Back to Journals » International Journal of General Medicine » Volume 17
Bioinformatics Analysis and Experimental Validation of Mitochondrial Autophagy Genes in Knee Osteoarthritis
Authors Tang K , Sun L, Chen L, Feng X, Wu J, Guo H, Zheng Y
Received 15 October 2023
Accepted for publication 6 February 2024
Published 23 February 2024 Volume 2024:17 Pages 639—650
DOI https://doi.org/10.2147/IJGM.S444847
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Woon-Man Kung
Kuihan Tang,1 Li Sun,2 Long Chen,2 Xiaobo Feng,1 Jiarui Wu,2 Hao Guo,2 Yong Zheng1
1Department of Orthopedics, Beijing Jishuitan Hospital Guizhou Hospital, Guiyang, 550014, People’s Republic of China; 2Department of Orthopedics, Guizhou Provincial People’s Hospital, Guiyang, 550000, People’s Republic of China
Correspondence: Yong Zheng, Email [email protected]
Background: Mitochondrial autophagy is closely related to the pathogenesis of osteoarthritis, In order to explore the role of mitochondrial autophagy related genes in knee osteoarthritis (KOA) and its molecular mechanism.
Methods: KOA-related transcriptome data were extracted from the Gene Expression Omnibus (GEO) database. Differentially expressed mitochondrial autophagy gene (DEMGs) were screened in patients with KOA by differential expression analysis. The STRING website was used to construct a protein-protein interaction (PPI) network among DEMGs. Molecular complex detection (MCODE) method in Cytoscape software was performed to identify hub DEMGs. Support vector machine recursive feature elimination (SVM-RFE) method was used to construct the hub DEMG diagnosis model. Genes with diagnostic value were identified as biomarkers by plotting receiver operating characteristic (ROC) curves and Expression validation. CIBERSORT algorithm was used to calculate the proportion of 22 immune cells in each sample in the GSE114007 dataset. Finally, biomarker expression was verified by qPCR.
Results: A total of 15 DEMGs were obtained and enrichment analyses showed that these DEMG strains were mainly enriched in the mitophagy-animal, shigellosis, autophagy-animal and FoxO signal pathways. The PPI network unveiled 13 DEMGs with interactions. In addition, 8 hub DEMGs (ULK1, CALCOCO2, MAP1LC3B, BNIP3L, GABARAPL1, BNIP3, FKBP8 and FOXO3) were obtained for KOA. And 5 model DEMGs (BNIP3L, BNIP3, MAP1LC3B, ULK1 and FOXO3) were screened. The ROC curves revealed that BNIP3 and FOXO3 has strong diagnostic value in these models of DEMG. Immune-infiltration and correlation analysis showed that BNIP3 and FOXO3 were significantly correlated with three different immune cells, including primary B cells, M0 macrophage and M2 macrophage. The cartilage tissue samples qPCR verification results show that FOXO3 and BNIP3 were all down-regulated in KOA (p < 0.01), and the validation results are consistent with the above analysis.
Conclusion: BNIP3 and FOXO3 have been identified as biomarkers for the diagnosis of KOA, which might supply a new insight for the pathogenesis and treatment of KOA.
Keywords: Gene Expression Omnibus, diagnostic, bioinformatics analysis, immune infiltration, biomarkers
Background
Knee Osteoarthritis (KOA) is a chronic and progressive disease characterized by loss of articular cartilage, subchondral sclerosis, and synovial and periarticular structural abnormalities that cause pain, swelling and even joint deformity in the surrounding tissues of the knee.1 Its pathogenic factors include abnormal biologic factors, genetic factors, cell aging and apoptosis, local inflammatory factors, free radicals and proteases, etc. Each of these risk factors may cause KOA, KOA is a degenerative, attritional disease,2 its pathology mainly involves articular cartilage and synovium, the key factor of KOA cartilage degeneration is chondrocyte death,3 which is caused by apoptosis and autophagy.4,5 Under normal conditions, articular chondrocytes maintain a dynamic balance between extracellular matrix (ECM) synthesis and degradation,6 which is broken in response to injury or inflammation and other stimuli, triggering chondrocyte stress and ECM degradation, it leads to abnormal accumulation of damaged proteins and dysfunction of organelles such as endoplasmic reticulum and mitochondria,7,8 thus, cartilage degradation, chondrocyte apoptosis, subchondral bone dysfunction, and gradually develop into knee osteoarthritis.9 According to the latest Global Burden of Disease study, about 250 million people worldwide suffer from osteoarthritis, and knee joint is the most common site of osteoarthritis.2 Due to the lack of early diagnostic indicators, patients with KOA often miss the opportunity for optimal treatment, resulting in a poor prognosis.
Mitochondria are membrane-coated organelles present in eukaryotic cells that regulate important cell functions and cell survival. Mitochondria have many important functions, including cell adenosine triphosphate (ATP) production, Ca2+ buffering, and being the main source of endogenous reactive oxygen species (ROS) under oxidative stress.10 Mitochondrial autophagy is a major intracellular protective mechanism, which can effectively promote the clearance of damaged mitochondria and thus maintain mitochondrial function.11,12 Mitochondria can protect chondrocytes and provide energy for cells, which is an important mechanism to maintain cell homeostasis and is closely related to the occurrence and development of cartilage degeneration in osteoarthritis.13 Inhibition of autophagy can lead to mitochondrial dysfunction and catabolic imbalance, leading to chondrocyte injury and apoptosis,14 thus leading to osteoarthritis.
In recent years, bioinformatics has provided powerful strategies for the screening of molecular markers, and cell classification tools have also facilitated the analysis of infiltration patterns of disease immune cells. Based on the role of mitochondrial autophagy in osteoarthritis, we explored the mechanism of mitochondrial autophagy gene in knee arthritis, attempted to identify diagnostic markers of KOA, and further explored the role of immune cell infiltration in KOA, providing theoretical possibilities for the development of new targeted drugs, which is of great significance for the study of the pathogenesis and prevention of KOA.
Materials and Methods
Data Extraction
RNA-sequencing (RNA-seq) data of KOA patients (GSE114007, GSE169077 and GSE51588) were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GSE114007 dataset, including cartilaginous tissues from 20 KOA samples and 18 normal samples, was used as the training set. The GSE169077 dataset, containing 6 KOA samples and 5 normal samples, was used as the validation set. The GSE51588 dataset (KOA: normal = 40:10) was also used as validation set. In addition, 88 mitochondrial autophagy gene (MGs) were downloaded for this study based on the relevant literature (Sun et al 2021).
Differential Expression Analysis
In the GSE114007 dataset, differentially expressed genes (DEGs) between KOA samples and normal samples were detected utilizing the “DESeq2” R package (version 1.34.0) with an adjusted p<0.05 and a |log2FC|>0.5. Clinical characteristics (gender and age) were also controlled for to exclude the effect of other confounding factors. Subsequently, the DEGs were intersected with MGs using the “VennDiagram” R package (version 1.6.20) (Xin et al 2022) to get differentially expressed MGs (DEMGs). Subsequently, The Gene Ontology (GO) and The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the DEMGs were performed using the “clusterProfiler” R package (Jahr et al 2019; Lazarou 2015).
PPI Network Construction and Hub DEMGs Screening
To explore the protein interactions among DEMGs, the STRING (https://string-db.org) website was used to construct a PPI network, and DEMGs with interaction scores greater than 0.4 were retained. Moreover, hub DEMGs were identified using MCODE algorithm in Cytoscape (Xin et al 2022).
Construction and Validation of a Diagnostic Model for DEMGs
Based on the identified hub DEMGs, the SVM-RFE method was applied to screened the model DEMGs using the “caret” R package (version 6.0–91, https://CRAN.R-project.org/package=caret). Subsequently, diagnostic value of the model DEMGs was validated in a validation set (GSE169077). In addition, we used the “pROC” R package (version 1.17.0.1) to map the ROC curve to assess their diagnostic value. Finally, the model DEMGs with diagnostic value were validated in the training and validation set.
Immune Cell Infiltration with CIBERSORT
To further explore the correlation between model DEMGs and different immune cells, the infiltration of 22 immune cells in the GSE114007 dataset was estimated by CIBERSORT algorithm. Then, the “psych” R package (version 2.2.3) was performed between model DEMGs and immune cells.
Experimental Verification
Cartilage tissue samples were obtained from KOA patients and normal samples with their knowledge and consent from Beijing Jishuitan Hospital Guizhou Hospital, and this study was approved by the Beijing Jishuitan Hospital Guizhou Hospital ethics committee, all methods were performed in accordance with relevant guidelines and regulations in the《Ethical considerations in the review of biomedical research involving human subjects》and the《Declaration of Helsinki》(approval number: LW20221206).
10 pairs of frozen cartilage tissue samples were divided into two groups, of which 10 samples were normal group (Normal) and the other 10 samples were KOA group (Case). Then, total RNA of samples was isolated and purified by TRIzol (Ambion) reagent following the instruction manual. Then, the extracted RNA was tested for concentration by NanoPhotometer N50. Next, reverse transcription via SureScript-First-strand-cDNA-synthesis-kit (Servicebio) by an ordinary PCR instrument. Reverse transcription product cDNA was diluted 5–20 times with ddH2O (RNase / DNase free). Subsequently, polymerase chain reaction (PCR) amplification reaction was performed by CFX96 real-time quantitative PCR instrument. 1 min at 95 °C (pre-denaturation), followed by at 95 °C for 20s (denaturation), 55 °C for 20s (annealing) and 72 °C for 30s (elongation). The above reactions were subjected to forty cycles. Primer sequences were showed in Table 1.
 |
Table 1 The Primer Sequences of Diagnostic Genes |
Statistical Analysis
All statistical analyses and visual plotting of the results were performed based on R software (https://www.r-project.org/, version 4.0.3, R Statistical Computing Project). Correlation of model DEMGs with immune cells using Spearman coefficient analysis.
Results
Identification of DEMGs
To screen DEMGs in KOA, we firstly screened DEGs between KOA samples and normal samples in the GSE114007 dataset. A total of 4279 DEGs, including 1969 down-regulated and 2310 up-regulated in KOA samples, were identified (Figure 1a and b). Comparison of differences in clinical characteristics revealed no significant effect. (Table S1). Then, 15 DEMGs were obtained for subsequent analysis by intersecting DEGs and MGs (Figure 1c).
GO and KEGG Analysis of the DEMGs
To explore the regulatory pathways and biological functions associated with the expression of DEMGs, we performed GO and KEGG analyses on these DMAEGs in the GSE114007 dataset. GO enrichment analysis suggested that in biological process (BP), these DEMGs were mainly involved in 5 items such as organelle disassembly, macroautophagy and cellular component disassembly; these DEMGs were mainly involved in 5 items such as mitochondrial outer membrane, organelle outer membrane and outer membrane in cellular component (CC); in molecular function (MF), these DEMGs are mainly involved in two items, ubiquitin protein ligase binding and ubiquitin-like protein ligase binding (Figure 2a). Moreover, KEGG pathway enrichment analysis indicated that these DEMGs were significantly associated with Mitophagy-animal, Shigellosis, Autophagy-animal, and FoxO signaling pathway pathways (Figure 2b).
Identification of the Hub DEMGs Through PPI Network
To explore the interaction between 15 DEMGs, the STRING website was applied to build a PPI network. Ultimately, a PPI network including 36 interactions and 13 nodes was obtained (Figure 3a). In addition, 8 hub DEMGs (ULK1, CALCOCO2, MAP1LC3B, BNIP3L, GABARAPL1, BNIP3, FKBP8, and FOXO3) were screened by the MCODE algorithm (Figure 3b).
Construction and Validation the DEMGs Diagnostic Model for KOA Patients
To further validation and selection of the DEMGs with significantly characteristic value of classifying KOA and normal samples from GSE114007 dataset, the SVM-RFE algorithm was performed to identify 5 model DEMGs (BNIP3L, BNIP3, MAP1LC3B, ULK1, and FOXO3) (Figure 4a). Meanwhile, we evaluated and validated the diagnostic value of the 5 model DEMGs in the GSE114007 and GSE169077 datasets, respectively. As shown in Figure 4b, the AUC values of these genes were above 0.9 in the training set, indicating that these genes have a strong diagnostic value. However, in the validation set, only BNIP3 and FOXO3 had AUC values, and its AUC values greater than 0.7 (Figure 4c). Additionally significant differences and consistent trends in BNIP3 and FOXO3 expression were found in the training and validation sets by expression analysis (Figure S1). Thus, these two genes were identified as having strong diagnostic value and were used for subsequent analysis.
Correlation Analysis of BNIP3 and FOXO3 in Immune Cells
To accurately assess the composition of immune cells in the KOA tissue microenvironment, the CIBERSORT algorithm was used to estimate the abundance of immune infiltrating cells of 22 different cell types and functional states between KOA and normal samples in the GSE114007 dataset. The results revealed that of the 22 immune cells only naive B cells, M0 Macrophages, and M2 Macrophages were significantly different between the KOA and normal samples (Figure 5a). Furthermore, the correlation analysis of these immune cells with BNIP3 as well as FOXO3 showed that BNIP3 was significantly negatively correlated with M0 Macrophages (|cor|>0.3 and p value<0.05); while FOXO3 was significantly negatively correlated with M0 Macrophages and showed a significant positive correlation with naive B cells (Figure 5b and c).
Quantitative Real-Time PCR (qPCR) Identification
Based on the qPCR verification results, it can be seen that FOXO3 and BNIP3 were all down-regulated in KOA (p < 0.01), and the validation results are consistent with the above analysis (Figure 6, Table 2).
 |
Table 2 qPCR Verification Results |
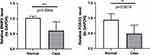 |
Figure 6 qPCR validation of the BNIP3 and FOXO3 between KOA (Case) and normal controls. **p < 0.01, ***p < 0.001. Abbreviation: NS, not significant. |
Discussion
Mitochondrial autophagy can protect human chondrocytes from a series of pathological effects caused by mitochondrial dysfunction and effectively slow down the progression of cartilage degeneration in KOA. In the past, it was thought that articular chondrocytes were in an ischemic and anoxic environment, and the role of mitochondria in the metabolic activities of chondrocytes was relatively limited. However, recent studies have shown that chondrocytes on the surface of cartilage are in a relatively aerobic state, and mitochondria-mediated aerobic respiration is still an important energy supply mode for articular chondrocytes.15 Mitochondria are involved in catabolism and anabolism, production of ROS, cell apoptosis and signal transduction. When the body is subjected to oxidative stress, endoplasmic reticulum stress, hypoxia ischemia, nutrient deficiency and other injuries, Mitochondrial autophagy disorders lead to mitochondrial dysfunction and catabolic imbalance.16 Mitochondrial damage can lead to the release of interleukin-1β (IL-1β), which further leads to chondrocyte death and ECM degradation. Overactivation of mitochondrial autophagy can lead to autophagy death and osteoarthritis.14,17
In this study, a total of 4279 DEGs were obtained from the GSE114007 dataset, 15 DEMGs were obtained by crossing DEGs and MGs. GO enrichment analysis showed that these DEMGs were mainly involved in organelle decomposition, macroautophagy and cell component decomposition in biological processes (BP). In the same type of cell, autophagy can be protective or harmful, depending on the type and degree of stimulation. Maintaining a healthy mitochondrial population is essential for cell survival. As a kind of autophagy, macrophage plays an important and complex role in bone metabolism. Eukaryotic cells can regulate molecular degradation and organelle renewal through macrophage, and play an important role in bone regeneration, bone metabolism, and bone degenerative diseases such as osteoarthritis.18 In addition, the main BP of KOA included inflammatory response regulation, collagen catabolic process, extracellular matrix decomposition, etc. When mechanical injury or inflammatory stimulation exceeds the compensatory capacity of articular cartilage, chondrocyte stress and ECM degradation are triggered, thus causing cartilage degeneration and gradually developing into KOA.19,20 KOA can be alleviated by inhibiting inflammation, chondro breakdown, and extracellular matrix degradation.21 In the cell component (CC), these DEMGs mainly involved 5 items such as mitochondrial outer membrane, organelle outer membrane and outer membrane. Mitochondrial outer membrane is a biological membrane located in the outermost layer of mitochondria. Under stimulation, Mitochondrial outer membrane permeabilization (MOMP) can permeabilization. ROS released by damaged chondrocytes promote the expression of matrix metalloproteinases such as MMP13. Meanwhile, excessive ROS generation leads to mitochondrial dysfunction and enhanced permeability of mitochondrial outer membrane.22 Mitochondrial proteins are released into the cytoplasm to mediate downstream reactions, Bnip3 is localized in the mitochondrial outer membrane, it induces mitochondrial autophagy in the presence of mitochondrial membrane permeability and Bax/Bak deficiency.23 In molecular function (MF), these DEMGs are mainly involved in ubiquitin-protein ligase binding and ubiquitin-like protein ligase binding. The ubiquitin-proteasome system is an important regulatory pathway to maintain protein homeostasis and function in cells, and abnormal regulation of ubiquitination can lead to metabolic bone disease. E3 ubiquitin ligases (E3) are key regulatory factors in the proliferation and differentiation of osteoblasts, play an important role in the regulation of bone formation related proteins and bone turnover, and have been shown to be ideal therapeutic targets for promoting bone formation and reducing bone loss,24 The degradation of BNIP3 by ubiquitination under severe hypoxia condition can lead to the inhibition of mitochondrial autophagy.25 KEGG pathway enrichment analysis showed that these DEMGs are mainly related to mitochondrial and autophagy signaling pathways, FoxO signaling pathway plays a function regulation role in bone cells, and is associated with bone metabolic diseases such as osteoarthritis and osteoporosis.26 FoxO transcription factor can prevent cellular and biological senescence, and its expression in OA cartilage is decreased with aging.27 Promote cartilage regeneration by up-regulating FOXO signaling pathway and alleviate the progression of osteoarthritis.28
In order to explore the interaction between DEMGs, we constructed a PPI network with the differential expression of mitochondrial autophagy genes using STRING, and used the MCODE algorithm to screen hub genes. Then, the SVM-RFE algorithm was used to verify the diagnostic markers of KOA, and the diagnostic value was evaluated and verified. Finally, BNIP3 and FOXO3 have been identified as potential biomarkers for the treatment of KOA patients, play a key role in the progression of KOA and can be used as therapeutic targets. BNIP3 is a mitochondrial outer membrane protein belonging to the Bcl-2 protein family, which regulates apoptosis, autophagy, mitochondrial transformation and cell protection.29 BNIP3 is an important mediator of autophagy.30 As a dual regulator, it has pro-apoptotic function.31 and controversial anti-apoptotic function.32,33 BNIP3 induces mitochondrial autophagy under various stress states. For example, under hypoxia conditions, BNIP3 promotes mitochondrial depolarization and directly activates mitochondrial autophagy by transporting mitochondria to autophagosomes.34,35 Bnip3 knockout reduces mitochondrial autophagy, resulting in accumulation of damaged mitochondria and increased production of reactive oxygen species.32 BNIP3 induced mitochondrial dysfunction mainly by activating Bax/Bak and the mitochondrial permeability transition pore (mPTP).36 Overexpression of BNIP3 inhibits HIF-1α knockout from enhancing H/R-induced apoptosis and ROS production, reverses the decline in mitochondrial autophagy during injury,37 and protects mitochondrial structure. The loss of BNIP3 leads to the accumulation of abnormal mitochondria,38 aggravates the damage of mitochondrial structure, significantly reduces mitochondrial autophagy, and thus promotes apoptosis.39 FOXO3 is a member of the Fox O transcription factor family, regulating cell survival, apoptosis, autophagy, oxidative stress and inflammatory response.40,41 It is a key transcription factor involved in pro-inflammatory cytokine induced chondrocyte metabolic disorders, and overexpression prevents IL-1β or TNF-α induced inflammation in various cell types.42,43 FOXO3 regulates apoptosis of chondrocytes44 and maintains meniscus and cartilage homeostasis,27,45 which may play an important role in the development of KOA. FOXO3 knockout mice resulted in more severe and earlier age-related OA-like changes at 18 months, and the primary cause appeared to be a decrease in protective genes (autophagy, antioxidants).27 Previous research found that FoxO1 can activate mitochondrial autophagy by activating the downstream target gene BNIP3.46 FOXO3 increases the expression of BNIP3 by binding to the upstream promoter region of BNIP3, thereby regulating mitochondrial function and integrity.47 In order to verify the expression of these autophagy related genes in KOA, we took clinical normal cartilage and KOA cartilage for qPCR verification, and the results showed that FOXO3 and BNIP3 were down-regulated in KOA cartilage, and the verification results were consistent with the above analysis. BNIP3 and FOXO3 were identified as biomarkers for the diagnosis of KOA, which may provide new insights into the pathogenesis of KOA.
The immune system plays a very important role in the development of osteoarthritis. Immune cell infiltration mediates the autoimmune response of osteoarthritis, induces the secretion of chemokines, pro-inflammatory cytokines and proteases, and thus disturbs the immune balance to accelerate cartilage erosion.48–50 To find specific diagnostic markers and analyze the infiltration pattern of KOA immune cells is of great significance for improving the prognosis of KOA patients. The results showed that naive B cells, M0 Macrophages and M2 Macrophages differ significantly between KOA and normal samples. The proportion of naive B cells, T cells CD8 and follicular helpers T-cells in the articular cartilage tissue of KOA patients was less. The memory resting T-cell CD4, M0 Macrophages and M2 Macrophages accounted for a larger proportion. Correlation analysis between BNIP3 and FOXO3 showed that BNIP3 and FOXO3 were negatively correlated with memory resting T-cell CD4, M0 Macrophages and M2 Macrophages. BNIP3 was positively correlated with T-cell CD8 and FOXO3 was positively correlated with B cells naive. M0 Macrophage was stimulated by interferon IFN-γ and lipopolysaccharide LPS to obtain M1-type macrophages and secrete a large number of pro-inflammatory factors. M0 Macrophage was stimulated by interleukin4 (IL-4) to obtain M2-type macrophages, which had anti-inflammatory and tissue repair effects.51 We speculate that BNIP3 and FOXO3 affect T-cell CD4 memory resting, M0 Macrophages and M2 Macrophages, and thus participate in the occurrence and progression of KOA. Previous studies have found that the inflammation of KOA is characterized by immune cell infiltration and cytokine secretion, but different from the inflammation of RA, the content of macrophages, T cells and B cells in the synovium of patients is higher than that of healthy patients but lower than that of RA patients.52 End-stage KOA articular-derived T cells polarize into CD3 + CD4 + CD8-T cell subsets, increasing the polarization of CD4 + T cells towards activated Th1 cells and cytokine secretion, thereby increasing local inflammation.53 The specific effects of these differentially expressed chemokines on immune infiltration of KOA remain to be further studied.
Conclusion
In this study, we found that BNIP3 and FOXO3 were diagnostic biomarkers of KOA. We also found that naive B cells, M0 Macrophages, and M2 Macrophages may play a key role in the occurrence and development of KOA, and further exploration of these immune cells may determine the target of KOA immunotherapy. However, our study also has certain limitations, most of the current studies remain theoretical, and the relationship between key genes and immune infiltration as well as their role in the development of KOA needs further research. In order to achieve the ultimate goal of KOA treatment, further clinical verification is needed.
Data Sharing Statement
The GSE114007, GSE169077 and GSE51588 datasets analyzed in this study were collected from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Ethics Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Beijing Jishuitan Hospital Guizhou Hospital ethics committee (approval number: LW20221206).
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent to Publish
The authors affirm that human research participants provided informed consent for publication of the images in Figure 6.
Acknowledgments
We thank the patients for their cooperation and continued support for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Disclosure
The authors have no relevant financial or non-financial interests to disclose for this work.
References
1. Primorac D, Molnar V, Rod E, et al. Knee osteoarthritis: a review of pathogenesis and State-Of-The-Art Non-Operative therapeutic considerations. Genes. 2020;11:854.
2. Fu K, Robbins SR, McDougall JJ. Osteoarthritis: the genesis of pain. Rheumatology. 2018;57(suppl_4):iv43–iv50. doi:10.1093/rheumatology/kex419
3. Charlier E, Relic B, Deroyer C, et al. Insights on molecular mechanisms of chondrocytes death in osteoarthritis. Int J Mol Sci. 2016;17(12):2146. doi:10.3390/ijms17122146
4. Shin HJ, Park H, Shin N, et al. Pink1-mediated chondrocytic mitophagy contributes to cartilage degeneration in osteoarthritis. J Clin Med. 2019;8(11):1849. doi:10.3390/jcm8111849
5. Almonte-Becerril M, Navarro-Garcia F, Gonzalez-Robles A, Vega-Lopez MA, Lavalle C, Kouri JB. Cell death of chondrocytes is a combination between apoptosis and autophagy during the pathogenesis of Osteoarthritis within an experimental model. Apoptosis. 2010;15(5):631–638. doi:10.1007/s10495-010-0458-z
6. Zhang Y, Vasheghani F, Li YH, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2015;74(7):1432–1440. doi:10.1136/annrheumdis-2013-204599
7. Wang T, He C. Pro-inflammatory cytokines: the link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018;44:38–50. doi:10.1016/j.cytogfr.2018.10.002
8. Uehara Y, Hirose J, Yamabe S, et al. Endoplasmic reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthritis Cartilage. 2014;22(7):1007–1017. doi:10.1016/j.joca.2014.04.025
9. Michael JW, Schlüter-Brust KU, Eysel P. The epidemiology, etiology, diagnosis, and treatment of osteoarthritis of the knee. Dtsch Arztebl Int. 2010;107(9):152–162. doi:10.3238/arztebl.2010.0152
10. Kurihara Y, Kanki T, Aoki Y, et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287(5):3265–3272. doi:10.1074/jbc.M111.280156
11. Kriegova E, Manukyan G, Mikulkova Z, et al. Gender-related differences observed among immune cells in synovial fluid in knee osteoarthritis. Osteoarthritis Cartilage. 2018;26(9):1247–1256. doi:10.1016/j.joca.2018.04.016
12. Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–1022. doi:10.1038/s41556-018-0176-2
13. Sun K, Jing X, Guo J, Yao X, Guo F. Mitophagy in degenerative joint diseases. Autophagy. 2021;17(9):2082–2092. doi:10.1080/15548627.2020.1822097
14. Xin R, Xu Y, Long D, et al. Mitochonic Acid-5 inhibits reactive oxygen species production and improves human chondrocyte survival by upregulating SIRT3-mediated, parkin-dependent mitophagy. Front Pharmacol. 2022;13:911716. doi:10.3389/fphar.2022.911716
15. Jahr H, Gunes S, Kuhn AR, Nebelung S, Pufe T. Bioreactor-controlled physoxia regulates TGF-β signaling to alter extracellular matrix synthesis by human chondrocytes. Int J Mol Sci. 2019;21(1):20. doi:10.3390/ijms21010020
16. Lazarou M. Keeping the immune system in check: a role for mitophagy. Immunol Cell Biol. 2015;93(1):3–10. doi:10.1038/icb.2014.75
17. Wu C, Zheng J, Yao X, et al. Defective autophagy in chondrocytes with Kashin-Beck disease but higher than osteoarthritis. Osteoarthritis Cartilage. 2014;22(11):1936–1946. doi:10.1016/j.joca.2014.08.010
18. Wang J, Zhang Y, Cao J, et al. The role of autophagy in bone metabolism and clinical significance. Autophagy. 2023;19(9):1–19.
19. Kraus VB, Blanco FJ, Englund M, Karsdal MA, Lohmander LS. Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthritis Cartilage. 2015;23(8):1233–1241. doi:10.1016/j.joca.2015.03.036
20. Malemud CJ. Matrix metalloproteinases and synovial joint pathology. Prog Mol Biol Transl Sci. 2017;148:305–325.
21. Zhang J, Fan F, Liu A, et al. Icariin: a potential molecule for treatment of knee osteoarthritis. Front Pharmacol. 2022;13:811808. doi:10.3389/fphar.2022.811808
22. Shang J, Li H, Wu B, et al. CircHIPK3 prevents chondrocyte apoptosis and cartilage degradation by sponging miR −30a-3p and promoting PON2. Cell Prolif. 2022;55(9):e13285. doi:10.1111/cpr.13285
23. Li S, Kuang M, Chen L, et al. The mitochondrial protein ERAL1 suppresses RNA virus infection by facilitating RIG-I-like receptor signaling. Cell Rep. 2021;34(3):108631. doi:10.1016/j.celrep.2020.108631
24. Jonason JH, Xiao G, Zhang M, Xing L, Chen D. Post-translational regulation of Runx2 in bone and cartilage. J Dent Res. 2009;88(8):693–703. doi:10.1177/0022034509341629
25. He YL, Li J, Gong SH, et al. BNIP3 phosphorylation by JNK1/2 promotes mitophagy via enhancing its stability under hypoxia. Cell Death Dis. 2022;13(11):966. doi:10.1038/s41419-022-05418-z
26. Ma X, Su P, Yin C, et al. The roles of FoxO transcription factors in regulation of bone cells function. Int J Mol Sci. 2020;22(1):21. doi:10.3390/ijms22010021
27. Matsuzaki T, Alvarez-Garcia O, Mokuda S, et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. 2018;10(428):eaan0746.
28. Kurakazu I, Akasaki Y, Hayashida M, et al. FOXO1 transcription factor regulates chondrogenic differentiation through transforming growth factor β1 signaling. J Biol Chem. 2019;294(46):17555–17569. doi:10.1074/jbc.RA119.009409
29. Maes H, Van Eygen S, Krysko DV, et al. BNIP3 supports melanoma cell migration and vasculogenic mimicry by orchestrating the actin cytoskeleton. Cell Death Dis. 2014;5(3):e1127. doi:10.1038/cddis.2014.94
30. Gong LJ, Wang XY, Gu WY, Wu X. Pinocembrin ameliorates intermittent hypoxia-induced neuroinflammation through BNIP3-dependent mitophagy in a murine model of sleep apnea. J Neuroinflammation. 2020;17(1):337. doi:10.1186/s12974-020-02014-w
31. Doerflinger M, Glab JA, Puthalakath H. BH3-only proteins: a 20-year stock-take. Febs j. 2015;282(6):1006–1016. doi:10.1111/febs.13190
32. Tang C, Han H, Liu Z, et al. Activation of BNIP3-mediated mitophagy protects against renal ischemia-reperfusion injury. Cell Death Dis. 2019;10(9):677. doi:10.1038/s41419-019-1899-0
33. Rikka S, Quinsay MN, Thomas RL, et al. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18(4):721–731. doi:10.1038/cdd.2010.146
34. Zhang T, Xue L, Li L, et al. BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J Biol Chem. 2016;291(41):21616–21629. doi:10.1074/jbc.M116.733410
35. O’Sullivan TE, Johnson LR, Kang HH, Sun JC. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity. 2015;43(2):331–342. doi:10.1016/j.immuni.2015.07.012
36. Gustafsson AB. Bnip3 as a dual regulator of mitochondrial turnover and cell death in the myocardium. Pediatr Cardiol. 2011;32(3):267–274. doi:10.1007/s00246-010-9876-5
37. Fu ZJ, Wang ZY, Xu L, et al. HIF-1α-BNIP3-mediated mitophagy in tubular cells protects against renal ischemia/reperfusion injury. Redox Biol. 2020;36:101671. doi:10.1016/j.redox.2020.101671
38. Liu K, Zhao Q, Sun H, et al. BNIP3 (BCL2 interacting protein 3) regulates pluripotency by modulating mitochondrial homeostasis via mitophagy. Cell Death Dis. 2022;13(4):334. doi:10.1038/s41419-022-04795-9
39. Zhang XB, Chen GP, Huang MH, et al. Bcl-2 19-kDa interacting protein 3 (BNIP3)-mediated mitophagy attenuates intermittent hypoxia-induced human renal tubular epithelial cell injury. Med Sci Monit. 2022;28:e936760. doi:10.12659/MSM.936760
40. Brandstetter B, Dalwigk K, Platzer A, et al. FOXO3 is involved in the tumor necrosis factor-driven inflammatory response in fibroblast-like synoviocytes. Lab Invest. 2019;99(5):648–658. doi:10.1038/s41374-018-0184-7
41. Song D, Ma J, Chen L, et al. FOXO3 promoted mitophagy via nuclear retention induced by manganese chloride in SH-SY5Y cells. Metallomics. 2017;9(9):1251–1259. doi:10.1039/C7MT00085E
42. Cui M, Huang Y, Tian C, Zhao Y, Zheng J. FOXO3a inhibits TNF-α- and IL-1β-induced astrocyte proliferation: implication for reactive astrogliosis. Glia. 2011;59(4):641–654. doi:10.1002/glia.21134
43. Kuo SJ, Liu SC, Huang YL, et al. TGF-β1 enhances FOXO3 expression in human synovial fibroblasts by inhibiting miR-92a through AMPK and p38 pathways. Aging. 2019;11(12):4075–4089. doi:10.18632/aging.102038
44. Zhao C, Li X, Sun G, et al. CircFOXO3 protects against osteoarthritis by targeting its parental gene FOXO3 and activating PI3K/AKT-mediated autophagy. Cell Death Dis. 2022;13(11):932. doi:10.1038/s41419-022-05390-8
45. Lee KI, Choi S, Matsuzaki T, et al. FOXO1 and FOXO3 transcription factors have unique functions in meniscus development and homeostasis during aging and osteoarthritis. Proc Natl Acad Sci U S A. 2020;117(6):3135–3143. doi:10.1073/pnas.1918673117
46. Huang C, Lin MZ, Cheng D, Braet F, Pollock CA, Chen XM. Thioredoxin-interacting protein mediates dysfunction of tubular autophagy in diabetic kidneys through inhibiting autophagic flux. Lab Invest. 2014;94(3):309–320. doi:10.1038/labinvest.2014.2
47. Lu P, Kamboj A, Gibson SB, Anderson CM. Poly(ADP-ribose) polymerase-1 causes mitochondrial damage and neuron death mediated by Bnip3. J Neurosci. 2014;34(48):15975–15987. doi:10.1523/JNEUROSCI.2499-14.2014
48. Faust HJ, Zhang H, Han J, et al. IL-17 and immunologically induced senescence regulate response to injury in osteoarthritis. J Clin Invest. 2020;130(10):5493–5507. doi:10.1172/JCI134091
49. Ma Y, Yang H, Zong X, et al. Artificial M2 macrophages for disease-modifying osteoarthritis therapeutics. Biomaterials. 2021;274:120865.
50. Chen Z, Ma Y, Li X, Deng Z, Zheng M, Zheng Q. The immune cell landscape in different anatomical structures of knee in osteoarthritis: a gene expression-based study. Biomed Res Int. 2020;2020:9647072. doi:10.1155/2020/9647072
51. Chen L, Yao Z, Zhang S, et al. Biomaterial-induced macrophage polarization for bone regeneration. Chin Chem Lett. 2023;34(6):107925. doi:10.1016/j.cclet.2022.107925
52. de Lange-Brokaar BJ, Ioan-Facsinay A, van Osch GJ, et al. Synovial inflammation, immune cells and their cytokines in osteoarthritis: a review. Osteoarthritis Cartilage. 2012;20(12):1484–1499. doi:10.1016/j.joca.2012.08.027
53. Rosshirt N, Hagmann S, Tripel E, et al. A predominant Th1 polarization is present in synovial fluid of end-stage osteoarthritic knee joints: analysis of peripheral blood, synovial fluid and synovial membrane. Clin Exp Immunol. 2019;195(3):395–406. doi:10.1111/cei.13230
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.