Back to Journals » Infection and Drug Resistance » Volume 16
Biofilm Formation and Aspartyl Proteinase Activity and Their Association with Azole Resistance Among Candida albicans Causing Vulvovaginal Candidiasis, Egypt
Authors Gerges MA , Fahmy YA, Hosny T , Gandor NH , Mohammed SY, Mohamed TMA , Abdelmoteleb NEM, Esmaeel NE
Received 23 May 2023
Accepted for publication 1 August 2023
Published 14 August 2023 Volume 2023:16 Pages 5283—5293
DOI https://doi.org/10.2147/IDR.S420580
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Marian A Gerges,1 Yasmin Ahmed Fahmy,1 Thoraya Hosny,2 Nessma H Gandor,2 Sherif Y Mohammed,2 Tahia Mohamed Ahmed Mohamed,2 Nabila Elsayed Mousa Abdelmoteleb,3 Noura E Esmaeel1
1Medical Microbiology and Immunology Department, Faculty of Medicine, Zagazig University, Zagazig, Egypt; 2Clinical Pathology Department, Faculty of Medicine, Zagazig University, Zagazig, Egypt; 3Gynecology and Obstetrics Department, Faculty of Medicine, Zagazig University, Zagazig, Egypt
Correspondence: Marian A Gerges, Medical Microbiology and Immunology Department, Faculty of Medicine, Zagazig University, Zagazig, 44519, Egypt, Tel +2 01003819530, Email [email protected]
Background: Candida albicans (C. albicans) is a major cause of vulvovaginal candidiasis (VVC), a condition that is commonly treated with azole agents. Biofilm formation and aspartyl proteinase production are important virulence factors that could be linked to azole resistance in C. albicans impeding therapy.
Aim: To find out the association of both factors with azole resistance among C. albicans isolated from VVC cases in Egyptian nonpregnant women of childbearing age.
Patients and Methods: In a cross-sectional study, C. albicans was isolated from nonpregnant females diagnosed clinically as having VVC during a 1-year study period. Susceptibility to azole agents was tested using the disc diffusion method. Biofilm formation and aspartyl proteinase production were assessed phenotypically. Additionally, two biofilm-related genes (ALS1 and HWP1) and three proteinase genes (SAP2, SAP4, and SAP6) were screened for using polymerase chain reaction (PCR).
Results: Among 204 C. albicans isolates, azole resistance ratios were as follows: voriconazole (30.4%), itraconazole (17.6%), fluconazole (11.3%) and econazole (6.4%). Biofilm-producing capacity was detected in 63.2% of isolates, and 63.2% were proteinase producers. The frequencies of ALS1 and HWP1 were 69.6% and 74.5%, respectively, while SAP2, SAP4, and SAP6 were 69.2%, 88.7%, and 64.7%, respectively. Biofilm formation was significantly associated with azole resistance (P < 0.001 for each tested azole agent) as was proteinase production (P < 0.001 for fluconazole, voriconazole, and econazole resistance and P = 0.047 for itraconazole).
Conclusion: Among nonpregnant Egyptian women of childbearing age, azole resistance in C. albicans causing VVC is significantly associated with biofilm formation and proteinase production. The development of new therapeutic agents that can target these factors is warranted.
Keywords: Candida albicans, vulvovaginal candidiasis, biofilm formation, aspartyl proteinase, azole resistance
Introduction
Vulvovaginal candidiasis (VCC) is a frequently encountered superficial fungal infection of the female lower genital tract characterized by itching, dyspareunia, soreness, and vaginal erythema and is caused by overgrowth of Candida species.1
Being ranked as the second most common vaginal infection, after bacterial vaginosis, it was estimated that approximately 70–75% of women of childbearing age can experience at least one episode of VVC during their lifetime with a global yearly prevalence of 3871 per100000women.2,3
Moreover, approximately 40% to 50% of VVC patients experience recurrence with 5% to 8% of them diagnosed with recurrent VVC ʺRVVCʺ (4 or more episodes per year),1,4 a condition associated with a high rate of therapeutic failure.5
The exact pathogenesis and risk factors of either VVC or RVVC are still to be elucidated as Candida species can colonize the lower genital tract of about 20% of healthy asymptomatic nonpregnant women forming part of vaginal microflora.6 However, and for reasons not fully understood, a transition to an infection state takes place. Factors that disrupt the vaginal microflora or interfere with the balanced host-Candida interplay can significantly increase the risk of VVC such as pregnancy, antibiotic usage, diabetes, and immunodeficiency.2
Candida albicans (C. albicans) continue to be the species most frequently (75–90%) recovered from cases of VVC.2,7–9 However, the frequency of non-albicans Candida (NAC), most notably C. glabrata, has been increasingly reported.10 Others include C. tropicalis, C. krusei, C. parapsilosis, and C. guilliermondii.11,12
C. albicans possess a large repertoire of virulence factors that enable them to cause serious health problems such as their capacity to adhere to epithelial cells, biofilm production, secretion of hydrolytic enzymes, phenotypic switching, and hyphal formation.13
The growth of C. albicans in biofilm directly contributes to virulence as the complex architecture of the biofilm and the composition of the extracellular matrix in which cells are embedded enable the organism to withstand host immune defenses and high concentrations of antifungal drugs compared to planktonic cells.14 Biofilm formation is a sequential process that starts with adherence of yeast cells to the vaginal epithelium. This is followed by formation of several layers of polymorphic cells that are engaged in a self-produced extracellular matrix material composed mainly of β-1,3 glucan, and, finally, dispersion of unbound yeast cells to seed other sites.15 Upon this finely tuned process, the expression of nearly 1000 genes is upregulated including those coding for cell wall proteins that mediate adhesion to vaginal epithelial cells and to other Candida cells. Agglutinin-like sequence (ALS), a family of glycoproteins, constitutes the largest family of adhesins.16 The hyphal wall protein 1 (HWP1), a mannoprotein attached to the β glucan of the hyphal cell wall, constitutes another key protein that anchors C. albicans to epithelial cells. Both ALS and HWP1 have a crucial role in mature biofilm formation, and their genes have been described to be upregulated during biofilm development.17,18
Apart from biofilm formation, secreted hydrolytic enzymes by C. albicans such as aspartyl proteinases, first identified by Staib in 1969,19 have been regarded as important virulence factors as they facilitate host tissue invasion. Furthermore, they could be engaged in immune evasion, adhesion, hyphal development, and biofilm formation.20 They include 10 members that are sequentially expressed at different stages of infection and are encoded by the aspartyl proteinases (SAP) gene family.21
The SAP2 protein has been widely studied and has been shown to play a crucial role in vaginal infection.22 SAP2 can degrade a wide spectrum of extracellular and host surface proteins such as keratin, collagen, and mucin.21 Interestingly and despite SAP2’s ability to degrade several host defense proteins such as immunoglobulins (Igs) and complement proteins, it can confer resistance against VVC and has been evaluated as a possible candidate for vaccine preparation.23
Among the antifungals used to treat VVC, azole agents are the drugs of choice because of their utmost efficacy, availability in oral form, and lesser toxicity.24 Azoles are five-membered heterocyclic compounds that exhibit their antifungal effect by binding to lanosterol 14-α-sterol demethylase, an important enzyme in ergosterol biosynthesis. This results in the synthesis of a fungistatic toxic sterol which, along with the ability of these agents to increase the level of reactive oxygen species, can inhibit fungal growth.25
Unfortunately, a significant rise in the frequency of azole resistance among C. albicans strains has been reported and is regarded as a serious health issue that can impede VVC therapy.6,26 This is because it intensifies the therapeutic challenges caused by NAC species that naturally exhibit intrinsic decreased susceptibility to azole agents such as C. krusei and C. glabrata.26 Furthermore, the development of resistance or tolerance to fluconazole in C. albicans, due to previous treatment, has been incriminated in the therapeutic failure of RVVC cases.5
This study aimed at studying the occurrence of biofilm formation and aspartyl proteinase production and their association with azole resistance in C. albicans causing VVC which is going to be of great utility in the proper understanding and management of VVC particularly resistant and/or RVVC cases.
Patients and Methods
This cross-sectional study was carried out over a period of one year (November 2021 to November 2022) at the Obstetrics and Gynecology Department, Zagazig University Hospitals, and Bacteriology and Molecular Biology Laboratories at the Medical Microbiology and Immunology and Clinical Pathology Departments, Faculty of Medicine, Zagazig University.
Sample Size Determination
The sample size was determined using open EPI assuming the expected frequency of Candida infection was 31.6% among the target population (women with vaginitis).27 At a 95% confidence interval and an effect size = 1, the estimated sample size was calculated to be 204 women.
Ethical Issues
The study protocol was approved by the Institutional Review Board of Zagazig University (approval no. IRB#: 9637). All patients completed informed consent, and all procedures involving human subjects were carried out in compliance with the updated 2013 Declaration of Helsinki.
Participants
Enrolled participants were recruited from patients attending the Gynecology Outpatient Clinics at Zagazig University Hospitals. Women (mean age 28.5 ± 6.13 years) suffering from symptoms of vulvovaginitis (burning, itching, curd-like discharge) were enrolled in this study. Pregnant women, those with cervical cancer, or were on antibiotics were excluded from the study.
Sampling and Identification of C. albicans
Two upper vaginal samples from each patient were collected using a sterile cotton swab. Within two hours, the samples were transferred and processed. Using one swab, a Gram-stained smear was prepared and examined to check for the presence of yeast cells, hyphae, and pseudohyphae and to exclude cases of bacterial vaginosis.28 The other swab was inoculated onto a Sabouraud Dextrose Agar (SDA) plate (Oxoid Ltd., Hampshire, UK), with 50 µg/mL chloramphenicol, and incubated at 37°C for 24–48 h. Isolates of C. albicans were identified by assessing germ tube and chlamydospore formation.29 This was confirmed by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometer (MALDI-TOF/MS) using the VITEK MS system (bioMérieux. Inc, Durham, USA). Pure C. albicans growths were preserved in sterile glycerol broth (15% V/V) and kept at −20°C.
Antifungal Susceptibility Testing
According to the Clinical and Laboratory Standards Institute,30 the susceptibility of C. albicans isolates to different antifungals was investigated by the disc diffusion method using cation-adjusted Mueller–Hinton agar (Oxoid, UK). The antifungal discs (Liofilchem, Italy) tested were amphotericin B (AMB, 100μg), fluconazole (FLU, 100 μg), voriconazole (VO, 1μg), itraconazole (ITC, 50 μg), caspofungin (CAS, 5μg), nystatin (NY, 100 μg), and econazole (ECN, 10 μg).
Phenotypic Detection of Biofilm Formation
This was done for all C. albicans isolates as previously described.31,32 Isolates were grown overnight on SDA plates, and fresh colonies were inoculated in Sabouraud dextrose broth (Oxoid, UK) and incubated for 24 h at 37°C and then adjusted (~1:100 dilution) to 0.5 McFarland standard using a fresh medium.32 Two hundred μL of the diluted cultures were added to each well of a sterile, polystyrene, flat-bottom microtiter plate (Techno Plastic Products, Switzerland), which was then covered with their lids and incubated for 48 h at 37°C. Sabouraud dextrose broth without C. albicans was used to inoculate negative control wells. The contents of the wells were then gently discarded by tapping the plate, and the wells were rinsed four times with 200 μL of phosphate-buffered saline (pH 7.2) to remove any free-floating planktonic cells. The plate was air-dried and 110 μL of 0.4% crystal violet solution (Fluka AG, Buchs, Switzerland) was added to each well to uniformly stain adherent cells. The plate was covered and kept for 45 minutes at room temperature before being thoroughly and repeatedly rinsed with distilled water. Stained biofilm with crystal violet was then solubilized in 200 μL of 95% ethanol, of which 100 μL was transferred to a new plate for reading. The optical density (OD) was measured using a microplate reader (Awareness Technologies stat fax 2100, CA, United States) at 595 nm. The isolates were tested in triplicate three times. The mean OD ± standard deviation (SD) was calculated for each tested isolate and for the control wells. The cut-off value of the OD (ODc) was calculated for each plate separately by adding the mean OD value of negative controls (mean ODnc) to three standard deviations of the negative controls (3 × SDnc) using the formula: 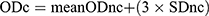 . A negative result (non-producer) was recorded if the mean OD of the tested isolate was equal to or less than the ODc. Positive results were categorized as either weak (ODc < OD ≤ 2 × ODc), moderate (2 × ODc < OD ≤ 4 × ODc), or strong biofilm producers (4 × ODc < OD).32
. A negative result (non-producer) was recorded if the mean OD of the tested isolate was equal to or less than the ODc. Positive results were categorized as either weak (ODc < OD ≤ 2 × ODc), moderate (2 × ODc < OD ≤ 4 × ODc), or strong biofilm producers (4 × ODc < OD).32
Phenotypic Detection of Aspartyl Proteinase Secretion
This was done according to Cassone et al with minor changes.33 Yeast cells were incubated overnight in YEPD medium (2% glucose, 1% yeast extract, and 2% Bacto peptone) and induced to secrete proteases by subculture on bovine serum albumin (BSA) containing medium. To prepare the medium, 1.17% yeast carbon base (Sigma-Aldrich, India), 0.01% yeast extract (NICE, Manimala Road, Edappally, Kerala, India), and 0.2% BSA (Sigma-Aldrich, USA) were first adjusted to a pH of 5.0 and sterilized by filtration before adding to a stock solution of autoclaved agar (2%). Ten μL suspension of each isolate at a concentration of 107 yeast cells/mL in YEPD medium was applied to the plate. A maximum of 6 isolates were tested for each 90-mm-diameter plate. The plates were incubated at 28°C for 7 days and examined daily for increased opacity around the growing fungi caused by precipitated albumin which subsequently cleared due to hydrolysis by acid proteinases. The results were evaluated as negative (–) for no clearance, mild activity (a lysis zone of 1–2 mm around the growth), and strong activity (a lysis zone of 3–5 mm around the growth). C. albicans ATCC 10231 was used as a positive control and the test was completed in triplicate.
Detection of Biofilm and Aspartyl Proteinase Genes
Polymerase chain reaction (PCR) was applied to screen C. albicans isolates for the biofilm-associated genes “ALS1 and HWP1” and the proteinase genes “SAP2, SAP4, and SAP6” using previously described primers.34–36 DNA was extracted using the DNA extraction Mini Kit (QIAGEN DNA Mini Kit). Each PCR reaction was conducted in a total volume of 25 μL. The thermal conditions were as follows: 95 °C for 5 minutes, followed by 40 amplification cycles of 95 °C for 15s, 60 °C for 30s, and 68 °C for 40s, then a final elongation at 68°C for 5 minutes. Two PCR reactions with the same cycling conditions were applied to each isolate, the first targeting HWP1, ALS1, and SAP2 and the second targeting SAP4 and SAP6. The amplified PCR products were visualized in 2% agarose gel stained with ethidium bromide (0.5g/mL) using UV transillumination. A 100 bp DNA ladder was used to detect the size (bp) of the amplified genes.
Statistical Analysis
Collected data were analyzed using SPSS version 22 software (SpssInc, Chicago, ILL Company). Categorical data were presented as numbers and percentages. The Chi-square test (χ2) was used to analyze categorical variables. P < 0.05 was considered significant.
Results
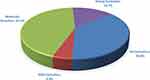
In this study, 204 non-duplicate C. albicans were isolated from an equal number of nonpregnant females suffering from vulvovaginitis. The susceptibility of the obtained C. albicans to the used antifungal agents is presented in Figure 1.
All isolates were susceptible to nystatin and amphotericin B, and 93.6% were susceptible to caspofungin. Among azole derivatives, the highest resistance ratio was recorded with voriconazole (30.4%), followed by itraconazole (17.6%), fluconazole (11.3%), then econazole (6.4%).
The in vitro ability of C. albicans isolates to produce biofilm is demonstrated in Figure 2. The results demonstrate that 63.2% of all isolates were biofilm producers where 14.7% (n = 30) were strong producers, 43.6% (n = 89) were moderate producers, and 4.9% (n = 10) were mild producers. However, 36.8% (n = 75) were non-producers.
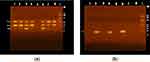
The biofilm-related genes ALS1 and HWP1 (Figure 3a) were detected in 69.6% and 74.5% of isolates with a significant association between both genes and moderate and strong biofilm production, respectively (P < 0.001 and 0.002, respectively) (Supplementary Table 1).
When the relationship between biofilm formation and azole resistance was investigated, the results demonstrated highly significant relations with all tested azole drugs (P < 0.001 for each) (Table 1).
 |
Table 1 Relationship Between Biofilm Formation and Azole Resistance in the Obtained C. albicans Isolates (n = 204) |
Among the obtained C. albicans isolates, 63.2% (n = 129) were able to produce aspartyl proteinases with 52.0% (n = 106) being mild producers and 11.3% (n = 23) being strong producers. The prevalence of three proteinase genes was tested and demonstrated that SAP4 was the most frequent protease gene detected among all isolates (88.7%, n = 181), followed by SAP2 (69.6%, n = 142), then SAP6 (64.7%, n = 132) (Figure 3a and b). Furthermore, highly significant associations were found between the three virulence genes and the ability to produce aspartyl proteinases in vitro (P < 0.001 for each). Additionally, SAP2 gene was found to be significantly associated with moderate and strong biofilm production (P < 0.001) (Supplementary Tables 2 and 3).
When the relationship between the production of proteinases and azole resistance was analyzed in the obtained isolates, highly significant associations with fluconazole, voriconazole, and econazole resistance (P < 0.001 for each) and a significant association with itraconazole resistance (P = 0.047) were found (Table 2).
 |
Table 2 Relationship Between Aspartyl Proteinase Production and Azole Resistance in the Obtained C. albicans Isolates (n = 204) |
Discussion
Despite being the drugs of choice for treating VVC, Candida species have evolved resistance to azole agents by different mechanisms, probably due to their fungistatic nature, resulting in therapeutic failures.37 C. albicans possesses a plethora of virulence factors and several studies have found that azole resistance could be linked to these factors.38–40 In this study, the capacity of C. albicans to produce biofilms and aspartyl proteinases as well as the association of both factors with azole resistance was assessed in VVC isolates.
Among a total of 204 C. albicans isolates, 30.4%, 17.6%, 11.3%, and 6.4% were resistant to voriconazole, itraconazole, fluconazole, and econazole, respectively. Varying levels of azole resistance have been reported previously in C. albicans. This has been shown to depend on the type and site of infection, additionally, on the history of former or prolonged exposure to these agents, particularly with fluconazole.41 Interestingly, it has been reported that invasive Candida members could be more susceptible to antifungals, including azoles, than those causing mucosal infection or colonization.42,43 In a previous Egyptian study, higher ratios of azole resistance were reported where 71% of C. albicans causing vulvovaginitis were resistant to voriconazole, 33% were resistant to itraconazole, and 27% were resistant to fluconazole.44 In other studies, and consistent with the current study, the resistance ratios for fluconazole and voriconazole were 22.44% and 18.89%, in Candida species isolated from different clinical specimens and 14% and 34% in C. albicans.45,46 However, lower ratios were reported elsewhere.47,48
In the current study, the susceptibility to antifungals other than azoles was tested due to their possible use as alternatives in resistant VVC cases.49–51 All isolates were susceptible to nystatin and amphotericin B and 93.6% were susceptible to caspofungin. This comes in accordance with previous studies.45,47,48
C. albicans cells have been shown to grow mostly in biofilms, on both biotic and abiotic surfaces, which provide ecological advantages to the fungus and further constitute a source of recalcitrant infections.52 The current study demonstrated that 63.2% of C. albicans isolates were biofilm-producers with 14.7%, 43.6%, and 4.9% being strong, moderate, and mild producers, respectively. The prevalence of the biofilm-related genes ALS1 and HWP1 was 69.6% and 74.5%, respectively, with a significant relation between both genes and moderate and strong biofilm production, respectively (P < 0.001 and 0.002).
A previous study reported that among 53 C. albicans isolated from systemic infection, 22.6% were strong biofilm producers, 1.9% were moderate, 17% were weak producers, and 58.5% were non-biofilm producers.53 The frequencies of ALS1 and HWP1 were 46.4% and 57.1% among 28 C. albicans causing catheter-associated candiduria in another study, where 32.1%, 21.4%, and 46.4% were strong, moderate, and weak biofilm producers, respectively. The authors documented that biofilm-forming isolates had a significantly higher prevalence of tested biofilm genes compared to the weak or non-biofilm-forming peers (P 0.049 and 0.001 for ALS1 and HWP1, respectively),54 which comes consistent with the current findings. Several factors could, however, influence in vitro biofilm production such as the isolation site of the tested strain, the initial inoculum size used in detection, and probably the substratum used whether silicone, polystyrene, polyvinyl chloride, or Teflon.55,56
Though not fully understood, biofilm formation has been firmly connected to antifungal resistance. Different mechanisms have been suggested to explain this complex phenomenon with the composition of the biofilm matrix being probably the most important owing to its sponge-like effect that sequesters azole agents.57 The present study recorded a highly significant association between biofilm formation and resistance to the tested azole agents (P < 0.001 for each agent). This comes consistent with previous reports that demonstrated a dramatic decrease in the susceptibility to antifungal agents of biofilm-associated Candida,58 and that also reported significant negative correlations (p < 0.05) between fluconazole and voriconazole minimal inhibitory concentrations (MICs) and biofilm production in C. albicans isolates obtained from vulvovaginal specimens.38 Similarly, significant relationships between fluconazole and itraconazole MICs and biofilm formation (P = 0.05 and P = 0.002, respectively) were reported in C. albicans isolated from other sites as nail samples.59
However, such a significant correlation between antifungal drug resistance and biofilm formation was not reported by other researchers in C. albicans isolated from VVC and bloodstream infection.60 Additionally, the in vivo ability to form biofilm was not directly connected to the level of fluconazole resistance in C. albicans clones isolated sequentially from an HIV-infected patient who received progressively increased fluconazole dosages over two years.61 This could be attributed to the different nature of the study and to the immune state of the studied patients which may interact with the organism in a complex way that affects the organism’s virulence and pathogenesis.62
Though remaining controversial,63 the secretion of extracellular tissue-damaging hydrolytic enzymes such as proteinases seems to have a significant role in both invasive Candida infection and in mucosal infections such as vulvovaginitis. C. albicans isolates causing vaginitis were shown to produce a significantly higher level of aspartyl proteinases than isolates recovered from asymptomatic vaginal carriers.34 In the current study, the ability to produce acid proteinases was demonstrated in 63.2% of C. albicans isolates with 11.3% being strong producers and 52.0% being mild producers. SAP4 was the most frequent proteinase gene detected (88.7%), followed by SAP2 (69.6%), then SAP6 (64.7%) with highly significant associations between the three virulence genes and the ability to produce aspartyl proteinases in vitro (P < 0.001 for each).
High rates of proteinase production were reported previously in C. albicans whether in cutaneous or systemic infections. This ranged from 100% and 72.85% of C. albicans isolated from cutaneous infection and onychomycosis, respectively, to 90.74% with C. albicans causing surgical site infection.47,59,64 High rates were also reported in isolates obtained from those with impaired immunity such as HIV-infected individuals (92%) and diabetic patients with chronic periodontitis (94.5%).65,66 Regarding VVC, proteinase production has been shown to be one of the most expressed virulence determinants in Candida species (45%) following the adherence ability (100%).67
The prevalence of proteinase genes in previous works demonstrated variable results. In C. albicans isolated from vulvovaginal colonization, SAP6 was found to be the most frequent gene (93.33%), while SAP7 was the most frequent (100%) in infection.68 However, using quantitative real-time RT-PCR, another study reported that SAP 2, SAP 4–6, and SAP 7 were the dominating proteinase genes expressed in both Candida carriers and patients with VVC and RVVC,69 which comes in line with the current study. SAP5 and SAP9 were the most frequently expressed genes in vivo during mucosal infection (oral and vaginal), while the SAP1 gene was the most prevalent in another study being found in 65% of C. albicans with a significant detection in biofilm-forming isolates (P 0.0001).48,70
These obvious variations could be attributed to the differences in the type and site of infection as well as the stage of infection which may influence the expression of virulence factors both quantitatively and qualitatively.71 Additionally, variations in the in vitro culture conditions used for proteinase detection such as medium composition, pH of the medium, and temperature of incubation were found to have a notable effect on the induction and the activity of these enzymes.21
When the relation between the production of proteinases and azole resistance was analyzed in the current study, highly significant associations were found with fluconazole, voriconazole, and econazole resistance (P < 0.001 for each) and a significant association with itraconazole resistance (P = 0.047) was recorded.
Similar results were reported previously where in one study all isolates resistant to antifungals were aspartyl proteinase producers.52 However, in previous work on C. albicans isolates from onychomycosis, such a significant relationship was not reported with fluconazole (P = 0.658) or itraconazole (P = 0.124).59 This could be attributed to the difference in the number of tested isolates, the different sites of infection, or probably the medium used for proteinase detection or the incubation conditions.72,73
Limitations of the Study
This study is not without limitations such as the lack of assessment of the expression level of the studied genes to reveal their exact contribution to virulence and the lack of assessing both biofilm and proteinase as well as azole resistance in C. albicans colonizing normal females to compare those causing VVC and those inhabiting the vagina as microflora.
Data Sharing Statement
All data and materials related to the study are included in the current manuscript and in the Supplementary Tables.
Ethical Approval and Consent to Participate
The study was approved by the Institutional Review Board (IRB), Faculty of Medicine, Zagazig University (Approval code 9637).
Disclosure
None of the authors have any conflicts of interest to declare for this work.
References
1. Sobel JD. Vulvovaginal candidosis. Lancet. 2007;369(9577):1961–1971. doi:10.1016/S0140-6736(07)60917-9
2. Gonçalves B, Ferreira C, Alves CT, Henriques M, Azeredo J, Silva S. Vulvovaginal candidiasis: epidemiology, microbiology and risk factors. Crit Rev Microbiol. 2016;42(6):905–927. doi:10.3109/1040841X.2015.1091805
3. Denning DW, Kneale M, Sobel JD, Rautemaa-Richardson R. Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis. 2018;18(11):e339–e347. doi:10.1016/S1473-3099(18)30103-8
4. Foxman B, Muraglia R, Dietz JP, Sobel JD, Wagner J. Prevalence of recurrent vulvovaginal candidiasis in 5 European countries and the United States: results from an internet panel survey. J Low Genit Tract Dis. 2013;17(3):340–345. doi:10.1097/LGT.0b013e318273e8cf
5. Arastehfar A, Kargar ML, Mohammadi SR, et al. A high rate of recurrent vulvovaginal candidiasis and therapeutic failure of azole derivatives among Iranian women. Front Microbiol. 2021;28(12):655069. doi:10.3389/fmicb.2021.655069
6. Kalia N, Singh J, Kaur M. Microbiota in vaginal health and pathogenesis of recurrent vulvovaginal infections: a critical review. Ann Clin Microbiol Antimicrob. 2020;19(1):5. doi:10.1186/s12941-020-0347-4
7. ElFeky DS, Gohar NM, El-Seidi EA, Ezzat MM, AboElew SH. Species identification and antifungal susceptibility pattern of Candida isolates in cases of vulvovaginal candidiasis. Alexandria J Med. 2016;52(3):269–277. doi:10.1016/j.ajme.2015.10.001
8. Rezaei-Matehkolaei A, Shafiei S, Zarei-Mahmoudabadi A. Isolation, molecular identification, and antifungal susceptibility profiles of vaginal isolates of Candida species. Iran J Microbiol. 2016;8(6):410–417.
9. Anh DN, Hung DN, Tien TV, et al. Prevalence, species distribution and antifungal susceptibility of Candida albicans causing vaginal discharge among symptomatic non-pregnant women of reproductive age at a tertiary care hospital, Vietnam. BMC Infect Dis. 2021;21(1):523. doi:10.1186/s12879-021-06192-7
10. Kennedy MA, Sobel JD. Vulvovaginal candidiasis caused by non-albicans Candida species: new insights. Curr Infect Dis Rep. 2010;12(6):465–470. doi:10.1007/s11908-010-0137-9
11. Bitew A, Abebaw Y. Vulvovaginal candidiasis: species distribution of Candida and their antifungal susceptibility pattern. BMC Womens Health. 2018;18(1):94. doi:10.1186/s12905-018-0607-z
12. Makanjuola O, Bongomin F, Fayemiwo SA. An update on the roles of non-albicans Candida species in vulvovaginitis. J Fungi. 2018;4(4):121. doi:10.3390/jof4040121
13. Ciurea CN, Kosovski IB, Mare AD, Toma F, Pintea-Simon IA, Man A. Candida and candidiasis-opportunism versus pathogenicity: a review of the virulence traits. Microorganisms. 2020;8(6):857. doi:10.3390/microorganisms8060857
14. Sandai D, Tabana YM, Ouweini AE, Ayodeji IO. Resistance of Candida albicans biofilms to drugs and the host immune system. Jundishapur J Microbiol. 2016;9(11):e37385. doi:10.5812/jjm.37385
15. Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18(5):310–321. doi:10.1016/j.micinf.2016.01.002
16. Cota E, Hoyer LL. The Candida albicans agglutinin-like sequence family of adhesins: functional insights gained from structural analysis. Future Microbiol. 2015;10(10):1635. doi:10.2217/fmb.15.79
17. Nobile CJ, Nett JE, Andes DR, Mitchell AP. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell. 2006;5(10):1604–1610. doi:10.1128/EC.00194-06
18. Nobile CJ, Schneider HA, Nett JE, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18(14):1017–1024. doi:10.1016/j.cub.2008.06.034
19. Staib F. Proteolysis and pathogenicity of Candida albicans strains. Myopathic Mycol Appl. 1969;37(4):345–348. doi:10.1007/BF02129881
20. Kulshrestha A, Gupta P. Secreted aspartyl proteases family: a perspective review on the regulation of fungal pathogenesis. Future Microbiol. 2023;18:295–309. doi:10.2217/fmb-2022-0143
21. Naglik J, Albrecht A, Bader O, Hube B. Candida albicans proteinases and host/pathogen interactions. Cell Microbiol. 2004;6(10):915. doi:10.1111/j.1462-5822.2004.00439.x
22. De Bernardis F, Cassone A, Sturtevant J, Calderone R. Expression of Candida albicans SAP1 and SAP2 in experimental vaginitis. Infect Immun. 1995;63(5):1887–1892. doi:10.1128/iai.63.5.1887-1892.1995
23. De Bernardis F, Boccanera M, Adriani D, Girolamo A, Cassone A. Intravaginal and intranasal immunizations are equally effective in inducing vaginal antibodies and conferring protection against vaginal candidiasis. Infection Immunol. 2002;70:2725–2729. doi:10.1128/IAI.70.5.2725-2729.2002
24. Burden AM, Hausammann L, Ceschi A, Kupferschmidt H, Weiler S. Observational cross-sectional case study of toxicities of antifungal drugs. J Glob Antimicrob Resist. 2022;29:520–526. doi:10.1016/j.jgar.2021.11.010
25. Bhattacharya S, Sae-Tia S, Fries BC. Candidiasis and mechanisms of antifungal resistance. Antibiotics. 2020;9(6):312. doi:10.3390/antibiotics9060312
26. Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. 2017;12:2173.
27. Salvi M. Prevalence of vulvovaginal candidiasis in females in the reproductive age group. Int J Reprod Contracept Obstet Gynecol. 2019;8(2):647–651. doi:10.18203/2320-1770.ijrcog20190299
28. Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29(2):297–301. doi:10.1128/jcm.29.2.297-301.1991
29. Coleman DC, Bennett DE, Sullivan DJ, et al. Oral Candida in HIV infection and AIDS: new perspectives/new approaches. Crit Rev Microbiol. 1993;19(2):61–82. doi:10.3109/10408419309113523
30. Clinical and Laboratory Standards Institute. Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts; Approved Guideline 2nd Ed, M44-A2. Wayne: Clinical and Laboratory Standards Institute; 2009.
31. Jin Y, Yip HK, Samaranayake YH, Yau JY, Samaranayake LP. Biofilm-forming ability of Candida albicans is unlikely to contribute to high levels of oral yeast carriage in cases of human immunodeficiency virus infection. J Clin Microbiol. 2003;41(7):2961–2967. doi:10.1128/JCM.41.7.2961-2967.2003
32. Stepanović S, Vuković D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–899. doi:10.1111/j.1600-0463.2007.apm_630.x
33. Cassone A, De Bernardis F, Mondello F, Ceddia T, Agatensi L. Evidence for a correlation between proteinase secretion and vulvovaginal candidosis. J Infect Dis. 1987;156(5):777–783. doi:10.1093/infdis/156.5.777
34. Naglik JR, Rodgers CA, Shirlaw PJ, et al. Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J Infect Dis. 2003;188(3):469–479. doi:10.1086/376536
35. Green CB, Cheng G, Chandra J, Mukherjee P, Ghannoum MA, Hoyer LL. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology. 2004;150(Pt 2):267–275. doi:10.1099/mic.0.26699-0
36. Naglik JR, Fostira F, Ruprai J, Staab JF, Challacombe SJ, Sundstrom P. Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J Med Microbiol. 2006;55(Pt 10):1323–1327. doi:10.1099/jmm.0.46737-0
37. Sobel JD, Sobel R. Current treatment options for vulvovaginal candidiasis caused by azole-resistant Candida species. Expert Opin Pharmacother. 2018;19(9):971–977. doi:10.1080/14656566.2018.1476490
38. El-Houssaini HH, Elnabawy OM, Nasser HA, Elkhatib WF. Correlation between antifungal resistance and virulence factors in Candida albicans recovered from vaginal specimens. Microb Pathog. 2019;128:13–19. doi:10.1016/j.micpath.2018.12.028
39. Monroy-Pérez E, Rodríguez-Bedolla RM, Garzón J, Vaca-Paniagua F, Arturo-Rojas Jiménez E, Paniagua-Contreras GL. Marked virulence and azole resistance in Candida albicans isolated from patients with periodontal disease. Microb Pathog. 2020;148:104436. doi:10.1016/j.micpath.2020.104436
40. Bohner F, Papp C, Gácser A. The effect of antifungal resistance development on the virulence of Candida species. FEMS Yeast Res. 2022;22(1):foac019. doi:10.1093/femsyr/foac019
41. Popp C, Ramírez-Zavala B, Schwanfelder S, Krüger I, Morschhäuser J, Berman J. Evolution of fluconazole-resistant Candida albicans strains by drug-induced mating competence and parasexual recombination. mBio. 2019;10(1):e02740–18. doi:10.1128/mBio.02740-18
42. Diekema D, Arbefeville S, Boyken L, Kroeger J, Pfaller M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn Microbiol Infect Dis. 2012;73(1):45–48. doi:10.1016/j.diagmicrobio.2012.02.001
43. Tan TY, Hsu LY, Alejandria MM, et al. Antifungal susceptibility of invasive Candida bloodstream isolates from the Asia-Pacific region. Med Mycol. 2016;54(5):471–477. doi:10.1093/mmy/myv114
44. Ali A, Azab M, Abdelrahman A. Distribution of secreted aspartyl protease (SAP) virulence genes and antifungal resistance genes at vulvovaginal candidiasis isolates. GSC Biol Pharm Sci. 2018;5(3):86–94. doi:10.30574/gscbps.2018.5.3.0149
45. Marak MB, Dhanashree B. Antifungal susceptibility and biofilm production of Candida spp. isolated from clinical samples. Int J Microbiol. 2018;10:7495218.
46. Yenisehirli G, Bulut N, Yenisehirli A, Bulut Y. In vitro susceptibilities of Candida albicans isolates to antifungal agents in Tokat, Turkey. Jundishapur J Microbiol. 2015;8(9):e28057. doi:10.5812/jjm.28057
47. Erum R, Samad F, Khan A, Kazmi SU. A comparative study on production of extracellular hydrolytic enzymes of Candida species isolated from patients with surgical site infection and from healthy individuals and their co-relation with antifungal drug resistance. BMC Microbiol. 2020;20(1):368. doi:10.1186/s12866-020-02045-6
48. Shrief R, Zaki ME, El-Sehsah EM, Ghaleb S, Mofreh M. Study of antifungal susceptibility, virulence genes and biofilm formation in Candida albicans. Open Microbiol J. 2019;13:241–248. doi:10.2174/1874285801913010241
49. Sobel JD. Candidal vulvovaginitis. Clin Obstet Gynecol. 1993;36(1):153–165. doi:10.1097/00003081-199303000-00021
50. Phillips AJ. Treatment of non-albicans Candida vaginitis with amphotericin B vaginal suppositories. Am J Obstet Gynecol. 2005;192(6):
51. Mendling W, Brasch J, Cornely OA, et al. Guideline: vulvovaginal candidosis (AWMF 015/072), S2k (excluding chronic mucocutaneous candidosis). Mycoses. 2015;58(Suppl 1):1–15. doi:10.1111/myc.12292
52. Wu X, Zhang S, Li H, et al. Biofilm formation of Candida albicans facilitates fungal infiltration and persister cell formation in vaginal candidiasis. Front Microbiol. 2020;11:1117. doi:10.3389/fmicb.2020.01117
53. Kadry AA, El-Ganiny AM, El-Baz AM. Relationship between Sap prevalence and biofilm formation among resistant clinical isolates of Candida albicans. Afr Health Sci. 2018;18(4):1166–1174. doi:10.4314/ahs.v18i4.37
54. Hamady A, Marei Y. Detection of als1 and hwp1 genes involved in biofilm formation in Candida albicans isolated from catheter-associated candiduria. Microbes Infect Dis. 2021;2:558–566.
55. Sadanandan B, Vaniyamparambath V, Lokesh KN, et al. Candida albicans biofilm formation and growth optimization for functional studies using response surface methodology. J Appl Microbiol. 2022;132(4):3277–3292. doi:10.1111/jam.15402
56. Estivill D, Arias A, Torres-Lana A, Carrillo-Muñoz AJ, Arévalo MP. Biofilm formations by five species of Candida on three clinical materials. J Microbiol Methods. 2011;86(2):238–242. doi:10.1016/j.mimet.2011.05.019
57. Silva S, Rodrigues CF, Araújo D, Rodrigues ME, Henriques M. Candida Species biofilms’ antifungal resistance. J Fungi. 2017;3(1):8. doi:10.3390/jof3010008
58. Chandra J, Mukherjee PK, Ghannoum M, Parsek M, Whiteley M, Mukherjee P. Candida biofilms: development, architecture, and resistance. Microbiol Spectr. 2015;3(4):10. doi:10.1128/microbiolspec.MB-0020-2015
59. Mohammadi F, Ghasemi Z, Familsatarian B, et al. Relationship between antifungal susceptibility profile and virulence factors in Candida albicans isolated from nail specimens. Rev Soc Bras Med Trop. 2020;7:53.
60. Tulasidas S, Rao P, Bhat S, Manipura R. A study on biofilm production and antifungal drug resistance among Candida species from vulvovaginal and bloodstream infections. Infect Drug Resist. 2018;11:2443–2448. doi:10.2147/IDR.S179462
61. Song NN, Qian GY, Zheng HL, et al. Biofilm alterations on the stepwise acquisition of fluconazole-resistant Candida albicans isolates. Int J Dermatol Venereol. 2022;5(3):132–139. doi:10.1097/JD9.0000000000000223
62. Czechowicz P, Nowicka J, Gościniak G. Virulence factors of Candida spp. and host immune response important in the pathogenesis of vulvovaginal candidiasis. Int J Mol Sci. 2022;23(11):5895. doi:10.3390/ijms23115895
63. Araújo Paulo de Medeiros M, de Melo AP V, de Sousa AM M, Silva-Rocha WP, Pipolo Milan E, Maranhão Chaves G. Characterization of virulence factors of vaginal and anal isolates of Candida albicans sequentially obtained from patients with vulvovaginal candidiasis in north-east Brazil. J Mycol Med. 2017;27(4):567–572. doi:10.1016/j.mycmed.2017.06.002
64. Ramos Lde S, Barbedo LS, Braga-Silva LA, Dos Santos AL, Pinto MR, Sgarbi DB. Protease and phospholipase activities of Candida spp. isolated from cutaneous candidiasis. Rev Iberoam Micol. 2015;32(2):122–125. doi:10.1016/j.riam.2014.01.003
65. de Paula Menezes R, de Melo Riceto ÉB, Borges AS, de Brito Röder DV, Dos Santos Pedroso R. Evaluation of virulence factors of Candida albicans isolated from HIV-positive individuals using HAART. Arch Oral Biol. 2016;66:61–65. doi:10.1016/j.archoralbio.2016.02.004
66. Sardi JC, Duque C, Höfling JF, Gonçalves RB. Genetic and phenotypic evaluation of Candida albicans strains isolated from subgingival biofilm of diabetic patients with chronic periodontitis. Med Mycol. 2012;50(5):467–475. doi:10.3109/13693786.2011.633233
67. Majumdar T, Mullick JB, Bir R, Roy J, Sil SK. Determination of virulence factors and biofilm formation among isolates of vulvovaginal candidiasis. J Med Sci. 2016;36(2):53–58. doi:10.4103/1011-4564.181521
68. Lima JS, Braga KRGS, Vieira CA, et al. Genotypic analysis of secreted aspartyl proteinases in vaginal Candida albicans isolates. J Bras Patol Med Lab. 2018;54(1):28–33.
69. Lian CH, Liu WD. Differential expression of Candida albicans secreted aspartyl proteinase in human vulvovaginal candidiasis. Mycoses. 2007;50(5):383–390. doi:10.1111/j.1439-0507.2007.01384.x
70. Naglik JR, Moyes D, Makwana J, et al. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology. 2008;154(Pt 11):3266–3280. doi:10.1099/mic.0.2008/022293-0
71. Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–128. doi:10.4161/viru.22913
72. Dostál J, Hamal P, Pavlícková L, et al. Simple method for screening Candida species isolates for the presence of secreted proteinases: a tool for the prediction of successful inhibitory treatment. J Clin Microbiol. 2003;41(2):712–716. doi:10.1128/JCM.41.2.712-716.2003
73. Bentubo HD, Gompertz OF. Effects of temperature and incubation time on the in vitro expression of proteases, phospholipases, lipases and DNases by different species of Trichosporon. Springerplus. 2014;3:377. doi:10.1186/2193-1801-3-377
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.