Back to Journals » Cancer Management and Research » Volume 13
Characterisation of Levonorgestrel-Resistant Endometrial Cancer Cells
Authors Dore M, Filoche S, Danielson K, Henry C
Received 6 July 2021
Accepted for publication 4 October 2021
Published 14 October 2021 Volume 2021:13 Pages 7871—7884
DOI https://doi.org/10.2147/CMAR.S327381
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Molly Dore,1 Sara Filoche,1 Kirsty Danielson,2 Claire Henry1
1Department of Obstetrics, Gynaecology & Women’s Health, University of Otago, Wellington, New Zealand; 2Department of Surgery and Anaesthesia, University of Otago, Wellington, New Zealand
Correspondence: Claire Henry
Department of Obstetrics, Gynaecology and Women’s Health, University of Otago, Wellington, New Zealand
Tel +64 22 092 6984
Email [email protected]
Background: Endometrial cancer (EC) is the most common gynaecologic malignancy in the developed world, and incidence is increasing in premenopausal women. The levonorgestrel intrauterine system (LNG-IUS) is gaining traction as an alternative treatment for hyperplasia and early-stage EC for women who are unable to undergo surgery. Thirty to 60% of the women do not respond to this treatment, making the unknown mechanisms of levonorgestrel (LNG) resistance a critical obstacle for the conservative management of EC. This study aimed to characterise LNG-IUS treatment resistance in early-stage endometrial cancer in cell-line models.
Methods: LNG-resistant endometrial cancer cell lines (MFE296R and MFE319R) and cultures from three early stage endometrial cancer patients were developed. The behavioural profile of MFE296R and MFE319R was analysed using proliferation, adhesion, migration (wound healing and transwell) and invasion (spheroid) assays. LNG-sensitive cell lines (MFE296S and MFE319S) were compared to LNGR cell lines (MFE296R and MFE319R). A literature search was conducted to identify possible candidate biomarkers of LNG resistance. RT-qPCR was used to analyse the mRNA expression of 17 candidate biomarkers in MFE296R and MFE319R. mRNA expression of the top differentially expressed genes was measured using RT-qPCR in primary cultures.
Results: LNG resistance did not affect proliferation or invasion in immortalised endometrial cancer cells. Transwell migration was significantly increased in MFE319R cells (p=0.03). Cellular adhesion significantly decreased in both MFE296R cells (p=0.012) and MFE319R cells (p=0.04). mRNA expression of KLF4 and SATB2 was significantly amplified in MFE296R and MFE319R cells. mRNA expression of KLF4 was significantly upregulated LNG-resistant primary cell lines.
Conclusion: LNG-resistant cells may have more oncogenic potential than their LNG-sensitive counterparts. Significant changes in the mRNA expression of KLF4 and SATB2 of LNG-resistant cells is a promising preliminary result in biomarker discovery for guiding LNG-IUS treatment of early stage endometrial cancer.
Keywords: endometrial cancer, biomarker, therapy, response, levonorgestrel-releasing intrauterine system
Background
Endometrial cancer (EC) is the most common gynaecologic malignancy globally contributing to 3.9% of the total cancers in women.1 EC is generally diagnosed at earlier stages, with 75% of EC cases being classed as low-grade endometrioid EC (EEC) histological subtypes.2,3 Currently, the standard treatment for early-stage EEC is a hysterectomy and a bilateral salpingo-oophorectomy4; however, up to 25% of EEC patients are premenopausal, with 5% of these women being under the age of 40.4,5 Furthermore, women with a high BMI are more likely to be diagnosed with EEC,6 however, up to 10% of these women are deemed inoperable.7 This data, alongside a shift in global demographics resulting in the rising prevalence of both ageing populations and obesity;8–10 has led to an increase in the incidence of EEC in inoperable women. This shift is increasing the demand for conservative management options for EEC.
Until recently, systemic progestogen therapy, such as medroxyprogesterone acetate (MPA), has been used for conservative management of EEC. MPA has proven to be efficacious in the treatment of hormone-sensitive tumours in women where surgery is not a treatment option.11 However, new evidence is shifting the conservative management of EC towards a long-acting reversible contraceptive device, the levonorgestrel intrauterine system (LNG-IUS). The LNG-IUS is currently used to treat women with abnormal and heavy bleeding (menorrhagia) as levonorgestrel (LNG) (a synthetic form of progesterone) suppresses endometrial proliferation through counteracting the effect of oestrogen.12 The evidence base for the use of the LNG-IUS when treating EEC appears promising, with treatment in this setting yielding response rates between 52% and 67%.13–16 Compared with oral progestogens, the LNG-IUS has a similar disease regression rate and is associated with fewer systemic adverse effects due to being placed locally in the uterus.17 Still, the evidence appears that there is inconsistency in response when using the LNG-IUS for treatment of early-stage EC in some women making the unknown mechanisms of LNG resistance a critical obstacle for the conservative management of EC.
Currently, the absence of predictive biomarkers for LNG-IUS treatment of early-stage EC limits the certainty of recommendation,18 with both response and monitoring of treatment depending on invasive biopsies every 3–6 months.4 In recent years, researchers have investigated a variety of approaches for predicting response to conservative treatment of EC; however, few papers investigate predictive biomarkers involved in LNG-IUS resistance explicitly,19 with only three looking at the effects of the LNG-IUS in women with EC. Our recent review comments on the current status of predictive biomarkers in LNG-IUS treatment of EC and hyperplasia19 and highlights the importance of biomarkers in LNG-IUS treatment of EC. Low protein expression of HE414 and progesterone receptorB20 alongside mRNA expression of FOXO121 have all been identified as possible predictive biomarkers of non-response to the LNG-IUS specifically. One of the largest-scale genetic-based studies done to date on the topic was conducted by Li et al, who observed upregulated mRNA expression of ANO1, SOX17, CHNL1, DACH1, RUNDC3B, SH3YL1 and CRISPLD1 in an Ishikawa progesterone resistant cell line.22 This study observes this occurrence in one commercial cell line and is based on MPA resistant cells, rather than LNG-resistant cells Additional research into genes implicated in progesterone resistance is clearly warranted and as currently, there are no predictive biomarkers used clinically in relation to LNG-IUS treatment.
This study aimed to identify differentially expressed genes (DEGs) in LNG-resistant cells compared to LNG-sensitive cells and to build a behavioural profile of LNG-resistant cell lines. These DEGs may then go on to serve as potential predictive biomarkers to be investigated further in patient cohorts.
Methods
Cell Culture
Endometrioid EC cell lines MFE296 and MFE319 were cultured as per the supplier’s recommendations: MFE296 in Minimum Essential Medium (MEM) (Gibco; Thermo Fisher Scientific #11095-080, CA, USA) containing 10% foetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific #10-091-148, CA, USA) and MFE319 in 1:1 MEM/Roswell Park Memorial Institute Medium (RPMI 1640) (Gibco; Thermo Fisher Scientific #11875-093, CA, USA) containing 20% FBS. All media were supplemented with 100U/mL penicillin/streptomycin (Gibco; Thermo Fisher Scientific, #15070-063, CA, USA). Cells were grown in 5% CO2 at 37 °C. Cells were confirmed negative for mycoplasma.
Ethics
The current study used primary cells derived from tissue samples of early-stage EC cultures donated by women as part of the Gynaecological Cancer Tissue Bank at Wellington hospital (Health and Disability Ethics Committees (HDEC) 15/CEN/143 and University of Otago health ethics committee H20/002). All patients provided written, informed consent prior to donation to the biobank.
Primary Cell Lines
The isolation of adenocarcinoma cells from cancer-associated stromal cells was carried out according to previous studies.23 A section of endometrial cancer tissue (5–20mm) was dissociated for 1 hour using collagenase type I (Sigma-Aldrich #C0130-500MG, NSW Australia) (10mg/mg) in TESCA buffer (50µM TES; 0.36µM CaCl) (Sigma-Aldrich #T1375-100, NSW Australia) diluted to 0.5mg/mL in PBS and DNAse 1 from bovine pancreas (Sigma-Aldrich #DN25, NSW Australia) diluted to 0.1mg/mL in PBS. Cell solution was passed through a 40µm cell strainer to separate cancer-associated stromal cells from adenocarcinoma cells. Cancer-associated stromal cells and adenocarcinoma cells were centrifuged and resuspended separately in Dulbecco’s Modified Eagle Medium (DMEM/F12) (Gibco; Thermo Fisher Scientific, CA, USA) medium containing 10% FBS and supplemented with 100U/mL penicillin/streptomycin. The adenocarcinoma cells were used in the current study. Cells were grown in 5% CO2 at 37 °C. Cells were passaged at 80% confluence up to four times.
Development of Resistant Cell Lines
The MFE319 and MFE296 cell lines were chosen for the current study due to their stability and their definitive EEC phenotypic classification.24 MFE319 and MFE296 cells have been reported as having relatively higher levels of hormone receptor (PR, PR-B and ER) levels, making them a suitable model for studying hormonal responses in cell culture.25 To develop a high-level laboratory model of resistance,26 the MFE296R and MFE319R cells were treated every 2 days with this concentration of LNG with no escalation. Resistance was then assessed via an IC50 and then determined through the observation of fold change. MFE296 and MFE319 cells were trypsinised, counted, and seeded on to a 96 well plate at a concentration of 3 × 105 cells/mL. Following a 24h incubation at 5% CO2 at 37 °C, cells were treated with escalating LNG (Sigma-Aldrich # 797-63-7) dissolved in dimethyl sulfoxide (DMSO, final concentration no greater than 0.001%) (Sigma-Aldrich #D5879, CA, USA) concentrations from 0–500µM and incubated for a further 24h. Plates were then analysed using the Cell Counting Kit-8 (CCK8) (Dojindo #CK04-11) according to manufacturer’s instructions. Readings at 450 nm were obtained after 3 hours using the Thermo Scientific TM Multiskan GO TM Microplate Spectrophotometer (Thermo Fisher Scientific, MA, USA). Any increase in absorbance indicated an increase in cell density. LNG treatment concentration for resistant clone development was identified as the point of 30% cell viability for each cell line.
LNG-resistant MFE319, MFE296 and primary cell lines were obtained from parental cells via continuous exposure to LNG dissolved in 0.001% DMSO. MFE296 cells were treated with 450µM LNG, MFE319 with 350µM LNG and primary cells with 100µM–200µM LNG. A DMSO control was created for each cell line at a dose of 0.001% dissolved in respective culture media.
A kill curve was used to confirm resistance. Following an LNG treatment period of 4 months, cells were trypsinised, counted, and seeded on to a 96 well plate at a concentration of 3 × 105 cells/mL for the LNG-sensitive cells and 6 × 105 cells/mL for the LNG-resistant cells of both MFE296 and MFE319 cell lines. Cells were incubated at 5% CO2 at 37 °C for 24h and then treated with escalating LNG concentrations from 0 to 2000µM and incubated for a further 24h. Following incubation plates were analysed using CCK8 according to manufacturer’s instructions.
Cell lines from here on will be referred to as MFE296R (LNG-resistant), MFE296S (LNG-sensitive) and MFE319R (LNG-resistant), MFE319S (LNG-sensitive).
Proliferation Assay
Cell proliferation was carried out according to previous studies.27 Cells were seeded in triplicate onto a 96-well plate at a concentration of 3 × 105 cells/mL (MFE296R, MFE296S and MFE319R, MFE319S) and incubated at 5% CO2 at 37 °C for 24 hours. Following incubation plates were analysed using CCK8 according to manufacturer’s instructions. Readings were obtained at 0-, 24-, 48- and 72-hour time points. All absorbance values were normalised to the T0 time point to give the normalised proliferation of each cell line.
Transwell Migration Assay
Cell migration was measured via the Boyden Chamber assay according to a previous study.27 Transwells (6.5-mm) with 8.0-μm pore polycarbonate membrane inserts (Sigma-Aldrich #CLS3422, NSW Australia) were used according to manufacturer’s instructions. Cells were seeded in transwell inserts at a concentration of 3×105 cells/mL for MFE296R and MFE296S and 1 × 106 cells/mL for MFE319R, and MFE319S and incubated for 48 hours at 5% CO2 at 37 °C. Following incubation, cells were washed twice with PBS and fixed with 100% ethanol at room temperature for 20 minutes and stained with 1% crystal violet at room temperature for 15 minutes. Following staining, transwells were washed twice with PBS and non-migrated cells were wiped off using a cotton swab. The membrane was then removed and mounted on a glass slide. Micrographs were taken of four quadrants of the membrane to accurately represent the entire membrane and the cell number was counted using ImageJ (Java Software,28 WI, USA). An average cell count of the four quadrants was used in statistical analysis.
Wound Migration Assay
Cell migration was measured via a wound healing assay. MFE296 and MFE319 cell lines were plated onto a six-well plate at a concentration of 1×106 cells/mL and incubated at 5% CO2 at 37 °C for 24 hours. Following incubation, a 10uL pipette tip was used to create a scratch through the centre of each well. Micrographs of the plates were taken at 0-, 24-, 48-, 72- and 96-hour time points using a 10× objective lens. Wound healing and the percentage of open area was measured using TScratch (CSElab software, ZH, Switzerland).29
Invasion (3D Tumour Spheroid)
Cell invasion was measured using a three-dimensional (3D) tumour spheroid invasion assay and carried out according to previous studies.30 Hanging drop cultures were prepared on a six-well plate using a cell concentration of 8×104 cells/mL for MFE296S and MFE296R, and 3×104 cells/mL for both MFE319S and MFE319R and incubated for 96 hours. Following incubation, once spheroids were visible they were embedded into a matrix of 3mg/mL type I collagen (rat tail) (Gibco; Thermo Fisher Scientific #A10483-01, NSW Australia) and 2.7 mg/mL matrigel (Corning Life Sciences #354234, MA, USA) matrix. Micrographs were taken to monitor invasion at 0, 24, 48, 72 and 96 hours following plating using the 20× objective lens. Spheroid invasion was then measured as the total area of spheroids using ImageJ (Java Software).28
2D Adhesion Assay
Cell adhesion was carried out according to previous studies.31 Adhesion was measured against collagen type I (rat tail) (Gibco; Thermo Fisher Scientific). Tissue culture plates were coated with collagen (10µg/mL) and 3% bovine serum albumin (BSA), as the negative control, in PBS. Coated plates were incubated for 24 h at 37 °C and then rinsed with 80% ethanol. Three percent BSA in PBS was added to each well and incubated for a further 30 min at 37 °C. After rinsing with PBS, concentrations of 5×105 cells/mL for all cell lines in serum-free media were added to the coated plates and left to adhere at 37 °C for 1 h. Following incubation, plates were washed 3 times with PBS and fixed with 100% ethanol before being stained with 0.1% crystal violet at room temperature for 30 min. Plates were then washed extensively with water to remove excess staining and then left to dry. Once dried, cells were lysed with 50% acetic acid. Absorbance was measured at 595nm using the Thermo Scientific TM Multiskan GO TM Microplate Spectrophotometer.
Literature Search
A literature search was carried out to identify a set of key genes involved in progesterone resistance when treating endometrial cancer or endometrial hyperplasia. SCOPUS, PubMed and Google Scholar were searched for the following keywords (“endometrial cancer*” OR “endometrial carcinoma*” OR “endometrial neoplasm*”) AND (IUD OR “intra-uterine-device*” OR IUS OR progesterone OR progestin OR Levornogestrel OR “intra-uterine device*” OR “intrauterine system*” OR “intra-uterine system*” OR Mirena) AND (“Biomarkers” OR “Marker” or “Predictive Marker*”) AND (“Response”)). Articles were critically assessed and genes were selected due to their implication in progesterone resistance in endometrial cancer (EC) and hyperplasia treatment. A review outlining these papers, has been written.19 From this, 13 genes were chosen for analysis: HOTAIR, HE4, ANO1, SOX17, CGNL1, DACH1, RUDC3B, SH3YL1, CRISPLD1, FOXO1, PR-B, ER, and MSX1. A further five genes were selected due to their relationship to EC: MDR1, DKK1, SATB2, CACNA2D3 and KLF4.
RNA Extraction
RNA extraction was carried out according to a previous study.32 LNG-resistant cells and LNG-sensitive controls for MFE319, MFE296, GB#13, GB#16 and GB#23 cells were harvested, pelleted, and the RNA from these cells extracted using the zymo Quick-RNA kit (Zymo cat# R1057, CA, USA) according to the manufacturer’s instructions. RNA quantification (in ng/µL) and purity was assessed using the NanoDrop spectrophotometer (Thermo Fisher Scientific). A 260/280 and 260/230 ratio of ~2.0nm was considered optimal “pure” RNA.
RT-qPCR
Conversion of RNA (1μg) to double-stranded cDNA was carried out using the QuantiTect® RT kit following the manufacturer’s instructions (Qiagen #205311). NCBI Primer Blast was used to create a list of potential primers for each of the genes (Supplementary Table 1). qPCR analysis was carried out according to previous studies.32 Twenty-five nanograms of cDNA, 100 nM of primers and 12.5μL SYBRGreen master mix (Qiagen # 204143) was used in each reaction. RT-qPCR cycling conditions were 95°C for 10min, (95°C for 15 seconds, 60°C for 30 seconds, 72°C for 40 seconds) for a total of 40 cycles and then 95°C for 60 seconds, followed by melt curve analysis. Ct values were analysed using the normalisation method against three reference genes: succinate dehydrogenase complex subunit (SDHA), 90-kDa heat shock protein 1 beta (HSPCB) and 60S ribosomal protein L13a (RPL13A) to calculate the ΔCt (ΔCt mRNA Ct – geomean endogenous reference genes). ΔΔCt was then calculated relative to sensitive controls (ΔCt(sample1) – ΔCt(sample2)). Fold change was then calculated.
Statistical Analysis
All values are represented as mean ± standard deviation (SD) unless otherwise stated. A f-test was used to determine variances in standard deviation prior to t-test analysis. An unpaired Student’s t-test was carried out for determination of resistance, transwell migration and adhesion assays to compare values between LNG-resistant and LNG-sensitive cell lines. An unpaired Student’s t-test was carried out on the mean of each time point for wound healing, migration and proliferation to give significance at each time point. Technical and biological triplicates (n=3) were carried out for each experiment. A paired Student’s t-test was performed on fold change values for differences in mRNA expression between LNG-resistant and LNG-sensitive MFE296, MFE319, GB#23, GB#52 and GB#67 cell lines. A p value of ≤0.05 was considered statistically significant.22 Analysis of TCGA mRNA expression data in endometrial cancer was conducted using UALCAN33 to investigate any relationships with clinical parameters.
Results
Development of LNG-Resistant MFE296 and MFE319 Cell Lines
Resistance was assessed via an IC50 and then determined through the observation of fold change. The IC50 values of MFE296S and MFE296R cells were 250µM and 1800µM, respectively, and the fold change increase in resistance from MFE296S and MFE296R was 6. The IC50 values of MFE319S and MFE319R cells were 250µM and 2000µM, respectively, and the fold change increase in resistance from MFE319S and MFE319R was 8 (Supplementary Figure 1).
LNG Resistance Impact on Cellular Proliferation in Immortalised Cell Lines
There was no significant difference in cell proliferation between LNG-resistant cells and LNG-sensitive control cells apart from a significant decrease in MFE319R absorbance and therefore, cell density at the 48-hour time point (MFE319S: 1.535 ± 0.12, MFE319R: 1.23 ± 0.07) (P<0.05). Proliferation increased at a similar rate in both cell lines over a period of 72 hours. This observation was conserved in both LNG-treated cells and controls (Figure 1A and B). Significance was determined using an unpaired Student’s t-test.
LNG Resistance Impact on Migration in Immortalised Cell Lines
A significant increase in wound migration was observed in the MFE296R cell line at the T72 hour time points (MFE296S: 9.33 ± 0.018, MFE296R: 4.13 ± 0.018) (p= 0.0248) and T96 hour time points (MFE296S: 6.65 ± 0.013, MFE296R: 1.44 ± 0.010) (p= 0.0106) compared to the MFE296S control represented by a smaller % open area. There was no significant difference in migration at any other time points (Figure 2A). There was no significant change in wound migration in the MFE319R cell line compared to the MFE319S control (Figure 2B). Representative images of wound healing can be seen in (Figure 2C and D). There was no significant difference in Boyden chamber transwell migration between MFE296R cells and MFE296S cells (Figure 3A). However, a significant increase in transwell migration was observed in the MFE319R cells compared MFE319S cells (MFE319S: 210.31 ± 9.24, MFE296R: 254.62 ± 31.77) (Figure 3B) (P =0.03). Representative images of transwell migration can be seen in (Figure 3C and D).
LNG Resistance Impact on Cellular Adhesion in Immortalised Cell Lines
The adhesive capacity of LNG-resistant and LNG-sensitive cells was evaluated through the ability of respective cell lines to adhere to collagen in 1 hour. Adhesion was significantly attenuated in the MFE296R cells (absorbance: 0.662 ± 0.11) compared to the MFE296S cells (absorbance: 0.308 ± 0.088) after 1 hour (p=0.012) (Figure 4A). Adhesion was also attenuated in the MFE319R cells (absorbance: 0.822 ± 0.18) compared to the MFE319S cells (absorbance: 0.502 ± 0.06) after 1 hour (p=0.04) (Figure 4B). BSA served as a negative control for cell adhesion.
LNG Resistance Impact on Invasion in Immortalised Cell Lines
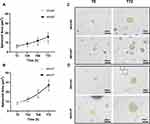
Cell invasion rate was measured via a 3D spheroid invasion assay embedded in an ECM matrix of collagen type I and matrigel. Spheroid size (µm2) was photographed every 24h for a total of 96h and then measured using Image J. There was no significant difference in spheroid size (µm2) in MFE296R cells compared to MFE296S (Figure 5A) or in MFE319R compared to MFE319S cells (Figure 5B). Representative images of MFE296 and MFE319 spheroids can be seen in Figure 5C and D.
LNG-Resistant Cells Express Different Levels of mRNA to LNG-Sensitive Cells in Immortalised Cell Lines
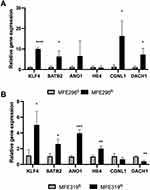
Four DEGs in the MFE296 resistant cell line were identified. Relative mRNA expression of KLF4, SATB2, CGNL1 and DACH1 were significantly upregulated in MFE296R compared to MFE296S cells (p= 0.0000, 0.0296, 0.0271, 0.0242 respectively (Figure 6A). Relative expression of these genes in MFE296R cells were KLF4: 10.1±0.7, SATB2: 6.3±2.7, DACH: 7.3±0.13 and CGNL1: 16.3±7.6. Non-significant data for MFE296 cell lines can be viewed in Supplementary Table 2. ΔCt mRNA expression of significant DEGs in the MFE296 cell line can be viewed in Supplementary Figure 2A.
Five DEGs were identified in the MFE319 cell line. Relative mRNA expression of KLF4, SATB2, ANO1, HE4 and DACH1 were significantly upregulated in the MFE319R compared to MFE319S cells (p= 0.0223, 0.0189, 0.0004, 0.0077, 0.0029 respectively) (Figure 6B). Relative expression of these genes in MFE319R cells were KLF4: 5.0±1.7, SATB2: 2.6±0.6, ANO1: 4.0±0.5, HE4: 2.0±0.3. Non-significant data for MFE319 cell lines can be viewed in Supplementary Table 3. ΔCt mRNA expression of significant DEGs in the MFE319 cell line can be viewed in Supplementary Figure 2B.
Relative mRNA expression of KLF4 was significantly amplified in primary LNGR cell lines compared to LNGS controls (p= 0.0213) (Figure 7). Relative expression of KLF4 in LNGR primary cells was 1.5±0.2. Relative mRNA expression of ER was significantly downregulated in primary LNGR cell lines compared to LNGS controls (p= 2e-6) Relative expression of ER in LNGR primary cells was 0.3±0.02. Non-significant data for primary cell lines can be viewed in (Supplementary Table 4).
Bioinformatic analysis of KLF4 and SATB2 in EC TCGA mRNA data using UALCAN33 showed that KLF4 mRNA expression is significantly decreased in EC primary tumours compared to healthy controls (Supplementary Figure 3A).There is no significant relationship between KLF4 and overall survival, weight or cancer stage (Supplementary Figure 3B–D). KLF4 mRNA expression is however significantly higher in patients aged between 40 and 60 years compared to patients between 60 and 100 years (Supplementary Figure 3E). SATB2 expression is significantly increased in EC primary tumours compared to healthy controls (Supplementary Figure 4A). There is no significant relationship between SATB2 and overall survival, age or weight; however, expression of SATB2 is significantly higher in stage 1, 2 and 3 cancers in comparison to stage 4 cancers (Supplementary Figure 4B–E).
Discussion
The current study aimed to build a behavioural profile of LNG-resistant cell lines and identify significant DEG’s in LNG-resistant cells compared to LNG-sensitive cell lines. LNG resistance did not affect proliferation or invasion in both MFE296R and MFE319R cell lines; however, it significantly increased transwell migration in MFE319R cells, increased wound healing migration at the 72- and 96-hour time points in MFE296R cells and decreased cellular adhesion to collagen in MFE29R cells and MFE319R cells. Transwell migration in MFE296R cells was unaffected by LNG resistance.
Increased proliferation, migration, invasion and decreased adherence are all implicated in the epithelial-mesenchymal transition (EMT) process. EMT is a physiological process involved in the early stage of carcinogenic dysregulation.34,35 During this process, cells begin to lose epithelial characteristics and acquire mesenchymal characteristics such as increased motility through the reprogramming of gene expression.34 Acquisition of EMT phenotype has been previously associated with drug resistance, which could make these tumours more likely to metastasise or recur following treatment.36 This is consistent with the results from the current study, showing that the LNG-resistant cells may be slightly more aggressive and, therefore, have more oncogenic potential than their LNG-sensitive counterparts. Clinically, this research is warranted as it is the first time the behavioural profile of LNG-resistant cells has been investigated. It is vital to understand if LNG-resistant cells behave differently as this could lead to alternative treatment pathways for women who are resistant to the LNG-IUS. This further demonstrates a need to identify women who will not respond to the LNG-IUS prior to treatment.
Seventeen potential biomarkers were identified in the literature and mRNA expression was investigated in LNG-resistant and LNG-sensitive cell lines. Expression of both KLF4 and SATB2 was significantly upregulated in resistant cell lines (MFE296R and MFE319R) compared to LNGS controls, suggesting that these genes have the potential to serve as predictive biomarkers for response to the LNG-IUS. In the current study, upregulation of ANO1 in MFE319R cell lines and upregulation of CGNL1 MFE296R cells compared to LNG-sensitive controls supports previous findings from Li et al,22 where upregulation of these genes was observed in MPA-resistant Ishikawa cell lines. The current study did not observe these genetic upregulations across both cell lines, nor did it observe upregulation of DACH1, RUNDC3B, SH3YL1 or CRISPLD1 in LNG-resistant cells compared to LNG-sensitive controls seen in the Li et al's22 study. The current study builds upon the Li et al’s study by identifying ANO1 and CGNL1 upregulation in further EEC immortalised cell lines and observing these genetic changes in relation to LNG treatment specifically. The findings of the current study do not support observations made by Orbo et al, and Behrouzi et al,14,37 who identified low protein expression of HE4 to be associated with a positive response to the LNG-IUS. In the current study, no significant change in HE4 ΔCt mRNA expression in immortalised or primary LNG-resistant cells compared to LNG-sensitive controls was detected. However, the current study only investigated mRNA expression of HE4, rather than changes in protein expression observed by previous studies14,37 and it is not yet clear whether changes in ΔCt mRNA expression of HE4 directly correlate to HE4 protein expression. The differential mRNA expression observed between the two cell lines and primary cell lines in the current study, alongside the cell lines and patient samples used in previous studies, can most likely be attributed to tumour heterogeneity. The degree of spatial diversity within an individual’s tumour is highly variable38 with research showing between 0 and 8000 heterogeneous coding mutations within primary tumours.39 This heterogeneity drives phenotypic variation, thus, posing a significant challenge to the diagnosis, management and treatment of cancer, alongside contributing to significant challenges when identifying biomarkers to guide clinical decision-making in cancer medicine.40 A panel biomarker approach with clinically validated cut-off points may be more appropriate to address these inter-tumour disparities. However, the current study serves as an exploratory investigation into genes that can be further investigated in a clinical setting.
This is the first time KLF4 and SATB2 have been investigated in the context of predictive biomarkers for EEC treatment and therefore serve as novel potential biomarkers that warrant further investigation. Of note, KLF4 appears to be the most promising potential biomarker due to its increase in mRNA expression in LNG-resistant cells conserved across both immortalised and primary cell lines. KLF4 is a member of the zinc finger containing Krüppel-like factor family.41 It is involved in the regulation of diverse physiological functions and cellular processes such as cellular growth, proliferation, differentiation and somatic cell reprogramming.41 KLF4 has been identified to play an important role in the pathogenesis of many cancers.41 KLF4 has been previously identified as playing a role in anti-cancer drug resistance in cancers including nasopharyngeal,42 prostate,43 colorectal,44 breast,45 thyroid46 and multiple myeloma.47 While KLF4 has not been investigated as a biomarker in EC, mRNA expression of KLF4 has previously been seen to increase in the presence of progesterone receptor agonists such as progesterone and dienogest (synthetic progestin) in endometrial epithelial cells.48 This is consistent with observed results in the current study. The molecular relevance of KLF4 in LNG resistance is unknown; however, it may be down to the role KLF4 plays in Wnt pathway antagonism,41 promotion of survival through suppression of BAX49 and TP5350 or its role as a stem cell factor promoting epithelial-mesenchymal transition (EMT).41 KLF4 has previously been shown to inhibit the MAPK pathway.41 Inhibition of the MAPK pathway has been shown to increase nuclear PR protein in endometriosis,51 and PR expression increases responsiveness to progesterone therapy.20,21,52,53 Another potential mechanism of resistance is transcriptional repression of FOXO1 by KLF4.54 While this relationship has only been investigated in glioblastoma, the relationship between FOXO1 and KLF4 is interesting as downregulation of FOXO1 mRNA levels has been shown to predict a negative response in EC and endometrial hyperplasia patients managed with the LNG-IUS.21 While the current study does not observe a significant decrease in FOXO1 mRNA expression, this relationship should be further investigated in the context of EC.
SATB2 is a nuclear matrix-associated transcription factor that is associated with abnormal expression in certain cancers but has not been reported in EC.55 Like KLF4, there is limited information available on the relevance of SATB2 in EC. Interestingly, SATB2 has been implicated as an upstream regulatory gene in endometrial leiomyomas through dysregulation of downstream genes associated with Wnt/beta catenin, TGF-beta growth factor signalling and retinoic acid signalling.56 In ovarian cancer, SATB2 is used to distinguish between ovarian tumour with mucinous features that may be of colorectal origin.57 No other research to date investigates the role of SATB2 in endometrial cancer or progesterone resistance. mRNA expression and relative protein expression of these genes should be analysed in further primary EEC samples to determine if they can serve as predictive biomarkers that can be used clinically.
Alongside the two DEGs identified in both MFE296 and MFE319 cell lines, the relative mRNA expression of hormone receptors ER and PR were measured in primary cell lines in the current study. Low expression of PR, both protein and mRNA, has been extensively shown in the literature to predict a negative response to progesterone treatment.20,21,52,53 Contrary to the literature, the current study observed no significant change in mRNA expression of PR in all three primary LNG-resistant cells. Research has shown that LNG-IUS treatment can lead to downregulation of nuclear PRs58 therefore, despite previous research showing PR to have the potential to predict negative response to treatment, it would not make an efficacious predictive biomarker due to its expression being somewhat reliant on treatment.58 Alongside this, the two main subtypes of PR, PR-A and PR-B have been said to have opposing roles in estrogen-induced endometrial proliferation.59 PR-A has been said to inhibit proliferation, while PR-B contributes to it. The two receptor subtypes are also very difficult to differentiate via immunohistochemistry (IHC) and the use of antibodies that recognise both receptor subtypes should be used to evaluate overall PR expression in conditions including endometrial cancer.60 The current study only investigated PR-B expression; therefore, it would be beneficial to use RT-qPCR to investigate both subtypes further.
Strengths and Limitations
Limitations of the current study include a candidate-based approach rather than a discovery-based one. The genes investigated in the current study were chosen due to being previously implicated in progesterone resistance or other tumorigenic pathways of EC. A discovery-based approach would be to do whole-transcriptome sequencing,61 which could be efficacious in identifying novel targets that have not been previously described in the literature. However, within the confines of this preliminary study, a candidate-based approach was most appropriate.
The high-level laboratory model of resistance adopted in the current study was the most appropriate method in this case; however, the level of resistance obtained in the short time-frame was on the lower side of resistance.26 It is important to note that there may be differences between induced resistance, developed in the laboratory, and spontaneous resistance, which occurs in patients that are intrinsically resistant to the LNG-IUS. Because of this, the pathways implicated in acquired resistance after prolonged treatment could differ from pathways that altered resistance in drug-naïve cells. This is why it is important to first identify potential markers using a high-level laboratory model, and then to validate those findings in samples from women treated with the LNG-IUS and relate to outcome.
Due to being a 2D monolayer model, these assays do not represent what would occur physiologically, due to the lack of effect of both gravity and other surrounding tissues. This can lead the assays to lack predictivity62; however, the methods used for the behavioural analysis of resistant cell lines are high throughput and well documented, allowing us to compare methodology and results to other examples in the literature.
Conclusion
The need for conservative approaches to endometrial cancer treatment is rising due to the increase in incidence in women who may chose not to have surgery. Despite advances in molecular research, no clinically relevant predictive biomarkers have been established. We have identified six potential biomarkers in immortalised and primary patient cell lines for the use of the LNG-IUS treatment of early-stage EC. These markers should go on to be investigated further in patient cohorts to develop clinically relevant tissue and blood-based tests.
Abbreviations
DEG, differently expressed gene; EC, endometrial cancer; EEC, endometrioid endometrial cancer; EMT, epithelial-mesenchymal transition; ER, estrogen receptor; IHC, immunohistochemistry; LNG, levonorgestrel; LNG-IUS, levonorgestrel intrauterine system; MPA, medroxyprogesterone acetate; PR, progesterone receptor.
Ethics Approval and Consent to Participate
The current study used primary cells derived from tissue samples of early-stage EC cultures donated by women as part of the Gynaecological Cancer Tissue Bank at Wellington hospital (Health and Disability Ethics Committees (HDEC) 15/CEN/143 and University of Otago health ethics committee H20/002). All patients provided written, informed consent prior to donation to the biobank.
Acknowledgments
We recognise and thank the women who donated tissue to gynaecological research. We acknowledge the pathologists at WSCL and clinicians involved in the gynaecological biobank.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
University of Otago research grant was provided to CH.
Disclosure
The authors declare that they have no competing interests.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Duska LR, Garrett A, Rueda BR, Haas J, Chang Y, Fuller AF. Endometrial cancer in women 40 years old or younger. Gynecol Oncol. 2001;83(2):388–393. doi:10.1006/gyno.2001.6434
3. Gottwald L, Pluta P, Piekarski J, et al. Long-term survival of endometrioid endometrial cancer patients. Arch Med Sci. 2010;6(6):937–944. doi:10.5114/aoms.2010.19305
4. Koh W-J, Abu-Rustum NR, Bean S, et al. Uterine neoplasms, version 1.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2018;16(2):170–199. doi:10.6004/jnccn.2018.0006
5. Pellerin GP, Finan MA. Endometrial cancer in women 45 years of age or younger: a clinicopathological analysis. Am J Obstet Gynecol. 2005;193(5):1640–1644. doi:10.1016/j.ajog.2005.05.003
6. McMahon MD, Scott DM, Saks E, Tower A, Raker CA, Matteson KA. Impact of obesity on outcomes of hysterectomy. J Minim Invasive Gynecol. 2014;21(2):259–265. doi:10.1016/j.jmig.2013.08.707
7. Acharya S, Esthappan J, Badiyan S, et al. Medically inoperable endometrial cancer in patients with a high body mass index (BMI): patterns of failure after 3-D image-based high dose rate (HDR) brachytherapy. Radiother Oncol. 2016;118(1):167–172. doi:10.1016/j.radonc.2015.12.019
8. Setiawan VW, Yang HP, Pike MC, et al. Type I and II endometrial cancers: have they different risk factors? J Clin Oncol. 2013;31(20):2607–2618. doi:10.1200/JCO.2012.48.2596
9. Christensen K, Doblhammer G, Rau R, Vaupel JW. Ageing populations: the challenges ahead. Lancet. 2009;374(9696):1196–1208. doi:10.1016/S0140-6736(09)61460-4
10. Wilson C, Tobin S, Young R. The exploding worldwide cancer burden. Int J Gynecol Cancer. 2004;14(1):1–11.
11. Mountzios G, Pectasides D, Bournakis E, et al. Developments in the systemic treatment of endometrial cancer. Crit Rev Oncol Hematol. 2011;79(3):278–292. doi:10.1016/j.critrevonc.2010.07.013
12. Beatty MN, Blumenthal PD. The levonorgestrel-releasing intrauterine system: safety, efficacy, and patient acceptability. Ther Clin Risk Manag. 2009;5(3):561–574.
13. Pal N, Broaddus RR, Urbauer DL, et al. Treatment of low-risk endometrial cancer and complex atypical hyperplasia with the levonorgestrel-releasing intrauterine device. Obstet Gynecol. 2018;131(1):109–116. doi:10.1097/AOG.0000000000002390
14. Behrouzi R, Ryan NAJ, Barr CE, et al. Baseline serum HE4 but not tissue HE4 expression predicts response to the levonorgestrel-releasing intrauterine system in atypical hyperplasia and early stage endometrial cancer. Cancers. 2020;12(2):276. doi:10.3390/cancers12020276
15. Westin SN, Fellman B, Sun CC, et al. Prospective Phase II trial of levonorgestrel intrauterine device: nonsurgical approach for complex atypical hyperplasia and early-stage endometrial cancer. Am J Obstet Gynecol. 2020;224(2):191–e1.
16. Janda M, Robledo KP, Gebski V, et al. Complete pathological response following levonorgestrel intrauterine device in clinically stage 1 endometrial adenocarcinoma: results of a randomized clinical trial. Gynecol Oncol. 2021;161(1):143–151. doi:10.1016/j.ygyno.2021.01.029
17. Gallos I, Alazzam M, Clark T, et al. Management of endometrial hyperplasia. Green-top guideline no. 67. In: RCOG/BSGE Joint Guideline. 2016. London: Royal College of Obstetricians and Gynaecologists;2016.
18. Barr CE, Crosbie EJ. Mirena coil is a suitable treatment of early-stage endometrial cancer in obese women. BJOG. 2020;127(8):1001.
19. Dore M, Filoche S, Danielson K, Henry C. Efficacy of the LNG-IUS for treatment of endometrial hyperplasia and early stage endometrial cancer: can biomarkers predict response? Gynecol Oncol Rep. 2021;36:100732. doi:10.1016/j.gore.2021.100732
20. Janzen DM, Rosales MA, Paik DY, et al. Progesterone receptor signaling in the microenvironment of endometrial cancer influences its response to hormonal therapy. Cancer Res. 2013;73(15):4697–4710. doi:10.1158/0008-5472.CAN-13-0930
21. Reyes HD, Carlson MJ, Devor EJ, et al. Downregulation of FOXO1 mRNA levels predicts treatment failure in patients with endometrial pathology conservatively managed with progestin-containing intrauterine devices. Gynecol Oncol. 2016;140(1):152–160. doi:10.1016/j.ygyno.2015.10.023
22. Li W, Wang S, Qiu C, et al. Comprehensive bioinformatics analysis of acquired progesterone resistance in endometrial cancer cell line. J Transl Med. 2019;17(1):58. doi:10.1186/s12967-019-1814-6
23. Barros FSV, Brosens JJ, Brighton PJ. Isolation and primary culture of various cell types from whole human endometrial biopsies. Bio-Protocol. 2016;6(22):e2028. doi:10.21769/BioProtoc.2028
24. Skok K, Maver U, Gradišnik L, Kozar N, Takač I, Arko D. Endometrial cancer and its cell lines. Mol Biol Rep. 2020;47(2):1399–1411. doi:10.1007/s11033-019-05226-3
25. Qu W, Zhao Y, Wang X, et al. Culture characters, genetic background, estrogen/progesterone receptor expression, and tumorigenic activities of frequently used sixteen endometrial cancer cell lines. Clinica Chimica Acta. 2019;489:225–232. doi:10.1016/j.cca.2018.08.013
26. McDermott M, Eustace AJ, Busschots S, et al. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: a practical guide with case studies. Front Oncol. 2014;4:40. doi:10.3389/fonc.2014.00040
27. Henry C, Llamosas E, Knipprath-Meszaros A, et al. Targeting the ROR1 and ROR2 receptors in epithelial ovarian cancer inhibits cell migration and invasion. Oncotarget. 2015;6(37):40310–40326. doi:10.18632/oncotarget.5643
28. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi:10.1038/nmeth.2089
29. Gebäck T, Schulz MMP, Koumoutsakos P, Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. BioTechniques. 2009;46(4):265–274. doi:10.2144/000113083
30. Vinci M, Box C, Eccles SA. Three-dimensional (3D) tumor spheroid invasion assay. J Vis Exp. 2015;99:e52686.
31. Henry C, Quadir A, Hawkins NJ, et al. Expression of the novel Wnt receptor ROR2 is increased in breast cancer and may regulate both β-catenin dependent and independent Wnt signalling. J Cancer Res Clin Oncol. 2015;141(2):243–254. doi:10.1007/s00432-014-1824-y
32. Henry C, Hacker N, Ford C. Silencing ROR1 and ROR2 inhibits invasion and adhesion in an organotypic model of ovarian cancer metastasis. Oncotarget. 2017;8(68):112727–112738. doi:10.18632/oncotarget.22559
33. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumour subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):659. doi:10.1016/j.neo.2017.05.002
34. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196.
35. Feitelson MA, Arzumanyan A, Kulathinal RJ, et al. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin Cancer Biol. 2015;35:S25–S54.
36. Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi:10.1038/onc.2010.215
37. Ørbo A, Arnes M, Lyså LM, Borgfelt C, Straume B. HE4 is a novel tissue marker for therapy response and progestin resistance in medium-and low-risk endometrial hyperplasia. Br J Cancer. 2016;115(6):725–730. doi:10.1038/bjc.2016.247
38. Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi:10.1038/nature12627
39. Stanta G, Bonin S. Overview on clinical relevance of intra-tumor heterogeneity. Front Med. 2018;5(85). doi:10.3389/fmed.2018.00085
40. Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi:10.1038/nature12625
41. Ghaleb AM, Yang VW. Krüppel-like factor 4 (KLF4): what we currently know. Gene. 2017;611:27–37. doi:10.1016/j.gene.2017.02.025
42. Lung HL, Kan R, Chau WY, et al. The anti-tumor function of the IKK inhibitor PS1145 and high levels of p65 and KLF4 are associated with the drug resistance in nasopharyngeal carcinoma cells. Sci Rep. 2019;12064(9):1.
43. Jiang Z, Zhang Y, Chen X, Wu P, Chen D. Long non-coding RNA LINC00673 silencing inhibits proliferation and drug resistance of prostate cancer cells via decreasing KLF4 promoter methylation. J Cell Mol Med. 2020;24(2):1878–1892. doi:10.1111/jcmm.14883
44. Deng X, Kong F, Li S, et al. A KLF4/PiHL/EZH2/HMGA2 regulatory axis and its function in promoting oxaliplatin-resistance of colorectal cancer. Cell Death Dis. 2021;485(12):1–3.
45. Jia Y, Zhou J, Luo X, et al. KLF4 overcomes tamoxifen resistance by suppressing MAPK signaling pathway and predicts good prognosis in breast cancer. Cell Signal. 2018;42:165–175. doi:10.1016/j.cellsig.2017.09.025
46. Lee SI, Kim DK, Seo EJ, et al. Role of Krüppel-like factor 4 in the maintenance of chemoresistance of anaplastic thyroid Cancer. Thyroid. 2017;27(11):1424–1432. doi:10.1089/thy.2016.0414
47. Schoenhals M, Kassambara A, Veyrune JL, et al. Krüppel-like factor 4 blocks tumor cell proliferation and promotes drug resistance in multiple myeloma. Haematologica. 2013;98(9):1442. doi:10.3324/haematol.2012.066944
48. Shimizu Y, Takeuchi T, Mita S, et al. Krüppel-like factor 4 mediates anti-proliferative effects of progesterone with G0/G1 arrest in human endometrial epithelial cells. J Endocrinol Invest. 2010;33(10):745–750. doi:10.1007/BF03346681
49. Ghaleb AM, Katz JP, Kaestner KH, et al. Krüppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene. 2007;26(16):2365–2373. doi:10.1038/sj.onc.1210022
50. Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptio na l repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7(11):1074–1082. doi:10.1038/ncb1314
51. Eaton JL, Unno K, Caraveo M, et al. Increased AKT or MEK1/2 activity influences progesterone receptor levels and localization in endometriosis. J Clin Endocrinol Metab. 2013;98(12):E1871–E18. doi:10.1210/jc.2013-1661
52. Akesson E, Gallos ID, Ganesan R, et al. Prognostic significance of estrogen and progesterone receptor expression in LNG-IUS (Mirena®) treatment of endometrial hyperplasia: an immunohistochemical study. Acta Obstet Gynecol Scand. 2010;89(3):393–398. doi:10.3109/00016340903556006
53. Fawzy M, Mosbah A, Zalata K, et al. Predictors of progestin therapy response in endometrial hyperplasia: an Immunohistochemical Study. Egypt J Fertil Steril. 2016;20(2):6–11. doi:10.21608/egyfs.2016.19528
54. Tang G, Liu D, Xiao G, et al. Transcriptional repression of FOXO1 by KLF4 contribute s to glioma progression. Oncotarget. 2016;7(49):81757–81767. doi:10.18632/oncotarget.13184
55. Uhlén M, Fagerberg L, Hallstrom BM. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. PubMed: 25613900. doi:10.1126/science.1260419
56. Sato S, Maekawa R, Tamura I, et al. SATB2 and NGR1: potential upstream regulatory factors in uterine leiomyo mas. J Assist Reprod Genet. 2019;36(11):2385–2397. doi:10.1007/s10815-019-01582-y
57. Moh M, Krings G, Ates D, Aysal A, Kim GE, Rabban JT. SATB2 expression distinguishes ovarian metastases of colorectal and appendiceal origin from primary ovarian tumors of mucinous or endometrioid type. Am J Surg Pathol. 2016;40(3):419–432. doi:10.1097/PAS.0000000000000553
58. Sletten ET, Smaglyukova N, Ørbo A, Sager G. Expression of nuclear progesterone receptors (nPRs), membrane progesterone receptors (mPRs) and progesterone receptor membrane components (PGRMCs) in the human endometrium after 6 months levonorgestrel low dose intrauterine therapy. J Steroid Biochem Mol Biol. 2020;202:105701. doi:10.1016/j.jsbmb.2020.105701
59. Patel B, Elguero S, Thakore S, Dahoud W, Bedaiwy M, Mesiano S. Role of nuclear progesterone receptor isoforms in uterine pathophysiology. Hum Reprod Update. 2015;21(2):155–173. doi:10.1093/humupd/dmu056
60. Mote PA, Johnston JF, Manninen T, Tuohimaa P, Clarke CL. Detection of progesterone receptor forms A and B by immunohistochemical analysis. J Clin Pathol. 2001;54(8):624–630. doi:10.1136/jcp.54.8.624
61. Yang X, Kui L, Tang M, et al. High-throughput transcriptome profiling in drug and biomarker discovery. Front Genet. 2020;11:19. doi:10.3389/fgene.2020.00019
62. Kapałczyńska M, Kolenda T, Przybyła W, et al. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci. 2018;14(4):910–919.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.