Back to Journals » Pathology and Laboratory Medicine International » Volume 7
BRAF G596R mutation in a slow-progressing melanoma
Received 11 May 2015
Accepted for publication 15 July 2015
Published 17 August 2015 Volume 2015:7 Pages 63—66
DOI https://doi.org/10.2147/PLMI.S88369
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Paul Zhang
Ling Gao, Deqin Ma
Department of Pathology, University of Iowa Hospitals and Clinics, Iowa City, IA, USA
Abstract: More than 50% of melanomas harbor a single V600E point mutation in the kinase domain of the BRAF gene, resulting in constitutive activation of the MAP kinase pathway. The kinase loop in BRAF begins with Asp-Phe-Gly (DFG; codons 594–596). Mutations in this region are rare, and the effect on tumorigenesis or progression is unknown. We present a slow-progressing metastatic melanoma with a p.G596R (c.1786G>C) mutation in an 84-year-old female. This mutation has only been reported in two cases of melanoma in the current literature, with no clinical information available. Mutations in the DFG motif are considered low-activating through indirect binding and allosteric activation of the CRAF protein. Our patient has carried a clinical diagnosis of melanoma for over 25 years, suggesting that the G596R mutation may be associated with her indolent clinical course. It is unknown whether patients carrying this mutation will benefit from vemurafenib therapy. Our patient declined medical treatment.
Keywords: melanoma, BRAF, V600E, G596R
Introduction
Mutations in the kinase domain of the BRAF gene drive tumorigenesis in melanoma through constitutive activation of the MAPK signaling pathway. The V600E mutation accounts for 80%–90% of BRAF mutations in melanomas, and patients with BRAF V600E-positive metastatic melanoma have shown increased overall survival when treated with vemurafenib, a US Food and Drug Administration-approved BRAF inhibitor.1,2
The BRAF kinase activation loop begins with Asp-Phe-Gly (DFG; codons 594–596) and ends with Ala/Pro/Glu (APE; codons 621–623).3 The DFG motif plays an important role in the regulation of kinase activity.3–5 The active conformation of BRAF is characterized by an open conformation of the activation loop, while a closed conformation of the activation loop represents the inactive form.3 The conformation of the DFG motif is also known to affect adenosine triphosphate-substrate binding and the catalytic competency of the kinase.6 Mutations in the DFG activation loop are rare, and information about their oncogenic effect or response to BRAF inhibitors is very limited. We report a G596R mutation in an 84-year-old female who had carried a diagnosis of melanoma for over 25 years.
Materials and methods
Histopathology and immunohistochemistry studies
Formalin-fixed, paraffin-embedded (FFPE) core biopsy tissue from the left level II neck mass was processed, and hematoxylin and eosin (H&E)-stained slides were examined by two pathologists. Immunohistochemistry studies were performed in our clinical immunohistochemistry laboratory using SOX10 (polyclonal antibody at 1:100, catalog number 383A-76, Cell Marque; Sigma-Aldrich Co, St Louis, MO, USA), HMB45 (monoclonal antibody at 1:100, clone HMB45, Dako; Agilent Technologies, Santa Clara, CA, USA), and melan A (monoclonal antibody at 1:25, catalog number M7196; Agilent Technologies). After antigen retrieval with high pH (9.0) solution, the slides were incubated with primary antibody at room temperature for 30 minutes followed by 15-minute incubation with the EnVision Flex Kit (catalog number K8000; Agilent Technologies). The slide was then stained with the chromogen 3,3″-diaminobenzidine for 5 minutes. No informed consent is required for a single case report according to the institutional research review committee of University of Iowa (Iowa City, IA).
DNA extraction and mutational analysis
One H&E-stained slide and ten unstained sections (6 μm in thickness) were cut from the FFPE block. Areas with the highest percentage of tumor and fewest nonneoplastic materials were marked on the H&E slide by a pathologist, and the corresponding areas from the unstained slides were manually microdissected using a razor blade. The paraffin flakes were placed in a 1.5 mL microcentrifuge tube, deparaffinized with 1,200 μL of xylene, vortexed, and centrifuged (16,000 g ×5 minutes). The tissue pellet was washed with 95% ethanol twice before proceeding with the extraction. Genomic DNA was isolated from the macrodissected FFPE sections using the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA). DNA concentration was measured using NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA).
Exon 15 of the BRAF gene was amplified by polymerase chain reaction (PCR) using the following primers: 5′-GATCCCTTTACTTACTACACCTCAG-3′ and 5′-GATCGTAACTCAGCAGCATCTC-3′. The PCR thermocycling conditions were as follows: 94°C for 2 minutes, followed by 30 cycles of 94°C for 15 seconds, 55°C for 15 seconds, and 72°C for 40 seconds. The amplification products were purified using the QIAquick PCR Purification Kit (Qiagen). Cycle sequencing was performed using the ABI Big Dye Terminator Mix, version 3.1. Postcycle sequencing products were purified using the Big Dye X Terminator Purification Kit from Applied Biosystems (Thermo Fisher Scientific) following the manufacturer’s instructions. The products were separated by capillary electrophoresis on an ABI 3130 Genetic Analyzer (Thermo Fisher Scientific).
Case report
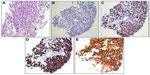
An 84-year-old female was first noted to have a pigmented lesion on her left forearm in 1988. The lesion was nodular and hyperpigmented, and was clinically diagnosed as malignant melanoma. The patient refused to have a biopsy or surgery, but instead elected to proceed with an alternative therapy using Cancell/Cantron. She had been considered “cured” from her disease until she noted a left submandibular lump in 2011. The mass enlarged with no other symptoms. She also noted a left subclavicular mass. Fine-needle aspiration was performed on both lesions at an outside hospital, and the materials were reviewed in our department. Both lesions were diagnosed as poorly differentiated malignant neoplasm with extensive necrosis. Immunohistochemistry studies showed that the tumor cells were negative for AE1/AE3, CK5/6, CK7, TTF1, p63, synaptophysin, and chromogranin. S-100 staining was noncontributory, due to the high background staining. Core biopsy of the left level II neck mass (5.3 cm) was performed at our hospital in 2013. Microscopically, sections showed a poorly differentiated neoplasm with highly pleomorphic malignant cells. The cells had abundant cytoplasm, markedly increased nuclear:cytoplasmic ratio, prominent nucleoli, and were mitotically active, with no glandular formation (Figure 1A). The neoplastic cells were in a background of lymphoplasmacytic cells with no skin identified, suggesting that the level II mass most likely represented a lymph node rather than a primary tumor site. By immunohistochemistry, the tumor cells were negative for AE1/AE3 (Figure 1B), and strongly and diffusely positive for SOX10, HMB45, and melan A (Figure 1C–E). The morphology and immunohistochemical phenotype were consistent with a diagnosis of metastatic malignant melanoma.
Mutation analysis by Sanger sequencing for BRAF exon 15 showed a G–C substitution at position 1786 (c.1786G>C ), which was predicted to cause an amino acid change from a glycine to arginine residue at codon 596 (p.G596R) (Figure 2). In addition to these lesions, she also developed a large, flat, and irregularly shaped hyperpigmented lesion on the left forearm, at approximately the same location as her previous melanoma. An ulcer with hyperpigmentation of the surrounding skin was also noted on her left leg. Positron emission tomography–computed tomography demonstrated two large lymph nodes in left cervical levels IB and IV, compatible with metastatic disease, and possible osseous erosion of the adjacent mandibular angle. The patient refused biopsy or further molecular testing on the additional lesions.
Discussion
V600E accounts for 90% of all the BRAF mutations in melanoma. The mutant protein is 500-fold more potent in activating the MAPK pathway and promoting cell proliferation.7 Other mutations with reduced oncogenic effects or impaired kinase activity within the DFG motif (codons 594–596) of the activation loop have also been identified by in vivo studies.4,5,7,8
Mutations in the DFG domain are rare. The G596R mutation has only been reported in two cases of melanoma by two different groups: an anorectal melanoma9 and a malignant melanoma of unknown site.10 Neither of these cases had available clinical information. This mutation has also been reported in colorectal, lung, and urinary carcinomas and cancer cell lines at a very low frequency.5,11,12 The lack of cytokeratin staining and strong positivity for three melanoma markers (SOX10, HMB45, and melan A), in addition to the morphology, provided convincing evidence that the lesion examined was a malignant melanoma. In vivo studies have shown that unlike the high-activity V600E mutant, which can directly phosphorylate and activate MEK, G596R alone is inactive but can activate MEK and MAPK phosphorylation indirectly by binding and allosterically activating the CRAF protein.4,5,7,8 In addition, the DFG domain is known to regulate BRAF kinase activity by changing the conformation of the kinase domain.6 The introduction of a bulky arginine residue at position 596 of the DFG motif is predicted to keep the conformation of the activating loop in an active conformation and mimic phosphorylation of the kinase.4 All the mutations identified in the DFG domain so far are considered to have low activity or impaired kinase activity. Our patient has carried a clinical diagnosis of malignant melanoma for over 25 years without receiving any treatment except the Cancell/Cantron alternative therapy. The G596R mutation in the DFG region most likely accounts for her indolent clinical course. It is unknown whether patients carrying this mutation will benefit from vemurafenib therapy; however, it is likely that patients carrying this mutation may not be responsive to the drug, since this mutation has lower activating activity for BRAF itself but instead can lead to impaired MEK activity. Our patient declined medical treatment.
There are conflicting findings regarding BRAF-mutation status in primary disease and subsequent metastasis. The reported concordance between primary and metastasis ranges from 68% to 96%.13 Our patient’s original skin lesion from 1988 was not available for testing. The G596R mutation in her metastatic disease may exist in the original melanoma. It may also be newly acquired and contributed to the sudden increase in tumor burden with multiple metastases.
In conclusion, we present a c.1786G>C, p.G586R mutation in the BRAF DGF motif in an 84-year-old female patient who had a slow-progressing malignant melanoma for 25 years. This case provided the first evidence of patient data in the literature that the G596R mutation, which has reduced BRAF oncogenic activity in melanoma, may be associated with the indolent clinical course of the patient.
Acknowledgment
The authors would like to thank Jacqueline Lekostaj, MD, PhD for critically reviewing the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. | |
Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. | |
Roskoski R Jr. RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399(3):313–317. | |
Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. | |
De Falco V, Giannini R, Tamburrino A, et al. Functional characterization of the novel T599I-VKSRdel BRAF mutation in a follicular variant papillary thyroid carcinoma. J Clin Endocrinol Metab. 2008;93(11):4398–4402. | |
Bollag G, Tsai J, Zhang J, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11(11):873–886. | |
Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20(6):963–969. | |
Moretti S, De Falco V, Tamburrino A, et al. Insights into the molecular function of the inactivating mutations of B-Raf involving the DFG motif. Biochim Biophys Acta. 2009;1793(11):1634–1645. | |
Ni S, Huang D, Chen X, et al. C-kit gene mutation and CD117 expression in human anorectal melanomas. Hum Pathol. 2012;43(6):801–807. | |
Beadling C, Heinrich MC, Warrick A, et al. Multiplex mutation screening by mass spectrometry evaluation of 820 cases from a personalized cancer medicine registry. J Mol Diagn. 2011;13(5):504–513. | |
Kinno T, Tsuta K, Shiraishi K, et al. Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Ann Oncol. 2014; 25(1):138–142. | |
Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. | |
Saroufim M, Habib RH, Gerges R, et al. Comparing BRAF mutation status in matched primary and metastatic cutaneous melanomas: implications on optimized targeted therapy. Exp Mol Pathol. 2014;97(3):315–320. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.