (K-B) methods. The whole-length ant(6) from EM clinical isolates was analyzed by polymerase chain reaction (PCR) and sequencing. The biochemical function of predictive ANT(6)FMS-007 from the FMS-007 whole genome was identified by 3D plate experiment and mass spectrometry analysis. Candidate active sites were predicted by multi-species sequence alignment and molecular docking, and other important sites were identified in the comparison of ant(6) genotypes and phenotypes of EM clinical isolates. Drug susceptibility test was used to verify the function of these sites.
Results: The predictive ANT(6)FMS-007 protein could inactivate STR by modifying STR with ATP to form STR-AMP. Four active sites (Asp-38, Asp-42, Lys-95, and Lys-213) of ANT(6)FMS-007 were identified. Thirty-one EM clinical isolates (74%) carried the ant(6) gene. Eight EM clinical isolates containing the ant(6) gene had MIC values (<=32μg/mL) lower by at least 16-fold than FMS-007 (512μg/mL) for STR, and N59H and K204Q were the common mutations in the ant(6) gene.
Conclusion: This assay verified the biochemical function of the predictive gene ant(6)FMS-007 and could provide an alternative method to study resistant gene function in multi-drug-resistant bacteria. The inconsistency between genotype and phenotype of resistant genes indicated that the combination of resistance gene detection and functional analysis could better provide precision medicine for clinical use.
Keywords: drug-resistance, aminoglycoside-6-adenyl transferase, characterization, active sites, Elizabethkingia meningoseptica
Introduction
In 1959, the first report of human infection due to EM was that 19 cases of meningitis cases in infants in the United States of America.1 Gram-negative bacteria of the genus Elizabethkingia emerged as an important conditional pathogen in hospital-acquired infections and were generally associated with multidrug resistance and high mortality.2,3 Clinical isolates of EM usually conferred resistance to multiple antibiotics, with a high rate of resistance to aminoglycosides.4,5
Aminoglycoside antibiotics (AGs) were broad-spectrum antibiotics extensively used to treat infections caused by aerobic bacterial pathogens.6 AGs could cause mistranslated proteins that disrupted the integrity of the bacterial cell membrane by targeting the regions of 16S rRNA on the 30S ribosomal subunit for aminoacyl-tRNA binding, resulting in cell death.7,8 The most prevalent resistance mechanism to AGs in the clinic was due to enzymatic modification that renders aminoglycosides of decreased affinity for their natural primary target, 16S rRNA.9 Aminoglycoside modifying enzymes (AMEs) included aminoglycoside phosphotransferases (APHs), aminoglycoside acetyltransferases (AACs), and aminoglycoside nucleoside transferases (ANTs).10 The ANTs family catalyzed the modification of hydroxyl groups, amino groups of 2-deoxystreptamine, or sugar moieties, which acted by adding AMP from an adenosine triphosphate (ATP) donor to hydroxyl groups at the 2″, 3″, 4′, 6 and 9 positions of aminoglycosides, including ANT(2″), ANT(3″), ANT(4′), ANT(6) and ANT(9).11
The genomes of EM from the Comprehensive Antibiotic Resistance Database and Virulence Factors of Pathogenic Bacteria Database were used to identify antibiotic-resistant genes associated with aminoglycosides, such as nucleoside transferase (ANT) and aminoglycoside N-acetyltransferase (AAC).12,13 In 2012, our team reported a multi-drug-resistant bacterial EM strain (FMS-007) from a patient with T-cell non-Hodgkin′s lymphoma (T-NHL), and the whole-genome sequence was obtained in one contig.14 In this study, a putative aminoglycoside-6-adenyl transferase gene (ant(6)EM) from FMS-007, encoding a 288 amino-acid protein, was identified by bioinformatics analysis. ANT(6) in Bacillus subtilis showed high specificity for streptomycin, and its structure consisted of two domains (294aa, 35.77 kDa, PDB entry 2pbe; New York SGX Research Center for Structural Genomics, unpublished work).15,16 The ant(6) gene (878 bp) also existed in streptomycin-resistant strains of Campylobacter jejuni.17 However, the active sites and molecular mechanism of ANT(6) were still unclear.
The ant(6)FMS-007 (864 bp) was cloned into pET15b and expressed in Escherichia coli BL21 (DE3). The results showed that ant(6)FMS-007 specifically mediated streptomycin resistance. Ten sites significantly affecting ANT(6)FMS-007 activity were also identified. This assay, especially the cloning/expression/transforming assay, could provide an alternative method for elucidating the drug-resistance mechanism of bacteria and aiding the development of new drugs.
Materials and Methods
Bacterial Strains
This study used 42 EM clinical isolates from three different hospitals in three Chinese cities between January 2015 and December 2023 and sorted the strains according to hospital source. FMS-007 was used as the EM standard strain. Sampling and isolation of bacterial strains were the routine hospital laboratory procedures, and identification was performed in the microbiology laboratory using the VITEK2 compact (BioMérieux, France) and the VITEK MS (BioMérieux, France) systems. All strains were stored at −80°C in 30% glycerol until use.
Antibiotic Susceptibility Test
The drug-resistant phenotype was screened by minimum inhibitory concentration (MIC) measurements using the broth microdilution (BMD) method at OD600 nm and the Kirby-Bauer (K-B) method according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI, 2021). A standard protocol was performed in a 96-well microplate (Bio-Kont, China). The following antimicrobial agents were used: streptomycin (STR), amikacin (AMK), gentamicin (GEN), kanamycin (KAN), and tobramycin (TOB) (Sangon Biotech, China). The discs used were STR (10 μg/disc), AMK (30 μg/disc), GEN (10 μg/disc), KAN (30 μg/disc), TOB (10 μg/disc) (Bio-Kont, China), and STR (300 μg/disc) (Binhe, China).
EM is a gram-negative non-Enterobacterales pathogen, but CLSI breakpoints have not been established for Elizabethkingia spp. The antimicrobial susceptibility results of EM were interpreted based on the breakpoints of the other non-Enterobacterales listed in the M100-S31 guidelines (CLSI, 2021), but those results without the breakpoints referred to Enterobacterales. The antimicrobial susceptibility results of all transformants (Escherichia coli) were interpreted according to the Enterobacterales in CLSI document M100-Ed31.
Cloning of ant(6)FMS-007 Gene
The genomic DNA of FMS-007 was obtained by the TIANamp Bacteria DNA Kit (Tiangen, China) and the primers used to amplify ant(6)FMS-007 were as follows: ANT(6)-F: 5′-GCCATATGAAAATTCGGGATGAAAAGCTC-3′, and ANT(6)-R: 5′-CCGGATCCCTAATTACAGGATTTATATACTGTTTTCA-3′. The primers contained BamHI and NdeI restriction sites, which allowed site-directed cloning into pET15b. The ant(6)FMS-007 products from FMS-007 and pET15b were treated with BamHI and NdeI (Takara, Japan) and ligated using T4 DNA ligase (Takara, Japan). The PCR products of clinical isolates were subjected to agarose gel electrophoresis and sequenced.
The recombinant plasmid ant(6)FMS-007/pET15b was transformed into E. coli TOP10 cells (Tiangen, China). Transformant was obtained by culturing cells on Luria-Bertani (LB) agar plates containing 100 μg/mL ampicillin (Sangon Biotech, China) for 18 h at 37 °C in a biochemical incubator (SPX-250B-Z, Boxun, China). The recombinant plasmid ant(6)FMS-007/pET15b was confirmed by sequencing (Sangon Biotech, China), extracted using a TIANprep Mini Plasmid Kit (Tiangen, China), and transformed into BL21 (DE3) competent E. coli cells (Tiangen, China) for expression.
Protein Expression and Purification
ANT(6)FMS-007 was expressed and purified. The ant(6)FMS-007/pET15b transformant was grown in 0.5 L LB broth containing 100 μg/mL ampicillin for 12 h at 37 °C. Expression was induced using 1 mM isopropyl β-d-thiogalactoside (IPTG) (Sigma-Aldrich, USA) for 12 h at 28 °C.
Cells were harvested by centrifugation at 3500 rpm for 10 min using a high-speed refrigerated centrifuge (Sorvall™ ST8R, ThermoFisher, USA). The cells were solubilized in 12 mL of lysis buffer (20 mM Tris, 300 mM NaCl, and 10 mM imidazole; pH 7.4) and lysed using an ultrasonic homogenizer (Scientz-IID; Xinzhi, Ningbo, China). Cell debris was removed by centrifugation at 12,000 rpm for 30 min at 4 °C. ANT(6)FMS-007 was purified by Ni-NTA affinity chromatography (GE Healthcare, USA). The supernatant was loaded onto a HisTrap column and incubated for 30 min at 4 °C. The column was washed with four column volumes of lysis and wash buffers (20 mM Tris, 300 mM NaCl, and 25 mM imidazole; pH 7.4). ANT(6)FMS-007 was eluted with two column volumes of elution buffer (20 mM Tris, 300 mM NaCl, and 250 mM imidazole; pH 7.4).
For the biochemical experiments, the elution buffer was replaced with 10 mM PBS by ultrafiltration through a filter with a molecular weight cut-off (MWCO) of 30 kDa (Millipore, USA) at 12,000 rpm for 5 minutes at 4 °C. The ultrafiltrate was washed six times with PBS at 12,000 rpm for 5 min at 4 °C. The concentration of ANT(6)FMS-007 was determined using a NanoDrop™ One spectrophotometer (ND-ONE-W; ThermoFisher, USA). The purity of ANT(6)FMS-007 was verified by SDS-PAGE analysis.
Three-Dimensional Experiment in a Microenvironment
The biological function of ANT(6)FMS-007 in vitro was evaluated using a modified three-dimensional (3D) experiment. According to the K-B method in the Clinical and Laboratory Standards Institute (CLSI, 2021), Mueller–Hinton (M-H) medium was inoculated with 0.5 McFarland’s E. coli BL21 (DE3) cells and an STR disc (300 μg/disc) was placed in the middle of the agar plate. Small slits were made 5 mm (within the zone of inhibition) from the disc by piercing the agar surface with a sterile pipette. The plate was kept upright for 5–10 min until the solution dried, and then 20 μL of different agents (left-side hole, 20 μL 10 mM PBS; The hole in the top, 5 μL 100 mM ATP+15 μL 10 mM PBS; right-side hole, 15 μL 2 μg/μL ANT(6)FMS-007+5 μL 10 mM PBS; The hole at the bottom: 15 μL 2 μg/μL ANT(6)FMS-007+5 μL 100 mM ATP) was added to four slits, respectively. The plates were then incubated for 24 hours at 37 °C. If the STR in the slits was inactivated, the E. coli BL21 (DE3) near the slits could grow.
Biochemical Function by Mass Spectrometry
Each reaction included 5 mM streptomycin in 50 mM NH4HCO3 reaction buffer mixed with 20 μg of ANT(6)FMS-007, 5 mM ATP, and 5 mM Mg2+ in a total volume of 100 μL for 12 h at 37 °C. The products were collected by ultrafiltration using a filter with an MWCO of 3 kDa (Millipore) at 12,000 rpm for 30 min at 4 °C. Ultrafiltrate was vacuum lyophilized (Labconco FreeZone 4.5 Liter, USA) overnight, and samples were resuspended in 15 µL of ultrapure water for MS analysis. The samples were profiled in positive ion reflector mode using an AXIMA MALDI-quadrupole ion trap TOF mass spectrometer (Shimadzu Corp., Japan) with a nitrogen pulsed laser (337 nm) and an acceleration voltage of 20 kV.
Structure Prediction and Molecular Docking
The 3D model of ANT(6)FMS-007 was predicted by online AlphaFold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb#scrollTo=kOblAo-xetgx) based on the ANT(6)FMS-007 amino acid sequence.
The molecular structures of ATP and STR were obtained from the PDB database (https://www.rcsb.org) as ligands for molecular docking. The ligands were optimized using Avogadro (http://avogadro.cc/), and molecular docking was performed using AutoDock Vina v.1.2.0.18 Data analysis and intermolecular force mapping were conducted using LigPlot+ v.2.2 (EMBL, Hinxton, UK). PyMOL 2.5 (Schrödinger, USA) was used for molecular structure mapping.
Candidate Sites Screening and Site-Directed Mutagenesis
Amino acid sequences of ANT(6) from multiple species were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/nuccore), and the secondary structure and sequence alignment of ANT(6) was analyzed by ESPript v.3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The candidate sites of ANT(6)FMS-007 were selected from conserved sites.
Three amino acid mutants with different charges were designed for each candidate site (Table S1). Primers of mutants were designed using QuickChange Primer Design (https://www.agilent.com.cn/store/primerDesignProgram.jsp?toggle=uploadNow&mutate=true&_requestid=204901) (Table S2). The recombinant plasmid ant(6)FMS-007/pET15b was used as the template for PCR amplification for the mutants. The PCR products were purified using an AxyPrepTM PCR Cleanup Kit (Corning, USA). The template was cleaved from the purified PCR products using the enzyme DnpI (New England Biolabs, USA). The mutant plasmids were confirmed by sequencing. Mutant plasmids were transformed into E. coli BL21 (DE3) cells for the detection of drug resistance.
Genotype Detection of Clinical Isolates
Genomic DNA was isolated from 43 EM isolates using the TIANamp Bacteria DNA Kit (Tiangen, China) and the primers used to amplify ant(6) were as follows: ANT(6)-F: 5′-GCCATATGAAAATTCGGGATGAAAAGCTC-3′ and ANT(6)-R: 5′-CCGGATCCCTAATTACAGGATTTATATACTGTTTTCA-3′. The PCR products of clinical isolates were subjected to agarose gel electrophoresis and sequenced.
Results
Drug Susceptibility of 42 EM Clinical Isolates to Aminoglycoside Antibiotics
Forty-two clinical isolates of EM were tested by BMD and K-B method for five aminoglycosides. The results of K-B method showed that all EM clinical isolates were resistant to TOB and KAN. The sensitivity rate to STR was 5%, and the sensitivity to AMK and GEN was less than 33%. The results of BMD method showed that all clinical strains were resistant to TOB and GEN, and the resistance rates to KAN and AMK were 97.6% and 66.7%, respectively (Table 1). Forty-two EM clinical isolates were highly resistant to aminoglycoside antibiotics, suggesting that EM bacteria may harbor aminoglycoside resistance genes. All predicted aminoglycoside resistance genes from FMS-007 genome sequence were screened, and one gene (ant(6)FMS-007) mediating STR resistance was found (Table S3).
 |
Table 1 Drug Susceptibility of 42 EM Clinical Isolates to Aminoglysamines |
ant(6)FMS-007 Antimicrobial Activity Testing
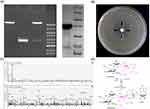
The ant(6)FMS-007 gene was cloned into pET15b (Figure 1A) and antimicrobial susceptibility was detected. FMS-007 was the positive control, and the pET15b transformant was the negative control. The ant(6)FMS-007/pET15b transformant exhibited resistance to STR and was sensitive to other aminoglycosides (AMK, TOB, GEN and KAN) (Table 2). Its MIC value (512 μg/mL) for STR reached the same MIC as FMS-007, a 128-fold increase compared with the pET15b transformant.
 |
Table 2 MIC and Inhibition Zone Diameters of ant(6)FMS-007 Transformants in E. coli BL21 |
Biochemical Function of ANT(6)FMS-007
Screening of antimicrobial susceptibility revealed that the ant(6)FMS-007/pET15b transformant showed a specific phenotype of STR resistance. ANT(6)FMS-007 was expressed and purified (Figure 1A) for 3D testing. The inhibition zone was sagittal around the bottom hole containing ANT(6)FMS-007 and ATP, while the inhibition zones of the other three holes were smooth arc (Figure 1B). The results indicated that ANT(6)FMS-007 could modify streptomycin to inactivate its antimicrobial activity, and ATP was also required.
To further investigate how ANT(6)FMS-007 modified STR, the products of the enzymatic reaction were analyzed by MS. Its molecular weight increased from 582.27 m/z (STR) or 600.28 m/z (STR-H2O) to 911.58 m/z (STR-AMP-H), 933.45 m/z (STR-AMP-Na), and 956.37 m/z (STR-AMP-Na2) (Figure 1C). The schematic diagram was shown by ChemDraw v.19.0 (PerkinElmer, Shanghai, China) (Figure 1D). This suggested that STR was combined with AMP by ANT(6)FMS-007 to form an inactivated STR-AMP.
Molecular Docking and Candidate Active Site Screening of ANT(6)EM
According to the ANT(6)FMS-007 multi-species amino acid sequence alignment (Figure 2A), eight conserved sites (Asp-38, Asp-42, Lys-95, Lys-148, Trp-162, Glu-191, Trp-199, and Lys-213) were selected as candidate sites, four of which were also action sites for molecular docking (Figure 2B, 2C). The function of these sites was identified by designing site-directed mutants (Table S2). STR susceptibility testing showed that the MIC values of mutants of D38H, D42E, K95A, K148D, W162R, E191R, W199R and K213D were at least 4-fold lower than those of the ant(6)FMS-007/pET15b transformant (Figure 2D).
ant(6) Genotype and Phenotype of Clinical EM Isolates Were Inconsistent
In order to study the relationship between genotype and phenotype of ant(6), 42 clinical EM isolates were performed. The result showed that 74% of EM clinical isolates carried the ant(6) gene (31/42), and 8 of them had MIC values (<=32μg/mL) significantly lower than FMS-007 (512μg/mL) for STR (Table 3). The ant(6) from the 8 strains were sequenced, and each contained three amino acid sites changed at least compared to ant(6)FMS-007, two of which were shared, His-59 (H59) and Gln-204 (Q204) (Table 4). Site-directed mutations at these two sites (N59H and K204Q) were performed on ant(6)FMS-007, respectively, and the STR resistance phenotype (64μg/mL) showed an 8-fold reduction compared with the wildtype (512μg/mL) (Figure 3).
 |
Table 3 ant(6)FMS-007 Genotype and Streptomycin Resistance Phenotype Relationship of EM Clinical Isolates |
 |
Table 4 Screening of EM ant(6) Changed Sites with Non-Obvious Drug Resistance Phenotypes |
Discussion
Part of the antimicrobial susceptibility testing conducted in this paper was not listed in CLSI, for example, the BMD method for streptomycin. EM is a gram-negative non-Enterobacterales pathogen for rare infections, but there is currently no reference standard for streptomycin in non-Enterobacterales. Those antimicrobial susceptibility results without breakpoints of EM in this study were referred to Enterobacterales listed in the M100-S31 guidelines (CLSI, 2021). The results generated from the antimicrobial susceptibility testing presented in this paper can only be regarded as a weak reference for clinical practice. More studies are required to generate data that matches the requirements for CLSI and clinical practice.
Antimicrobial resistance was highly noticeable among EM, and clinical isolates resistant to multiple or almost all available antibiotics have been consistently emerging.19,20 In this study, a streptomycin resistance gene, ant(6)FMS-007, was predicted and verified from a multi-drug-resistant EM clinical isolate, FMS-007. The low streptomycin MIC values of 8 EM strains carrying the ant(6) gene were due to point mutations, indicating that the detected drug-resistant gene alone did not represent a bacterial drug-resistant phenotype. Yang et al reported that the genotype and phenotype of aadS (Kanamycin kinase) from 20 EM clinical isolates displayed good consistency in aminoglycosides.21 However, another study about Mycobacterium tuberculosis indicated that mutations are an important factor in the inconsistency between the results of phenotype and genotype drug susceptibility tests for ofloxacin.22 Therefore, in clinical bacterial drug resistance detection, in addition to PCR detection of drug resistance genes, it was also necessary to observe the base changes of drug resistance gene sequencing, which would improve our understanding of the mechanism of drug resistance and provide a basis for clinically precise treatment.
The BMD method and the K-B method remained the classical methods to evaluate the resistance phenotype of bacteria.23,24 There were other ways to observe the function of drug resistance. Nuclear magnetic resonance spectroscopy (NMR) could be applied in analyzing the different metabolites of two P. aeruginosa strains (antibiotic susceptible vs resistant), and the result showed that the level of uridine monophosphate (UMP) in the resistance strain was lower, which could play an important role in sustaining virulence, biofilm formation, and antibiotic resistance.25 Corzana et al reported the molecular recognition and conformational selection phenomena of Bacillus subtilis ANT(6) to STR by NMR and MS.26 In this study, the biochemical function of ANT(6)FMS-007 was studied at the molecular level by MS. ANT(6)FMS-007 was shown to modify STR using ATP to form STR-AMP (inactive). In brief, MS would provide a more intuitive and reliable method for showing the modification of antibiotics caused by resistance enzymes.
There was a correlation between drug-resistant enzymatic activity and the active sites, and the enzyme activity could be promoted or lost when the active sites changed or fluctuated.27,28 The active sites of the ANT family were different.9,29 Stern et al reported that ANT(3″)(9) from Salmonella enterica catalyzed the transfer of an adenylyl group from ATP to position 3″ of streptomycin and position 9 of spectinomycin and its active sites were Trp-173, Asp-178, and Glu-87.30 Glu-145 and Lys-149 participated in the ANT(4′) modification of kanamycin.9 The active sites of ANT(6) have not been reported. In this study, the possible active sites of ANT(6)FMS-007 (Asp-38, Asp-42, Lys-95, and Lys-213) were effectively predicted by multi-species sequence alignment and molecular docking methods. The method established in this study could effectively predict the active sites of drug-resistant enzymes.
The rapid and ongoing spread of antibiotic resistance poses a serious threat to global public health.31 Bacterial drug resistance was prominent in clinical infections, and the detection methods of the drug resistance phenotype included the BMD method and the K-B method.32 With the development of technology, sequence-independent methods of resistance determination were also being developed, including matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS), Raman spectroscopy, fluorescence in situ hybridization (FISH) and microfluidics-based techniques, which could reduce antimicrobial susceptibility testing (AST) to 4 h in one study.33,34 Bacterial antimicrobial resistance is usually genetically encoded, so gene testing and whole-genome sequencing could facilitate rapid antimicrobial resistance gene identification and characterization.35 With the continuous increase in microbial genome and drug-resistant gene database data, new drug-resistant genes appeared.36,37
Although the novel genes could be characterized by whole-genome sequencing, functional verification was still needed. Polymerase chain reaction (PCR) detection of drug-resistant genes was fast, but there could be inconsistencies between genotype and phenotype. This study provides an alternative method for the identification and detection of drug-resistant genes with unknown functions.
Conclusion
The present study concluded that ANT(6)FMS-007 could specifically inactivate STR by modifying STR with ATP to form STR-AMP. Thirty-one EM clinical isolates carried the ant(6) gene. The genotype and phenotype of eight clinical isolates carrying the ant(6) gene were inconsistent, which could be due to amino acid mutations (N59H and K204Q). Ten sites affecting the activity of ANT(6)FMS-007 were identified, which may provide targets for the identification of gene function and the development of new drugs. This whole cloning/expressing/transforming protocol could establish an alternative method for the verification of known or unknown functional resistant genes.
Data Sharing Statement
All the reported data are available within the article.
Ethics Statement
This study was approved by the Medical Ethics Committee of Zhejiang Chinese Medical University (ethical number 20220310-6). Written informed consent was obtained from all participants. This study was performed in line with the principles of the Declaration of Helsinki. All the methods were performed in accordance with the relevant institutional ethical committee guidelines.
Acknowledgments
This work was supported by Key Laboratory of Medical Molecular Virology (MOE & NHC),School of Basic Medical Sciences, Shanghai Medical College, Fudan University under Grant [FDMV-2021003].
Disclosure
The authors declare that the research was conducted in the absence of any commercial funding, and no conflict of interest exists.
References
1. King EO. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am J Clin Pathol. 1959;31(3):241–247. doi:10.1093/ajcp/31.3.241
2. Zajmi A, Teo J, Yeo CC. Epidemiology and characteristics of Elizabethkingia spp. Infections in Southeast Asia. Microorganisms. 2022;10(5):882. doi:10.3390/microorganisms10050882
3. Lau SK, Chow WN, Foo CH, et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep. 2016;6(1):26045. doi:10.1038/srep26045
4. Lee YL, Liu KM, Chang HL, et al. The evolutionary trend and genomic features of an emerging lineage of elizabethkingia anophelis strains in Taiwan. Microbiol Spectr. 2022;10(1):e0168221. doi:10.1128/spectrum.01682-21
5. Wang M, Gao H, Lin N, et al. The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate. Microbiologyopen. 2019;8(11):e804. doi:10.1002/mbo3.804
6. Jospe-Kaufman M, Siomin L, Fridman M. The relationship between the structure and toxicity of aminoglycoside antibiotics. Bioorg Med Chem Lett. 2020;30(13):127218. doi:10.1016/j.bmcl.2020.127218
7. Jaimee G, Halami PM. Emerging resistance to aminoglycosides in lactic acid bacteria of food origin-an impending menace. Appl Microbiol Biotechnol. 2016;100(3):1137–1151. doi:10.1007/s00253-015-7184-y
8. Ramirez MS, Tolmasky ME. Amikacin: uses, resistance, and prospects for inhibition. Molecules. 2017;22(12):2267. doi:10.3390/molecules22122267
9. Zarate SG, De la Cruz Claure ML, Benito-Arenas R, Revuelta J, Santana AG, Bastida A. Overcoming aminoglycoside enzymatic resistance: design of novel antibiotics and inhibitors. Molecules. 2018;23(2):284. doi:10.3390/molecules23020284
10. Wachino JI, Doi Y, Arakawa Y. Aminoglycoside resistance: updates with a focus on acquired 16s ribosomal RNA methyltransferases. Infect Dis Clin North Am. 2020;34(4):887–902. doi:10.1016/j.idc.2020.06.002
11. Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407(6802):340–348. doi:10.1038/35030019
12. Breurec S, Criscuolo A, Diancourt L, et al. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci Rep. 2016;6(1):30379. doi:10.1038/srep30379
13. Teo J, Tan SY, Liu Y, et al. Comparative genomic analysis of malaria mosquito vector-associated novel pathogen Elizabethkingia anophelis. Genome Biol Evol. 2014;6(5):1158–1165. doi:10.1093/gbe/evu094
14. Sun G, Wang L, Bao C, Li T, Ma L, Chen L. Complete genome sequence of Elizabethkingia meningoseptica, isolated from a T-cell non-Hodgkin’s lymphoma patient. Genome Announc. 2015;3(3):10–128 doi:10.1128/genomeA.00673-15 doi:.1
15. Latorre M, Penalver P, Revuelta J, Asensio JL, Garcia-Junceda E, Bastida A. Rescue of the streptomycin antibiotic activity by using streptidine as a “decoy acceptor” for the aminoglycoside-inactivating enzyme adenyl transferase. Chem Commun. 2007;27(27):2829–2831. doi:10.1039/b704785a
16. Chen Y, Nasvall J, Wu S, Andersson DI, Selmer M. Structure of AadA from Salmonella enterica: a monomeric aminoglycoside (3”)(9) adenyltransferase. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 11):2267–2277. doi:10.1107/S1399004715016429
17. Hormeno L, Ugarte-Ruiz M, Palomo G, et al. ant(6)-I genes encoding aminoglycoside O-nucleotidyltransferases are widely spread Among streptomycin resistant strains of campylobacter jejuni and Campylobacter coli. Front Microbiol. 2018;9:2515. doi:10.3389/fmicb.2018.02515
18. Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model. 2021;61(8):3891–3898. doi:10.1021/acs.jcim.1c00203
19. Larkin PMK, Mortimer L, Malenfant JH, et al. Investigation of phylogeny and drug resistance mechanisms of Elizabethkingia anophelis isolated from blood and lower respiratory tract. Microb Drug Resist. 2021;27(9):1259–1264. doi:10.1089/mdr.2020.0263
20. Exner M, Bhattacharya S, Christiansen B, et al. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control. 2017;12:Doc05. doi:10.3205/dgkh000290
21. Yang C, Liu Z, Yu S, Ye K, Li X, Shen D. Comparison of three species of Elizabethkingia genus by whole-genome sequence analysis. FEMS Microbiol Lett. 2021;368(5):fnab018. doi:10.1093/femsle/fnab018
22. Chernyaeva E, Fedorova E, Zhemkova G, Korneev Y, Kozlov A. Characterization of multiple and extensively drug resistant Mycobacterium tuberculosis isolates with different ofloxacin-resistance levels. Tuberculosis. 2013;93(3):291–295. doi:10.1016/j.tube.2013.02.005
23. Orszulik ST. Quality and suitability of antimicrobial discs: theoretical and practical sources of error and variability. Expert Rev Mol Diagn. 2020;20(3):277–283. doi:10.1080/14737159.2020.1719070
24. Kowalska-Krochmal B, Dudek-Wicher R. The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens. 2021;10(2):165. doi:10.3390/pathogens10020165
25. Mielko KA, Jablonski SJ, Pruss L, et al. Metabolomics comparison of drug-resistant and drug-susceptible Pseudomonas aeruginosa strain (Intra- and Extracellular Analysis). Int J Mol Sci. 2021;22(19):10820. doi:10.3390/ijms221910820
26. Corzana F, Cuesta I, Bastida A, et al. Molecular recognition of aminoglycoside antibiotics by bacterial defence proteins: NMR study of the structural and conformational features of streptomycin inactivation by Bacillus subtilis aminoglycoside-6-adenyl transferase. Chemistry. 2005;11(17):5102–5113. doi:10.1002/chem.200400941
27. Zhang H, Ma G, Zhu Y, et al. Active-site conformational fluctuations promote the enzymatic activity of NDM-1. Antimicrob Agents Chemother. 2018;62(11). doi:10.1128/AAC.01579-18
28. Salahuddin P, Kumar A, Khan AU. Structure, function of serine and metallo-beta-lactamases and their inhibitors. Curr Protein Pept Sci. 2018;19(2):130–144. doi:10.2174/0929866524666170724160623
29. Matesanz R, Diaz JF, Corzana F, Santana AG, Bastida A, Asensio JL. Multiple keys for a single lock: the unusual structural plasticity of the nucleotidyltransferase (4’)/kanamycin complex. Chemistry. 2012;18(10):2875–2889. doi:10.1002/chem.201101888
30. Stern AL, Van der Verren SE, Kanchugal PS, Nasvall J, Gutierrez-de-teran H, Selmer M. Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase from Salmonella enterica. J Biol Chem. 2018;293(29):11481–11490. doi:10.1074/jbc.RA118.003989
31. Fodor A, Abate BA, Deak P, et al. Multidrug Resistance (MDR) and collateral sensitivity in bacteria, with special attention to genetic and evolutionary aspects and to the perspectives of antimicrobial peptides-a review. Pathogens. 2020;9(7):522. doi:10.3390/pathogens9070522
32. Yin D, Guo Y, Li M, et al. Performance of VITEK 2, E-test, Kirby-Bauer disk diffusion, and modified Kirby-Bauer disk diffusion compared to reference broth microdilution for testing tigecycline susceptibility of carbapenem-resistant K. pneumoniae and A. baumannii in a multicenter study in China. Eur J Clin Microbiol Infect Dis. 2021;40(6):1149–1154. doi:10.1007/s10096-020-04123-z
33. Yuan S, Chen Y, Lin K, et al. Single cell raman spectroscopy deuterium isotope probing for rapid antimicrobial susceptibility test of Elizabethkingia spp. Front Microbiol. 2022;13:876925. doi:10.3389/fmicb.2022.876925
34. Boolchandani M, D’Souza AW, Dantas G. Sequencing-based methods and resources to study antimicrobial resistance. Nat Rev Genet. 2019;20(6):356–370. doi:10.1038/s41576-019-0108-4
35. Wu SH, Xiao YX, Hsiao HC, Jou R, She RC. Development and assessment of a novel whole-gene-based targeted next-generation sequencing assay for detecting the susceptibility of mycobacterium tuberculosis to 14 drugs. Microbiol Spectr. 2022;10(6):e0260522. doi:10.1128/spectrum.02605-22
36. Alcock BP, Huynh W, Chalil R, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023;51(D1):D690–D699. doi:10.1093/nar/gkac920
37. Doster E, Lakin SM, Dean CJ, et al. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020;48(D1):D561–D569. doi:10.1093/nar/gkz1010
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.