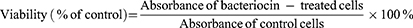Back to Journals » Journal of Inflammation Research » Volume 15
Bacteriocin BacSp222 and Its Succinylated Forms Exhibit Proinflammatory Activities Toward Innate Immune Cells
Authors Śmiałek J , Bzowska M , Hinz A, Mężyk-Kopeć R , Sołtys K, Mak P
Received 23 February 2022
Accepted for publication 12 July 2022
Published 12 August 2022 Volume 2022:15 Pages 4601—4621
DOI https://doi.org/10.2147/JIR.S362066
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Adam D Bachstetter
Justyna Śmiałek,1,* Monika Bzowska,2,* Alicja Hinz,2 Renata Mężyk-Kopeć,2 Kamilla Sołtys,2 Paweł Mak1
1Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Kraków, Poland; 2Department of Cell Biochemistry, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Kraków, Poland
*These authors contributed equally to this work
Correspondence: Paweł Mak, Department of Analytical Biochemistry, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Gronostajowa 7 St., Kraków, 30-387, Poland, Tel +48 12 664 6511, Fax +48 12 664 6902, Email [email protected]
Purpose: The zoonotic opportunistic pathogen Staphylococcus pseudintermedius 222 produces BacSp222 – an atypical peptide exhibiting the features of a bacteriocin, a virulence factor, and a molecule modulating the host inflammatory reaction. The peptide is secreted in an unmodified form and, additionally, two forms modified posttranslationally by succinylation. This study is a comprehensive report focusing on the proinflammatory properties of such molecules.
Methods: The study was performed on mouse monocyte/macrophage-like and endothelial cell lines as well as human neutrophils. The following peptides were studied: BacSp222, its succinylated forms, the form deprived of formylated methionine, and a reference bacteriocin – nisin. The measurements of the nitric oxide (NO) level, induced NO synthase (iNOS) expression, the profile of secreted cytokines, NF-kappa-B activation, reactive oxygen species (ROS) biosynthesis, and the formation of extracellular traps were conducted to evaluate the proinflammatory activity of the studied peptides.
Results: BacSp222 and its succinylated forms effectively induced NO production and iNOS expression when combined with IFN-gamma in macrophage-like cells. All natural BacSp222 forms used alone or with IFN-gamma stimulated the production of TNF-alpha, MCP-1, and IL-1-alpha, while the co-stimulation with IFN-gamma increased IL-10 and IL-27. Upregulated TNF-alpha secretion observed after BacSp222 exposition resulted from increased expression but not from membrane TNF-alpha proteolysis. In neutrophils, all forms of bacteriocin upregulated IL-8, but did not induce ROS production or NETs formation. In all experiments, the activities of deformylated bacteriocin were lower or unequivocal in comparison to other forms of the peptide.
Conclusion: All naturally secreted forms of BacSp222 exhibit proinflammatory activity against monocyte-macrophage cells and neutrophils, confirming that the biological role of BacSp222 goes beyond bactericidal and cytotoxic effects. The atypical posttranslational modification (succinylation) does not diminish its immunomodulatory activity in contrast to the lower antibacterial potential or cytotoxicity of such modified form established in previous studies.
Keywords: cytokines, neutrophils, NF-kappa-B activation, nisin, nitric oxide, posttranslational modifications, Staphylococcus pseudintermedius 222
Introduction
Pathogenic bacteria produce a broad spectrum of molecular factors that facilitate colonization of the host organism, combat its immune response, and help acquire nutrients. These compounds are called virulence factors and include cytolytic exotoxins, enzymes degrading host tissues, adhesion molecules facilitating physical attachment of bacteria, fimbriae or pili granting motility for evading microorganisms, extracellular factors that enable biofilm formation, chelators of iron, pro-apoptotic factors or modulin molecules causing inflammatory reactions, and a variety of molecules inhibiting phagocytosis, chemotaxis, or oxygen-dependent bactericidal mechanisms. These virulence factors are mainly polypeptides, ie, peptides, proteins, or glycoproteins. However, others are polysaccharides, lipopolysaccharides, lipoteichoic acids, or low-molecular-weight compounds such as siderophores or pigments.1–3 Due to such a highly diverse and multipotent nature of virulence factors, particular symptoms of infectious disease result from both direct host tissue damage and over- or under-reaction of the host immune response. The inflammatory response manifesting in the increased production of factors related to inflammation (such as NO, proinflammatory cytokines or adhesion molecules) is the normal host response to invading pathogens. However, the overproduction of proinflammatory agents might lead to immune-inflammatory diseases. Moreover, the pathogenicity of opportunistic microorganisms has a limited or conditional character because they delicately evade different host immune response mechanisms and are able to cause disease only in immunocompromised but not in immunocompetent hosts.4
Opportunistic pathogens are an integral part of the human and animal skin microbiota and their composition varies significantly depending on anatomical localization, moist, dry, or sebaceous habitats, age, external physicochemical factors, and host health status. Alterations in microbiome profiles result in local inflammatory reactions and/or systemic diseases.5 The growth and composition of the microbiome are affected by the host immune response and two important microbial mechanisms: the quorum sensing phenomenon and the production of bacteriocins. Quorum sensing relies on communication between bacterial cells living in a particular community. It is controlled by the secretion of diffusible signaling molecules regulating the population density and coordinating gene expression. This mechanism ensures that the growth rate of cells, their biological status (such as horizontal gene transfer competencies, formation of capsules or biofilms), and production of key molecules (virulence factors, secretion systems, stress-response molecules) are coordinated and give benefits to the whole bacterial population.6
On the other hand, bacteriocins are ribosomally synthesized polypeptides able to kill or inhibit the growth of similar or closely related bacterial strains. The production on ribosomes is essential because it distinguishes bacteriocins from classical polypeptide antibiotics, ie, non-ribosomally synthesized products of secondary metabolism.7 Bacteriocins are exceptionally widespread among different microorganisms, and some authors suggest that virtually all bacterial species can produce such molecules. Moreover, many species produce different bacteriocins, allowing them to effectively eliminate competitors in a polymicrobial environment.8 Thus, bacteriocins appear to be critical molecular factors influencing the proper microbiome composition of the skin, mucous membranes, and gastrointestinal and urogenital tracts.5,9
Bacteriocins are peptides or proteins with various sizes, structures, mechanisms of action, and organization of genes. For example, bacteriocins produced by staphylococci – widespread residents and opportunistic pathogens of the skin and mucous membranes of humans and many animals – comprise four groups: 1) lantibiotics (small single-chain posttranslationally modified peptides, such as epidermin, gallidermin, or BacCH91), 2) unmodified single-chain peptides (such as aureocin A53, epidermicin NI01, and BacSp222), 3) unmodified multi-chain peptides (such as staphylococcin C55 and aureocin A70), and 4) bacteriolytic enzymes (such as lysostaphins LSs and LSp222 as well as endopeptidase ALE-1).10,11 Among these factors, one staphylococcal bacteriocin has attracted our special attention in recent years, namely the BacSp222 peptide, as this interesting molecule combines the features of a virulence factor, a bacteriocin, and a molecule that modulates the host inflammatory reaction.
BacSp222 is produced by a commensal bacterium Staphylococcus pseudintermedius strain 222.12 This particular strain was initially isolated from dog skin, but in general S. pseudintermedius is a typical zoonotic bacterium of canine origin able to infect humans.13 BacSp222 is a 50-amino acid long peptide encoded on a 15 kb p222 plasmid. Nuclear magnetic resonance (NMR) studies have shown that the peptide is globular and forms a bundle composed of four helices, as in the case of the other staphylococcal subclass IId of bacteriocins: aureocin A53, lacticin Q, entrocin 7A/JSA, and enterocin 7B/JSB.14,15 However, despite the significant similarities in the structure of these molecules, their size, amino acid composition, formylation at the N-terminus, and physicochemical properties, all these bacteriocins have quite different amino acid sequences. In addition, our recent study has shown that BacSp222 is produced in three forms: one unmodified and two posttranslationally modified, named suc-K20-BacSp222 and suc-K11/K20-BacSp222. Both these latter forms are succinylated at the epsilon-amino group of lysine residues K20 or K20 and K11, and such modifications are unique and not seen previously in known bacteriocins. Our studies revealed that the production of such modified forms occurs in response to environmental changes, protects the cells of the producing bacteria from autotoxicity of the secreted bacteriocin and limits the pathogenicity of the strain.15 The principal biological activity of BacSp222 is bactericidal action against Gram-positive bacteria, especially staphylococci. However, in the context of the present work, it is particularly interesting that BacSp222 also demonstrates some features of a virulence factor and an immunomodulatory peptide. At concentrations similar to bactericidal ones, it exhibits cytotoxicity toward many eukaryotic cells, including murine monocytes and macrophages as well as human skin fibroblasts and keratinocytes. Moreover, at much lower nanomolar concentrations, in macrophage-like cell lines BacSp222 efficiently enhances interferon gamma-induced release of nitric oxide (NO) - an essential molecule in various inflammatory pathways.12
The current work is a comprehensive study focusing on the proinflammatory properties of BacSp222 and their succinylated forms (suc-K20-BacSp222 and suc-K11/K20-BacSp222). Previous studies have revealed that such modified forms of bacteriocin exert lower toxicity against bacteria and eukaryotic cells. Therefore, we wanted to investigate differences in their proinflammatory activities. In addition, due to the significant role of bacterial formyl peptides in inflammatory reactions, we also studied the form of a bacteriocin deprived of N-terminal formyl-methionine (-fM-BacSp222).
Macrophages, neutrophils, and endothelial cells are part of the cellular arm of innate immunity, which is activated immediately after pathogen invasion, while nitric oxide (NO) represents molecules produced by innate immune cells in response to various biological (bacterial components or cytokines) or physical factors (eg, shear stress). Independently, inducible NO synthase (iNOS) is an enzyme responsible for NO production during inflammation.21 The expression of iNOS can be induced by bacteria-derived molecules, such as LPS, or by cytokines induced at the site of inflammation. Although IFN-γ-induced transcription factors have been shown as essential for iNOS expression, the much more potent expression of iNOS occurs when IFN-γ is combined with LPS or other proinflammatory cytokines.22,23
In the presented study the experiments were performed using mouse monocyte/macrophage-like and endothelial cell lines as well as human neutrophils. The study documented the influence of mentioned earlier peptides on NO biosynthesis and the expression of induced NO synthase (iNOS). We also determined the profile of cytokines produced after stimulation of cells with the peptides and their influence on reactive oxygen species (ROS) biosynthesis and on the formation of extracellular traps by neutrophils. The majority of the present experiments were conducted together with a comparable widely applied commercial bacteriocin – nisin A.
Materials and Methods
Peptides and Protein Chemistry Techniques
BacSp222 and its succinylated forms (suc-K20-BacSp222 and suc-K11/K20-BacSp222) were purified from bacterial post-culture medium by reversed-phase high-pressure liquid chromatography (RP-HPLC) essentially as described earlier.15 The bacteriocin deprived of N-terminal formyl-methionine (-fM-BacSp222) was obtained after overnight digestion of BacSp222 using cyanogen bromide and 70% (v/v) trifluoroacetic acid (TFA). After the reaction, the modified peptide was purified using RP-HPLC as described above. Nisin A was purified from a commercially available preparation (Sigma, St. Louis, MO, USA) according to the modified procedure described originally by Taylor et al.16 In brief, nisin A was extracted from the commercial solid by shaking in methanol; the mixture was then spun down, the deposit was discarded, and the clear supernatant was evaporated to dryness and redissolved in 0.1% (v/v) TFA. The obtained solution was subjected to two consecutive purification steps by RP-HPLC using two buffers: A, containing 0.1% (v/v) TFA and B, containing 0.07% TFA and 80% (both v/v) acetonitrile. The first separation was performed using a Discovery Bio Wide Pore C5 10 × 250 mm column (Sigma, St. Louis, MO, USA), the linear gradient 10% to 100% of buffer B for 30 min, and a flow rate of 1.5 mL/min. The second purification step was performed using a Discovery Bio Wide Pore C8 10 × 250 mm column (Sigma, St. Louis, MO, USA), linear-gradient 40% to 70% of buffer B, and a flow rate of 1.5 mL/min. The concentration of all peptides in the solutions was determined by an amino acid analysis performed as described earlier.17 The purity (estimated at over 99%), homogeneity, and identity of all peptides used were checked by analytical RP-HPLC and mass spectrometry (Supplementary Figures 1 and 2). Each peptide used in this study was essentially free of lipopolysaccharide (LPS), as assayed by an E-TOXATE kit (Sigma, St. Louis, MO, USA).
Cell Culture Conditions
Murine monocyte/macrophage RAW 264.7 cells (ATCC TIB-71) and murine monocyte/macrophage P388.D1 cells (ATCC CCL-46) were obtained from the American Type Culture Collection (Manassas, VA, USA). Murine brain endothelial (MBE, passage 18–19) cells were a kind gift from R. Auerbach (University of Wisconsin, Madison, WI, USA). The Animal Welfare Committee approved their use in the experiments at the Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, in accordance with the Polish and European legal acts.
Citrated human blood from healthy volunteers was purchased from the Regional Center of Blood Donation and Treatment in Kraków, Poland. The Regional Center of Blood Donation and Treatment deidentifies blood materials appropriately for the assurance of the confidentiality of human subjects. Thus, this study complies with relevant exclusions from the approval of human subjects. Human polymorphonuclear neutrophils (hPMNs) were isolated from the blood by centrifugation in a Ficoll-Paque Plus (GE Healthcare, Chicago, IL, USA) density gradient.
The cells were cultured in an incubator at 5% CO2, 37°C, and >95% humidity in DMEM + Glutamax (4,5 g/l D-glucose, 110 mg/l sodium pyruvate) medium (GIBCO, Paisley, UK, catalog number 10569–010) containing 5% (v/v) fetal bovine serum (FBS, GIBCO, Paisley, UK) (RAW 264.7, P388.D1), or in DMEM + Glutamax containing 10% (v/v) FBS (MBE), or in RPMI (LONZA, Basel, Switzerland, catalog number BE12-702F) medium containing 10% (v/v) FBS (neutrophils).
Analysis of NO Production by the Cells
RAW 264.7 and P388.D1 cells were seeded on 96-well plates in 100 μL DMEM supplemented with 5% (v/v) FBS at the density of 2 × 104 cells/well. After 16 h, the medium was replaced with fresh DMEM with 2% (v/v) FBS. The cells were stimulated for 24 h with 1) 100 ng/mL lipopolysaccharide from E. coli (LPS, Sigma, St. Louis, MO, USA), 2) 10 ng/mL mouse interferon γ (IFN-γ, Biolegend, San Diego, CA, USA), 3) 1 μM BacSp222, 4) 1 μM suc-K20-BacSp222, 5) 1 μM suc-K11/K20-BacSp222, 6) 1 μM -fM-BacSp222, 7) 1 μM nisin A, or 8) INF-γ combined with LPS or with bacteriocins at concentrations specified above.
MBE cells were seeded on 96-well plates (density of 4 × 105 cells/well, 100 µL 10% (v/v) FBS in DMEM). After 24 h, the medium was discarded and replaced with 100 µL 2% (v/v) FBS in DMEM. After 24 h, the medium was again replaced with fresh DMEM. The MBE cells were stimulated/co-stimulated with 1) 10 ng/mL human tumor necrosis factor α (TNFα PromoKine, Kibbutz Beit-Haemek, Israel), 2) 10 ng/mL IFN-γ, 3) 10 ng/mL human interleukin 1β (IL-1β, Biolegend, San Diego, CA, USA), 100 ng/mL LPS, 4) 10 ng/mL TNFα and IFN-γ or IL-1β or 100 ng/mL LPS, 5) 10 ng/mL IFN-γ and IL-1β or LPS, 6) 10 ng/mL IL-1β and LPS, 7) 10 ng/mL TNFα and IFN-γ and IL-1β, 8) 1 µM BacSp222, 9) 1 µM BacSp222 and combination with all the cytokines specified above, 10) 1 µM -fM-BacSp222, or 11) 1 µM -fM-BacSp222 and combination with all the cytokines specified above.
Nitrate levels were measured using the Griess assay.18,19 For this purpose, 100 μL of the cultured medium was incubated for 10 min with 100 μL of a solution of 1% (w/v) sulfanilic acid (Sigma, St. Louis, MO, USA)/0.1% (w/v) N-(1-naphtyl) ethylenediamine dihydrochloride (Sigma, St. Louis, MO, USA) in 2.5% (v/v) H3PO4. A sodium nitrate (Sigma, St. Louis, MO, USA) solution in the concentration range from 1.95 to 250 μM was used to prepare the calibration curve. Then, absorbance was measured at 545 nm with a Synergy H1 Hybrid plate reader controlled by Gene5 version 2.00.18 software (both from BIOTEK Instruments, Winooski, VT, USA).
Analysis of Cell Viability
RAW 264.7, P388.D1, or MBE cells were cultured and stimulated as described in the previous section. The MTT assay was applied to measure the metabolic activity of cells indicating their viability. The assay was performed according to the standard protocol and the absorbance was measured at 545 nm using a Synergy H1 Hybrid plate reader controlled by Gene5 version 2.00.18 software (BIOTEK Instruments, Winooski, VT, USA). The cell viability was calculated according to the formula:
Human polymorphonuclear neutrophils were cultured and stimulated with -fM-BacSp222 for 4h. The LDH release and the level of ATP were determined according to the protocol described earlier.15
Analysis of the iNOS mRNA Level in Murine Monocyte-Macrophage Cells
RAW 264.7 and P388.D1 cells were seeded on 12-well plates in 1 mL DMEM supplemented with 5% (v/v) FBS at the density of 3 × 105 cells/well. After 16 h, the medium was replaced with fresh DMEM with 2% (v/v) FBS, and the cells were stimulated for 6 h with 1) 100 ng/mL LPS, 2) 10 ng/mL IFN-γ, 3) 1 μM BacSp222, 4) 1 μM suc-K20-BacSp222, 5) 1 μM suc-K11/K20-BacSp222, 6) 1 μM -fM-BacSp222, 7) 1 μM nisin A, or 8) 10 ng/mL IFN-γ combined with 100 ng/mL LPS or 1 μM bacteriocins. Then, the post-cultured medium was removed. Total cellular RNA was isolated according to the method proposed by Chomczynski and Sacchi.20 The reverse transcription used 500 ng of total RNA and was performed using a procedure provided by the M-MLV transcriptase manufacturer (Promega, Madison, WI, USA). RT-qPCR was performed using RT PCR Mix SYBR (A&A Biotechnology, Gdansk, Poland) and the Eco Real-Time PCR System (Illumina, San Diego, CA, USA). The samples were tested in duplicates; two reference genes, PolR2b and eEF2, were used. The relative quantification of gene expression was determined using the ΔΔCq method, a built-in function of EcoStudy Version 5.0.4890 software (Illumina, San Diego, CA, USA). The sequences of the primers used are as follows:
iNOS Forward: 5’-AAGGCCAAACACAGCATACC-3’.
iNOS Reverse: 5’-CTGAAGCACTAGCCAGGGAC-3’.
eEF2 Forward: 5’-CCACGGCAAGTCCACGCTGAC-3’.
eEF2 Reverse: 5’-AGAAGAGGGAGATGGCGGTGGATT-3’.
PolR2b Forward: 5’-GGATTCTGGGAACGTCGGAG-3’.
PolR2b Reverse: 5’-CCGGAGTGATCTCATCGTCG-3’.
Analysis of Cytokine Production by RAW 264.7 and P388.D1 Cells Exposed to Bacteriocins
RAW 264.7 and P388.D1 cells were seeded on a 96-well plate (3 × 104 cells per well in 100 µL DMEM, 5% (v/v) FBS). After 16 h, the medium was replaced with DMEM supplemented with 2% (v/v) FBS and 1) 100 ng/mL LPS, 2) 10 ng/mL IFN-γ, 3) 1 μM BacSp222, 4) 1 μM suc-K20-BacSp222, 5) 1 μM suc-K11/K20-BacSp222, 6) 1 μM -fM-BacSp222, 7) 1 μM nisin A, and 8) 10 ng/mL INF-γ combined with 100 ng/mL LPS or 1 μM bacteriocins. After 24-h stimulation, the cultured media were collected, and cytokine concentrations (IL-1α, IL-1β, IL-6, IL-10, IL-12p70, IL-17α, IL-23, IL-27, MCP-1, IFN-β, IFN-γ, TNFα, and GM-CSF) were determined using a LEGENDplex Mouse Inflammation Panel kit (Biolegend, San Diego, CA, USA) and a BD LSRFortessa Cell Analyzer flow cytometer (BD Bioscience, Franklins Lake, NJ, USA). The results were analyzed using LEGENDplex software (Biolegend, San Diego, CA, USA).
Analysis of TNFα Production by Murine Monocyte-Macrophage Cells
RAW 264.7 and P388.D1 cells were cultured and stimulated as described in the section “Analysis of NO production by the cells”. After 6-h stimulation, the level of TNFα secreted to the culture medium was determined using ELISA MAXTM Standard Set Mouse TNFα (Biolegend, San Diego, CA, USA) according to the manufacturer’s protocol. Absorbance was measured at 450 nm using a Synergy H1 Hybrid plate reader controlled by Gene5 version 2.00.18 software (BIOTEK Instruments, Winooski, VT, USA).
Analysis of TNFα Proteolysis
P388.D1 cells were seeded on a 96-well plate at the density of 3 × 104 cells/well in 100 µL DMEM containing 5% (v/v) FBS. After 16 h, the medium was replaced with fresh DMEM supplemented with 2% (v/v) FBS, and the cells were stimulated with 100 ng/mL LPS for 6 h to induce TNFα expression. Then, the medium was again replaced with 100 µL of fresh DMEM or DMEM supplemented with metalloproteinase inhibitors: 1 μM galardin (Tocris Bioscience, Bristol, UK) or 1 mM phenanthroline (Bioshop, Burlington, ON, Canada). After 30 minutes, the cells were stimulated with 1) phorbol 12-myristate 13-acetate (PMA, Sigma, St. Louis, MO, USA), 2) 1 µM BacSp222, or 3) 1 µM -fM-BacSp222 for 45 minutes, and the supernatants were used to determine the concentration of released soluble TNFα.
Analysis of NF-κB Activation in P388.D1 Cells
P388.D1 cells were seeded on a 6-well plate at the density of 1 × 106 cells/well in 3 mL DMEM containing 5% (v/v) FBS. After 16 h, the medium was replaced with fresh DMEM with 2% (v/v) FBS, and the cells were stimulated with 1) 100 ng/mL LPS, 2) 1 μM BacSp222, 3) 1 μM suc-K20-BacSp222, or 4) 1 μM -fM-BacSp222 for 30 minutes. Then, the medium was removed, and the cells were washed with cold phosphate-buffered saline (PBS) and 150 μL RIPA buffer with Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, USA) and phosphatase inhibitors (20 μM NaF, 10 μM Na3VO4, 50 mM β-glycerol phosphate). The solutions were centrifuged, and the total protein concentration was measured using a bicinchoninic acid assay (BCA). Then, Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions was performed using Mini-Protean Precast Gels (Bio-Rad, Hercules, CA, USA). After electrophoresis, the proteins were transferred onto a 0.4 μm pore size polyvinylidene difluoride (PVDF) membrane (Millipore, Burlington, MA, USA) using 25 mM Tris/192 mM glycine buffer containing 10% (v/v) methanol. Then, the membrane was blocked in 5% (w/v) skimmed milk in Tris-buffered saline containing 0.05% (v/v) Tween-20 (TBST) and incubated for 1h with rabbit anti-phosphorylated-NF-κB p65 (Ser536) (Cell Signalling Technology, Danvers, MA, USA) or with mouse anti-NF-κB p65 antibodies (Santa Cruz Biotechnology, Dallas, TX, USA) dissolved in 5% (w/v) skimmed milk or in 5% (w/v) BSA in TBST. Then, the membrane was incubated for 1 h with HRP-conjugated goat anti-rabbit IgG (Sigma, St. Louis, MA, USA) or with HRP-conjugated goat anti-mouse Ig (BD Pharmingen, San Diego, CA, USA) in 5% milk in TBST. Immobilon Western Chemiluminescence HRP Substrate (EMD Millipore Corporation, Burlington, MA, USA) and luminescence imaging platform Fusion FX (Vilber Lourmat, Eberhardzell, Germany) were used to detect the signal. The images were merged in Fusion Capt Advance software (Vilber Lourmat, Eberhardzell, Germany) and the intensity of the bands corresponding to NF-κB was quantitated by the densitometric analysis (ImageJ software v. 1.53c, National Institute of Health). The results are presented relative to control (unstimulated cells).
Analysis of IL-8 Production
Freshly isolated hPMNs were transferred onto a 12-well plate (20 × 106 cells per well in 1 mL 2% (v/v) FBS in RPMI) and stimulated with 1) 100 ng/mL LPS, 2) 1 µM BacSp222, 3) 1 µM suc-K20-BacSp222, or 4) 1 µM -fM-BacSp222. After 17 h exposure to the stimulants, the concentrations of IL-8 cytokine were measured in post-cultured media using Human IL-8/CXCL8 DuoSet ELISA (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instruction.
Analysis of Reactive Oxygen Species (ROS) Production
Freshly isolated hPMNs were transferred onto a black-walled, clear-bottom 96-well plate at the density of 1 × 105 cells per well in 100 µL DMEM without phenol red (GIBCO, Paisley, UK) containing 10% (v/v) FBS and fluorescent probes: 25 µM dichloro-dihydro-fluorescein diacetate (DCF-HDA, Sigma, St. Louis, MO, USA), or 10 µM dihydrorhodamine (DHR, EMD Millipore, Darmstadt, Germany), or 10 µM dihydroethidium (DHE, EMD Millipore, Darmstadt, Germany). At the same time, the media were supplemented with the following stimulants: 1) 100 nM PMA, 2) 1 μM BacSp222, 3) 1 μM suc-K20-BacSp222, 4) 1 μM suc-K11/K20-BacSp222, 5) 1 μM -fM-BacSp222, or 6) 1 μM nisin A. Then, kinetic fluorescent measurements were conducted for 14 h (DCF-HDA, DHR) or 3 h (DHE) using a Synergy H1 Hybrid plate reader and Gene5 version 2.00.18 software (BIOTEK Instruments, Winooski, VT, USA). The intensity of fluorescence was measured in 15-minute intervals. The excitation and emission wavelengths for the individual probes were as follows: DCF 470 nm/535 nm, DHR 485 nm/525 nm, and DHE 490 nm/590 nm.
Analysis of Neutrophil Extracellular Traps (NETs) Formation by Detection of Released DNA
hPMNs were seeded on a 96-well plate (1x 105 cells/well, 100 μL, 10% (v/v) FBS/DMEM without phenol red) and stimulated with 1) 100 nM PMA, 2) 1 μM BacSp222, 3) 1 μM suc-K20-BacSp222, 4) 1 μM suc-K11/K20-BacSp222, 5) 1 μM -fM-BacSp222, or 6) 1 μM nisin for 4 h. Then, DNase (A&A Biotechnology, Gdansk, Poland) was added to the final concentration of 10 U/mL. After 20-minute incubation at 37°C, the DNA digestion was stopped by adding an EDTA solution (final concentration 5 mM). Next, the cells were centrifuged (10 minutes, 800 g), and 50 µL of the post-cultured medium containing released and digested DNA was mixed with 50 µL of a 0.5 μM SYTOX™ Green Nucleic Acid Stain solution (Thermo Fisher Scientific, Waltham, MA, USA) in DMEM without phenol red on a new black 96-well plate. After 10 minutes of incubation in the dark at RT, the fluorescence intensity was measured at 465 nm/525 nm (Ex/Em) using a Synergy H1 Hybrid plate reader and Gene5 version 2.00.18 software (BIOTEK Instruments, Winooski, VT, USA).
Analysis of NET Formation by Fluorescence Microscopy
hPMNs (2 × 106 cells per sample) were seeded on poly-L-lysine-coated (Sigma, St. Louis, MO, USA) glass coverslips placed in a 12-well culture plate. The cells were stimulated with 1) 100 nM PMA, 2) 1 μM BacSp222, or 3) 1 μM -fM-BacSp222 for 3 h and fixed in a 4% (v/v) solution of methanol–free formaldehyde (Thermo Fisher Scientific, Waltham, MA, USA) in PBS for 15 min at RT. After washing (3× in PBS), the cells were blocked and permeabilized in the blocking buffer (5% (v/v) FBS, 0.3% (v/v) Triton X-100 in PBS) for 1 h at RT. Then, the cells were incubated overnight at 4°C in a humidified light-protected chamber with recombinant anti-myeloperoxidase antibody (Abcam, Cambridge, UK) diluted in the blocking buffer. After the incubation, the cells were washed three times with PBS. Then, they were incubated with Alexa Fluor-647 goat anti-rabbit antibodies (Invitrogen, Waltham, MA, USA) diluted in blocking buffer for 75 min at RT in the dark. After washing with PBS, the nuclei were stained with DAPI (Thermo Fisher Scientific, Waltham, MA, USA), and then the cells were again washed with PBS. The samples were mounted onto slides in ProLong Glass Antifade Mountant (Thermo Fisher Scientific, Waltham, MA, USA) and stored for 24 h in the dark. Finally, the samples were observed using a Leica DMI600B inverted widefield fluorescence microscope (Leica Microsystems, Wetzlar, Germany). Images were acquired using a 40x objective and Leica Application Suite X (3.6.0.20104) software. The A4 (DAPI detection) and Y5 (Alexa-647 detection) filter sets (Leica Microsystems) were used for imaging. Final image adjustments were performed using ImageJ 1.53c (National Institutes of Health, Bethesda, MD, USA).
Data Presentation and Statistical Analysis
All experiments were performed with at least two independent replications. The data were presented as mean ± standard error of the mean (SEM). Statistical analysis of the results for cytokines levels, NO concentration, DNA release assay, or ROS detection assay was performed using Statistica software (Tibco Software, version 13.3). Statistical significance of differences between the particular result and control was calculated by one-way ANOVA with Dunnett’s post-hoc test and shown in Figures as asterisks: *, ** or *** for p < 0.05, p < 0.01, p < 0.001, respectively.
Results
All Natural Forms of Bacteriocin BacSp222 Enhance IFN-γ-induced NO Production by Murine Macrophage-like Cell Lines
We have shown recently that unmodified BacSp222 significantly upregulates NO synthesis when applied in combination with IFN-γ.12 Here, we extended our research to include the other recently discovered and naturally occurring forms of BacSp222: suc-K20-BacSp222, suc-K11/K20-BacSp222, and BacSp222 with chemically removed formylated methionine -fM-BacSp222. We analyzed the impact of these molecules on iNOS expression and NO production in P388.D1 and RAW 264.7 cells.
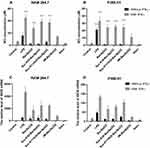
We noticed that BacSp222, suc-K20-BacSp222, and suc-K11/K20-BacSp222 needed IFN-γ co-stimulation to significantly induce iNOS expression, upregulating NO production in P388.D1 and RAW 264.7 cells. Interestingly, the level of NO production induced by both suc-K20-BacSp222 and suc-K11/K20-BacSp222 forms in the presence of IFN-γ was comparable to the level induced by the combination of IFN-γ and LPS (the measured NO concentrations were 39 µM, 37 µM, 45 µM for RAW 264.7 and 25 µM, 24 µM, 33 µM for P388.D1 respectively). On the other hand, the potential of -fM-BacSp222 to generate NO production was significantly lower (the measured NO concentrations were 14 µM for RAW 264.7 and 11 µM for P388.D1) (Figure 1). It is also worth underlining here that, at the concentrations used, all forms of BacSp222 did not affect the viability of the cells (Supplementary Materials Figure 3). Furthermore, in contrast to all forms of BacSp222, nisin A, which is a bacteriocin produced by Lactococcus lactis, did not influence the NO production and iNOS expression regardless of the presence or absence of IFN-γ.
As known, proinflammatory cytokines (TNFα, IL-1β, IFN-γ) or LPS induce iNOS expression in macrophages and endothelial cells.24,25 Given this, we analyzed the impact of BacSp222 and -fM-BacSp222 on NO production in endothelial cells. We used mouse brain endothelial cells (MBE) as a model. However, our results revealed that the MBE cells did not produce NO after stimulation with any of the bacteriocins studied, even if the bacteriocins were combined with IFN-γ, TNFα, or IL-1β (Supplementary Materials Figure 4).
Altogether, our results show that, in addition to BacSp222, the other naturally occurring forms of BacSp222 possess the potential to induce NO production when combined with IFN-γ. However, their activity was limited to macrophage-like cells.
All Natural Forms of Bacteriocin BacSp222 Activate NF-κB – a Major Transcription Factor Controlling the Inflammatory Process
To better understand the mechanism that underlines the bacteriocin-induced iNOS expression in the presence of IFN-γ, we asked whether activation of NF-κB is involved in this phenomenon. We asked this question based on the assumption that the transcription of the iNOS gene requires activation of both IFN-γ-induced transcription factors and NF-κB.26
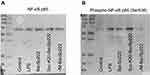
To this end, we analyzed the impact of three forms of bacteriocin BacSp222 on the activation of this transcription factor. Inactive NF-κB consists of a p50 and p65 subunit complex bound to the IκBα inhibitor. The activation of NF-κB requires phosphorylation of its p65 subunit and dissociation of the IκBα inhibitor.27 Thus, to analyze the activation of NF-κB, we assayed the level of the phosphorylated p65 subunit. The P388.D1 cells were incubated for 30 minutes with bacteriocins BacSp222, suc-K20-BacSp222, -fM-BacSp222, or LPS as a positive control for potent NF-κB activation, and an extract of intracellular proteins was prepared. We used antibodies specific to phosphorylated Ser536 p65 and Western blotting analysis. The results showed a higher level of phosphorylated p65 in the cells exposed to bacteriocin BacSp222 and its modified form suc-K20-BacSp222. However, -fM-BacSp222 did not induce p65 phosphorylation (Figure 2A). At the same time, the level of unphosphorylated p65 was the same in all experimental groups (Figure 2B). The quantitative densitometric results of described Western Blot images are presented in Supplementary Materials Figure 5.
These results indicate that in P388.D1 cells stimulated by BacSp222 bacteriocin the transcription factor that undergoes activation is NF-κB.
All Forms of Bacteriocin BacSp222 Increase the Production of Proinflammatory Cytokines by Murine Macrophage-Like Cell Lines
The expression of genes of many proinflammatory factors (especially cytokines) in response to bacterial PAMPs (pathogen-associated molecular patterns) undergoes robust amplification when the cells are simultaneously stimulated with interferons.28 Therefore, we were interested in seeing a similar phenomenon in the studied bacteriocins. We investigated the effect of all forms of BacSp222 on the cytokine production by RAW 264.7 cells. Measurements were performed after stimulation only with bacteriocins or co-stimulation with bacteriocins and IFN-γ.
The concentrations of cytokines in the culture media were analyzed after 24 h of exposure of the cells to the aforementioned factors. None form of BacSp222 or nisin A affected the production of such cytokines as IFN-γ, IL-12p, IL-17A, GM-CSF, IL-6, IL-1β, and IL-23 (Supplementary Figures 6 and 7). However, as shown in Figure 3, BacSp222, suc-K20-BacSp222, and suc-K11/K20-BacSp222 markedly stimulated the TNFα, MCP-1, and IL-1α production (bacteriocins used alone or with IFN-γ). The lowest increase in cytokine concentrations was observed for -fM-BacSp222. On the other hand, a significant increase in cytokines IL-10 and IL-27 was observed for BacSp222, suc-K20-BacSp222, and suc-K11/K20-BacSp222, but only after the co-stimulation with IFN-γ (Figure 3). In contrast, the IFN-β production was increased after the exposure of the cells to all forms of bacteriocin in the presence of IFN-γ. It is worth underlining that these changes in cytokine concentrations after the stimulation with LPS or the co-stimulation with LPS and IFN-γ were similar to the concentration profiles determined after the stimulation with bacteriocins BacSp222 or the co-stimulation with bacteriocins BacSp222 and IFN-γ, respectively. In contrast, nisin A did not cause any changes in the production of the cytokines studied.
We also analyzed the profile of cytokine secretion by P388.D1 cells stimulated with different forms of BacSp222 and nisin A. The levels of TNFα, IL-27 were increased in samples stimulated with the tested forms of BacSp222 (Supplementary Figure 8), whereas none of the bacteriocins affected the levels of IL-1α, IL-1β, IL-10, IL-6, IL-17A, GM-SCF, IL-23, IFN-β, and IFN- γ (Supplementary Figures 9 and 10).
Altogether, these results show that all forms of bacteriocin BacSp222 potentiate the production of selected cytokines by murine macrophage-like cells.
All Forms of Bacteriocin BacSp222 Stimulate the Expression but Not the Proteolysis of Membrane TNFα
TNFα is a major proinflammatory cytokine with broad pleiotropic activity. Various bacterial factors influence the expression of TNFα, and LPS is the best studied factor. TNFα has two biologically active forms: membrane (mTNFα) and soluble (sTNFα). Soluble TNFα is generated exclusively through proteolysis of mTNFα by a disintegrin and metalloprotease 17 (ADAM17 metalloprotease). After binding to TLR4, selected components of bacterial origin, such as LPS, can indirectly stimulate the activity of different sheddases and the process of ectodomain proteolysis.29–31 Therefore, we investigated whether the increased sTNFα secretion observed after the treatment of the cells with the bacteriocins resulted from the upregulated cytokine expression or rather from the activated proteolysis of mTNFα.
First, we confirmed that all forms of bacteriocin BacSp222 increased sTNFα secretion by the RAW 264.7 cells and, to a somewhat lesser degree, by the P388.D1 cells (Figure 4A and B) just after 6 hours of incubation. We then investigated the effect of bacteriocin BacSp222 on mTNFα proteolysis. The cells were stimulated with LPS for 6 hours to induce mTNFα expression. Next, the medium was collected to remove sTNFα secreted during the incubation with LPS, and selected groups of cells were treated with metalloprotease inhibitors (galardin and phenanthroline). The cells were then stimulated with BacSp222 and -fM-BacSp222 or with PMA for 45 minutes to activate protein ectodomain shedding (PMA is an activator of ADAM17, the essential mTNFα convertase in monocyte/macrophage cells). The concentration of sTNFα secreted by the stimulated cells during this time and reflecting the intensity of mTNFα proteolysis was determined using ELISA. As shown in Figure 4C, increased sTNFα concentrations were confirmed only for the PMA-stimulated cells. Therefore, our results unambiguously demonstrate that the increase in sTNFα secretion observed after the stimulation with bacteriocins resulted exclusively from the increased cytokine expression and not from the mTNFα proteolysis.
Bacteriocin BacSp222 and Its Forms Upregulate the Production of IL-8 by hPMNs but Do Not Induce ROS Production or NETs Formation by hPMNs
Our studies on murine monocyte-macrophage cells revealed that bacteriocin BacSp222 and its forms activated the NF-κB transcription factor, leading to increased expression of proteins associated with inflammation, such as iNOS and TNFα. Next to monocytes and macrophages, PMNs are the other essential cells involved in the innate immune response.32,33 The expression of many receptors specific to molecules of bacterial origin allows PMNs to be sensitive sensors detecting pathogens and activating mechanisms leading to their elimination. One of the most important biochemical pathways stimulated in activated PMNs is the NF-κB pathway.34 NF-κB is responsible for the expression of IL-8, one of the most critical proinflammatory cytokines produced by activated PMNs. As demonstrated previously in this work, the natural forms of bacteriocin BacSp222 activated NF-κB in monocytes and macrophages, so we decided to explore whether hPMNs produce IL-8 after stimulation with these peptides. The lack of toxicity of the different forms of bacteriocin BacSp222 (at 1 µM concentration) against hPMNs was confirmed in our previous studies,15 and additionally we have analyzed the toxicity of -fM-BacSp222 (Supplementary Figure 11). Then, the cells were incubated with LPS or with the analyzed BacSp222 forms, and the concentration of IL-8 was measured with ELISA. As shown in Figure 5, BacSp222, suc-K20-BacSp222, and -fM-BacSp222 effectively upregulated the level of IL-8 secreted by neutrophils – similarly to LPS. Therefore, we confirmed that bacteriocin BacSp222 and its succinylated form stimulated cytokine production by the mouse monocyte-macrophage cells and hPMNs. Interestingly, the same effect was noticed in the case of -fM-BacSp222. We also confirmed simultaneously that the nisin-treated hPMNs did not produce IL-8 (Supplementary Materials Figure 12).
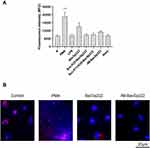
The production of ROS by hPMNs treated by studied bacteriocins was verified using three fluorescent probes: 2’-7ʹdichlorofluorescin diacetate (DCF-HDA), dihydrorhodamine (DHR), and dihydroethidium (DHE). After treatment of cells by different forms of BacSp222, nisin or PMA the fluorescence intensity was measured for 3 h (DHE-stained cells) or 14 h (DHR- and DCF-HDA-stained cells). This approach allowed monitoring the possible production of ROS in the longer term after the addition of the bacteriocins. However, as shown in Figure 6 and Supplementary Figure 13, the increased ROS levels were observed only after the stimulation with PMA, a known inducer of ROS production in cells.
Furthermore, we investigated the formation of NETs by neutrophils after the stimulation with the different forms of bacteriocin using two complementary methods. The neutrophils were first incubated with bacteriocins, LPS, or PMA for 3 hours, and the DNA released from the cells was stained using a fluorescent dye. As shown in Figure 7A, increased fluorescence intensity, corresponding to the increased DNA released, was observed only in the PMA-treated cells (positive control of NET formation). Therefore, the NET formation was not confirmed for cells treated with either bacteriocins or LPS. In the next experiment, NETs formation was observed using a fluorescence microscope. hPMNs were incubated with PMA or bacteriocins for 3 hours and then stained with DAPI (to visualize DNA) as well as antibodies specific to myeloperoxidase (MPO). As demonstrated in Figure 7B and Supplementary Figure 14, the extracellular DNA and MPO were detected only after the treatment with PMA, a known inducer of NOX2 activity. Based on these observations, it can be clearly stated that all the studied forms of bacteriocin BacSp222 do not induce NETs formation.
Discussion
During the characterization of the biological activity of bacteriocin BacSp222 produced by Streptococcus pseudintermedius 222, we observed that the peptide significantly increases NO production by mouse monocytic-macrophage cells treated simultaneously with IFN-γ.12 The objective of the present study focused on evaluating the immunomodulatory activity of various forms of BacSp222: BacSp222, suc-K20-BacSp222, and suc-K11/K20-BacSp222. Simultaneously, we compared their activity with nisin A, one of the best-known and widely commercially applied bacteriocins.37 Because eukaryotic cells are known to recognize formylated peptides precisely and effectively,38,39 we also included -fM-BacSp222 (BacSp222 chemically deprived of formyl-Met) in our research.
Our previous studies specified the bactericidal activity and cytotoxicity of all the tested forms of BacSp222; thus, in the present experiments, the cells were incubated with bacteriocin at a non-toxic concentration.15 We confirmed that bacteriocin BacSp222 and its two succinylated forms, suc-K20-BacSp222 and suc-K11/K20-BacSp222, significantly increased NO production and iNOS expression in the IFN-γ-stimulated RAW 264.7 and P388.D1 cells. Furthermore, the weakest induction of iNOS expression was observed for -fM-BacSp222, indicating that the lack of formylated methionine decreases the stimulatory potential of bacteriocin. At the same time, nisin A did not affect the iNOS expression alone or in co-stimulation with IFN-γ.
Our study revealed that the increase in the NO production in cells exposed to BacSp222 and suc-K20-BacSp222 (but not -fM-BacSp222) resulted from the activation of the most crucial transcription factor inducing iNOS transcription in cells, ie, NF-κB. However, the activation of NF-κB alone is not sufficient for iNOS promoter stimulation. When cells are additionally treated with IFN-γ, the activation of the signal transducer and activator of transcription 1 (STAT1) allows cooperating with NF-κB in the iNOS promoter binding. Therefore, co-stimulation with bacteriocins and IFN-γ results in much more efficient NO production. iNOS expression is induced primarily in immune cells but is not limited only to these cells. Numerous studies have confirmed that iNOS undergoes expression in endothelial cells, smooth muscle cells, astrocytes, fibroblasts, or hepatocytes.22,40,41 The most potent stimulators of iNOS expression are LPS and proinflammatory cytokines: IFN-γ, TNFα, and IL-1β. Combining these stimulators results in a synergistic effect; however, such bacterial molecules as LPS appear to stimulate iNOS expression in immune cells best.24,42,43 Our present research confirmed this phenomenon. BacSp222 enhanced the expression of the iNOS gene only in the macrophages but not in the endothelial cells; therefore, bacteriocin BacSp222 and its succinylated form could be recognized as a novel peptide inducer of NO production by immune cells. To date, the best-known bacterial stimulator of iNOS expression is LPS. However, pneumolysin from Streptococcus pneumoniae, CpG containing DNA, or formyl-methionyl-leucyl-phenylalanine (fMLP) have also been indicated as NO production stimulators.44–48 It is worth underlining that there is no research analyzing the influence of the other bacteriocins on NO production by iNOS; even the well-characterized and commonly used bacteriocin nisin A has not been previously verified in this respect. According to the current knowledge, the only bacteriocin analyzed for induction of iNOS expression is the circular bacteriocin AS-48 produced by Enterococcus faecalis. Cebrian et al observed that bacteriocin AS-48 alone did not induce NO synthesis regardless of the concentration but, contrary to our studies, their experiments did not verify the effect of AS-48 combined with IFN-γ. Interestingly, AS-48 used in some concentrations (2.7 µM and 13.5 µM) decreased the NO production in response to LPS, whereas this phenomenon was not observed in other concentrations (0.27 µM and 27 µM).49
The activation of NF-κB in immune cells increases the expression of cytokines/chemokines involved in the inflammatory response, such as TNFα, MCP-1, or IL-1β.49–52 Our studies revealed that bacteriocin BacSp222 and its posttranslationally modified form suc-K20-BacSp222 led to NF-κB activation and, consequently, increased the secretion of cytokines involved in the inflammatory response. Simultaneously, all natural forms of BacSp222 individually or combined with IFN-γ enhanced the expression of TNFα, MCP-1, IFN-β, IL-10, IL-27, and IL-1α in the murine monocyte-macrophage cells. On the other hand, nisin A had no impact on the cytokine production. Interestingly for bacteriocin BacSp222 and its modified forms, the profiles of upregulated cytokines were similar to the profiles observed upon exposure to LPS or LPS co-stimulated with IFN-γ. The augmented expression of inflammatory genes, including genes coding for cytokines, observed after the co-stimulation with LPS and IFN-γ, results from the integration of intracellular signals generated independently by LPS and IFN-γ. The activation of the STAT1 transcription factor by IFNs and NF-κB by LPS provides transcriptional complexes enabling robust and synergistic upregulation of many genes related to inflammation.53,54 Therefore, our results indicate that a similar phenomenon might concern bacteriocin BacSp222 and IFN-γ. The present studies have revealed that BacSp222 regulates the production of cytokines, pointing to BacSp222 as a peptide with proinflammatory activity. TNFα, IL-1, and chemokine MCP-1 are factors activating local and systemic responses responsible for the resolution of inflammation.55 At the same time, IL-27 and IFN-β are considered cytokines with unique pro-and anti-inflammatory properties, while IL-10 has anti-inflammatory activity.54,56 It is known that activation of immune cells by LPS induces secretion of proinflammatory cytokines (TNFα, MCP-1), but it is always followed by upregulation of anti-inflammatory cytokines, such as IL-10, to precisely control the inflammatory response.57 In our opinion, a similar mechanism may be responsible for the increase in the IL-10 concentration observed after 24 hours of incubation of the RAW 264.7 cells with bacteriocin BacSp222 and its modified forms.
Neutrophils are the most abundant innate immune response cells exerting multiple functions.33 These cells with high sensitivity are able to detect pathogen-associated molecular patterns (PAMPs), rapidly migrate to the place of infection, and secrete proinflammatory cytokines such as IL-8, IL-1β, and IFN-γ, to signal, communicate, attract, and stimulate the other populations of immune system cells involved in elimination of pathogens. Moreover, neutrophils are also masters of phagocytosis, degranulation, ROS production, and extracellular trap formation. However, the excessive activation of neutrophils can lead to acute or chronic inflammation, resulting in tissue damage.33,58 Our studies have shown that each form of bacteriocin BacSp222 stimulates human neutrophils to produce IL-8. Simultaneously, we have not confirmed increased expression of this chemokine in the case of nisin A-treated neutrophils. These results align with our observations that BacSp222 and suc-K20-BacSp222 activate NF-κB and stimulate the expression of proinflammatory cytokines in murine monocytic-macrophage cells. Furthermore, numerous studies indicate that, similarly to monocytes and macrophages, NF-κB controls the expression of proinflammatory cytokines in neutrophils.34 The proinflammatory chemokine IL-8 (also known as CXCL8) is a chemotactic agent recruiting leukocytes, primarily neutrophils, and monocytes but also cytotoxic T cells to infected or inflamed tissues.59 The increased IL-8 expression is induced primarily via activation of PAMP receptors such as TLRs and FPRs.36,60 However, cytokines and other molecules associated with inflammation are also potent stimulators of IL-8 expression.61,62 The production of cytokines by immune cells exposed to bacteriocins is poorly understood; the published reports are mostly focused on nisin, but some of them are contradictory. Aranha et al did not observe an effect of nisin A on the production of TNFα, IL-6, IL-8, IL-10, and GM-CSF by rabbit cells isolated from cervicovaginal tissue explants.63 Another team showed an increase in the production of IL-1β by porcine peripheral blood mononuclear cells (PBMC) only in response to a very high concentration of partially purified nisin A. However, at the same time, the same preparation of nisin decreased the LPS-induced secretion of IL-6.64 Mouritzen et al observed that post-culture media collected from PBMC stimulated simultaneously with LPS and nisin A and found a decrease in the TNFα level, compared to cells exposed only to LPS.65 Similar observations indicating suppression of LPS activity were reported for nisin Z. Simultaneously, a stimulating effect of nisin Z on the production of chemokine MCP-1 and IL-8 by PBMC was observed.66
The production of ROS and formation of neutrophil extracellular traps (NETs) are essential functions of neutrophils in the first line of defense against invading pathogens. ROS production is triggered by activating several receptors that bind molecules of bacterial origin or even some host proteins (such as adhesion molecules or antibodies). The stimulation of the intracellular signaling cascade eventually leads to assembling a multiprotein NADPH oxidase complex (NOX2) responsible for ROS production.35 Large amounts of ROS can induce damage to DNA and cellular structures and effectively participate in pathogen elimination. Furthermore, ROS production is also necessary for activating granular proteases, resulting in the formation of NETs consisting of DNA and proteins from granules, including elastase and myeloperoxidase (MPO). Numerous observations indicate that ROS also play a key role in intracellular signal transmission, leading to the activation of selected transcription factors, thereby indirectly participating in the regulation of different gene expression. Some data indicate that endogenous ROS produced by NOX2 by activated neutrophils may also be potent inducers of IL-8 expression. ROS-dependent IL-8 expression was already observed in response to fMLP or LPS.36,67,68 Since we confirmed that the bacteriocin and its different forms stimulated the production of this cytokine by hPMNs, we were interested whether studied bacteriocins also induces ROS formation in these cells. However, our studies did not prove upregulation of ROS synthesis in the neutrophils exposed to either BacSp222 or nisin A. ROS production in neutrophils often increases after stimulation of various receptors. Signaling transmitted by integrin, Fc receptors, and FPRs can stimulate this process by direct activation of the NOX2 complex. However, ligand binding to the other receptors, including TLRs, primes neutrophils and makes NOX2 more susceptible to activation by the secondary stimulus.35 Perhaps, bacteriocin BacSp222 needs an additional stimulating factor to induce ROS formation in neutrophils. However, verifying this hypothesis requires separate, more detailed research.
Finally, our research has confirmed that bacteriocin BacSp222, its modified forms, and nisin A do not induce NET formation. Because the detection of NETs was carried out using two different methods and a few hours after the addition of the stimulants, we excluded all forms of NETs: vital, mitochondrial (occurring up to one hour after the exposure to the stimulants), and suicidal (occurring a few hours after the exposure to the stimulants).69 These results appear consistent with the results of experiments that also excluded ROS production after stimulation of neutrophils with bacteriocin BacSp222, its modified forms, and nisin A. This is because various observations indicate that the classical intracellular pathway triggering NET formation depends just on endogenous ROS production. For example, ROS produced in a NOX2-dependent or NOX2-independent manner are required for peptidyl arginine deaminase 4 (PAD4), a key enzyme involved in histone decondensation.70,71 The NET formation was also observed for various stimuli such as whole bacteria or bacteria-derived components, fungi, viruses, complexes of antibodies with antigens, autoantibodies, or cytokines.72
The potential effect of the other bacteriocins on NET formation is very poorly understood. Begde et al showed that nisin A at a concentration of 75 or 150 μM induced NET formation by human neutrophils in a NOX2-dependent manner. However, these studies have some limitations since the authors did not study the viability of neutrophils treated with such high concentrations of nisin A, nor did they provide detailed information about the purity or homogeneity of the nisin A used in the experiments.73 In our experiments, the concentration of bacteriocins (various forms of BacSp222 and nisin A) was much lower (1 μM) than the cytotoxic one, and we used highly purified preparations devoid of other potential components that could stimulate immune cells (such as LPS).
In summary, our studies indicate that all natural forms of BacSp222 A exhibit proinflammatory activity against murine (monocyte-macrophages cells) and hPMNs. Simultaneously, we have excluded this activity in the comparable and widely applied commercial bacteriocin – nisin A. The proinflammatory effect of bacteriocin BacSp222 primarily consists in NF-κB activation, increased iNOS expression, and upregulated secretion of several cytokines regulating the inflammatory response. However, this proinflammatory effect does not involve ROS production and NET formation by neutrophils, limiting the potentially harmful effects on the host cells. Our studies significantly extend the knowledge of the biological importance of bacteriocins, because the details of their interactions with eukaryotic cells, particularly with immune cells, are poorly documented. Moreover, the results of the inflammatory activities of many bacteriocins – especially nisin – are often contradictory. In our opinion, these discrepancies in observations most probably result from using nisin preparations with insufficient or varying degrees of purity. Our research is one of the first comparative and comprehensive studies of the interactions of the studied bacteriocins used in low concentrations. It was performed using highly purified peptide preparations, which were additionally verified for the presence of possible contamination by LPS. The results of the experiments performed with the mouse and human immune cells have revealed increased production of NO and selected cytokines and confirmed the proinflammatory activity of bacteriocin BacSp222. We have also demonstrated that the posttranslational modifications of this bacteriocin (succinylation) do not diminish its immunomodulatory activity, contrary to the significantly lower antibacterial properties or cytotoxic activity of such modified forms of this peptide against eukaryotic cells.15 Currently, we do not know the detailed mechanism of interactions of BacSp222 forms with host cells. Particularly, a question arises whether and through which receptor bacteriocin BacSp222 interacts with cells. TLRs and FPRs are receptors through which various cells recognize PAMPs. The biological activity of BacSp222 manifested in the activation of NF-κB, the stimulation of NO production, and cytokine expression, especially in cooperation with IFN-γ, may indicate interaction with one of the TLRs. In our study, we used BacSp222 deprived of formylated methionine to compare its activity with its natural forms. However, we did not obtain conclusive results indicating significantly lower activity of -fM-BaccSp222. The -fM-BacSp222 form was less likely to increase NO production and activate NF-κB, but simultaneously stimulated the production of the selected cytokines to a similar degree as the formylated forms of BacSp222. Thus, BacSp222 binding to FPRs also seems debatable. Other researchers analyzing bacteriocin interactions with eukaryotic cells have not yet attempted to identify potential receptors through which bacteriocins interact with cells.
Conclusion
After contact with the peptide, the mouse and human immune cells mobilized NF-κB and exhibited increased production of NO as well as selected cytokines, confirming the proinflammatory activity of bacteriocin BacSp222. The atypical posttranslational modification of bacteriocin, ie succinylation, does not diminish such biological activities. On the other hand, the experiments with BacSp222 deprived of formylated methionine gave no unequivocal results. Therefore, the question of the type of the cellular receptor involved in bacteriocin recognition is still open.
Acknowledgments
The work was financed by National Science Centre, Poland, in the framework of the project No 2018/31/B/NZ3/01226 (to P.M.). The authors thank Joanna Bereta, Oliwia Bocheńska, Grażyna Braś, Urszula Jankowska, Justyna Karkowska-Kuleta, Jakub Kochan and Joanna Kozieł for their precious help and tips. We acknowledge Proteomics and Mass Spectrometry Core Facility of the Malopolska Centre of Biotechnology, Jagiellonian University, for peptide mass determination.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Webb SAR, Kahler CM. Bench-to-bedside review: bacterial virulence and subversion of host defences. Crit Care. 2008;12(6):234. doi:10.1186/CC7091/TABLES/1
2. Casadevall A, Pirofski LA. Virulence factors and their mechanisms of action: the view from a damage–response framework. J Water Health. 2009;7(S1):S2–S18. doi:10.2166/WH.2009.036
3. Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA. Mechanisms of bacterial pathogenicity. Postgrad Med J. 2002;78(918):216–224. doi:10.1136/PMJ.78.918.216
4. Johnson DI. Bacterial virulence factors. In: Bacterial Pathogens and Their Virulence Factors,
5. Bay L, Ring HC. Human skin microbiota in health and disease: the cutaneous communities’ interplay in equilibrium and dysbiosis: the cutaneous communities’ interplay in equilibrium and dysbiosis. APMIS. 2021. doi:10.1111/APM.13201
6. Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17(6):371–382. doi:10.1038/s41579-019-0186-5
7. Simons A, Alhanout K, Duval RE. Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorg. 2020;8(5):639. doi:10.3390/MICROORGANISMS8050639
8. Cintas LM, Casaus MP, Herranz C, Nes IF, Hernández PE. Review: bacteriocins of Lactic Acid Bacteria. Food Sci Technol Int. 2001;7(4):281–305. doi:10.1106/R8DE-P6HU-CLXP-5RYT
9. Cesa-Luna C, Alatorre-Cruz JM, Carreño-López R, Quintero-Hernández V, Baez A. Emerging applications of bacteriocins as antimicrobials, anticancer drugs, and modulators of the gastrointestinal microbiota. Polish J Microbiol. 2021;70(2):143–159. doi:10.33073/PJM-2021-020
10. Mak P. Staphylococcal Bacteriocins. In: Vincenzo S editor. Pet-to-Man Travelling Staphylococci: A World in Progress. Academic Press; 2018:161–171. doi:10.1016/B978-0-12-813547-1.00013-3
11. Bonar E, Bukowski M, Chlebicka K, et al. Human skin microbiota-friendly lysostaphin. Int J Biol Macromol. 2021;183:852–860. doi:10.1016/J.IJBIOMAC.2021.04.154
12. Wladyka B, Piejko M, Bzowska M, et al. A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci Rep. 2015;5:14569. doi:10.1038/srep14569
13. Somayaji R, Priyantha MAR, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis. 2016;85(4):471–476. doi:10.1016/J.DIAGMICROBIO.2016.05.008
14. Nowakowski M, Jaremko Ł, Wladyka B, Dubin G, Ejchart A, Mak P. Spatial attributes of the four-helix bundle group of bacteriocins – the high-resolution structure of BacSp222 in solution. Int J Biol Macromol. 2018;107:2715–2724. doi:10.1016/j.ijbiomac.2017.10.158
15. Śmiałek J, Nowakowski M, Bzowska M, et al. Structure, biosynthesis, and biological activity of succinylated forms of bacteriocin BacSp222. Int J Mol Sci. 2021;22(12):6256. doi:10.3390/IJMS22126256
16. Taylor TM, Davidson PM, Zhong Q. Extraction of Nisin from a 2.5% commercial nisin product using methanol and ethanol solutions. J Food Prot. 2007;70(5):1272–1276. doi:10.4315/0362-028X-70.5.1272
17. Wladyka B, Wielebska K, Wloka M, et al. Isolation, biochemical characterization, and cloning of a bacteriocin from the poultry-associated Staphylococcus aureus strain CH-91. Appl Microbiol Biotechnol. 2013;97(16):7229–7239. doi:10.1007/S00253-012-4578-Y
18. Griess P. Weselsky und Benedikt „Ueber einige Azoverbindungen” [Remarks on the treatise of HH. Weselsky and Benedikt “On some azo compounds”]. Berichte der Dtsch Chem Gesellschaft. 1879;12(1):426–428. doi:10.1002/CBER.187901201117
19. Hakobyan L, Monforte-Gómez B, Moliner-Martínez Y, Molins-Legua C, Campíns-Falcó P. Improving sustainability of the Griess reaction by reagent stabilization on PDMS membranes and ZnNPs as reductor of nitrates: application to different water samples. Polym. 2022;14(3):464. doi:10.3390/POLYM14030464
20. Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581–585. doi:10.1038/nprot.2006.83
21. Kobayashi Y. The regulatory role of nitric oxide in proinflammatory cytokine expression during the induction and resolution of inflammation. J Leukoc Biol. 2010;88(6):1157–1162. doi:10.1189/JLB.0310149
22. Kleinert H, Schwarz PM, Förstermann U. Regulation of the expression of inducible nitric oxide synthase. Biol Chem. 2003;384(10–11):1343–1364. doi:10.1515/BC.2003.152/MACHINEREADABLECITATION/RIS
23. Kleinert H, Pautz A, Linker K, Schwarz PM. Regulation of the expression of inducible nitric oxide synthase. Eur J Pharmacol. 2004;500(1–3):255–266. doi:10.1016/J.EJPHAR.2004.07.030
24. Bereta J, Cohen MC, Bereta M. Stimulatory effect of ouabain on VCAM-1 and iNOS expression in murine endothelial cells: involvement of NF-kappa B. FEBS Lett. 1995;377(1):21–25. doi:10.1016/0014-5793(95)01301-6
25. Bereta J, Bereta M, Allison AC, Kruger PB, Koj A. Inhibitory effect of di-catechol rooperol on VCAM-1 and iNOS expression in cytokine-stimulated endothelium. Life Sci. 1997;60(4–5):325–334. doi:10.1016/S0024-3205(96)00633-9
26. Mir M, Tolosa L, Asensio VJ, Lladó J, Olmos G. Complementary roles of tumor necrosis factor alpha and interferon gamma in inducible microglial nitric oxide generation. J Neuroimmunol. 2008;204(1–2):101–109. doi:10.1016/J.JNEUROIM.2008.07.002
27. Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-κB and IκB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30(1):43–52. doi:10.1016/J.TIBS.2004.11.009
28. Cavaillon JM. Exotoxins and endotoxins: inducers of inflammatory cytokines. Toxicon. 2018;149:45–53. doi:10.1016/J.TOXICON.2017.10.016
29. Yan I, Schwarz J, Lücke K, et al. ADAM17 controls IL-6 signaling by cleavage of the murine IL-6Rα from the cell surface of leukocytes during inflammatory responses. J Leukoc Biol. 2016;99(5):749–760. doi:10.1189/JLB.3A0515-207R
30. Flynn CM, Garbers Y, Lokau J, et al. Activation of Toll-like Receptor 2 (TLR2) induces Interleukin-6 trans-signaling. Sci Rep. 2019;9(1):1–11. doi:10.1038/s41598-019-43617-5
31. Glenn G, van der Geer P. Toll-like receptors stimulate regulated intramembrane proteolysis of the CSF-1 receptor through Erk activation. FEBS Lett. 2008;582(6):911–915. doi:10.1016/J.FEBSLET.2008.02.029
32. Fine N, Barzilay O, Sun C, et al. Primed PMNs in healthy mouse and human circulation are first responders during acute inflammation. Blood Adv. 2019;3(10):1622–1637. doi:10.1182/BLOODADVANCES.2018030585
33. Rosales C. Neutrophils at the crossroads of innate and adaptive immunity. J Leukoc Biol. 2020;108(1):377–396. doi:10.1002/JLB.4MIR0220-574RR
34. Cloutier A, Ear T, Blais-Charron E, Dubois CM, McDonald PP. Differential involvement of NF-κB and MAP kinase pathways in the generation of inflammatory cytokines by human neutrophils. J Leukoc Biol. 2007;81(2):567–577. doi:10.1189/JLB.0806536
35. Nguyen GT, Green ER, Mecsas J. Neutrophils to the ROScue: mechanisms of NADPH oxidase activation and bacterial resistance. Front Cell Infect Microbiol. 2017;7:373. doi:10.3389/FCIMB.2017.00373/BIBTEX
36. Hidalgo MA, Carretta MD, Teuber SE, et al. FMLP-Induced IL-8 release is dependent on NADPH oxidase in human neutrophils. J Immunol Res. 2015;2015:120348. doi:10.1155/2015/120348
37. Hansen JN, Sandine WE. Nisin as a model food preservative. Crit Rev Food Sci Nutr. 1994;34(1):69–93. doi:10.1080/10408399409527650
38. He HQ, Ye RD. The formyl peptide receptors: diversity of ligands and mechanism for recognition. Mol. 2017;22(3):455. doi:10.3390/MOLECULES22030455
39. Weiß E, Kretschmer D. Formyl-peptide receptors in infection, inflammation, and cancer. Trends Immunol. 2018;39(10):815–829. doi:10.1016/J.IT.2018.08.005
40. Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2(10):907–916. doi:10.1038/ni1001-907
41. Korhonen R, Lahti A, Kankaanranta H, Moilanen E. Nitric oxide production and signaling in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4(4):471–479. doi:10.2174/1568010054526359
42. Geller DA, Nussler AK, Di Silvio M, et al. Cytokines, endotoxin, and glucocorticoids regulate the expression of inducible nitric oxide synthase in hepatocytes. Proc Natl Acad Sci. 1993;90(2):522–526. doi:10.1073/PNAS.90.2.522
43. Sparrow JR, Nathan C, Vodovotz Y. Cytokine regulation of nitric oxide synthase in mouse retinal pigment epithelial cells in culture. Exp Eye Res. 1994;59(2):129–139. doi:10.1006/EXER.1994.1091
44. Braun JS, Novak R, Gao G, Murray PJ, Shenep JL, Fischetti VA. Pneumolysin, a protein toxin of streptococcus pneumoniae, induces nitric oxide production from macrophages. Infect Immun. 1999;67(8):3750–3756. doi:10.1128/IAI.67.8.3750-3756.1999
45. Malley R, Henneke P, Morse SC, et al. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci. 2003;100(4):1966–1971. doi:10.1073/PNAS.0435928100
46. BenMohamed F, Medina M, Wu Y-Z, et al. Toll-like receptor 9 deficiency protects mice against pseudomonas aeruginosa lung infection. PLoS One. 2014;9(3):e90466. doi:10.1371/JOURNAL.PONE.0090466
47. Lee SH, Lee JG, Kim JR, Baek SH. Toll-like receptor 9-mediated cytosolic phospholipase A2 activation regulates expression of inducible nitric oxide synthase. Biochem Biophys Res Commun. 2007;364(4):996–1001. doi:10.1016/J.BBRC.2007.10.111
48. Lee H, Park C, Cho IH, et al. Double-stranded RNA induces iNOS gene expression in Schwann cells, sensory neuronal death, and peripheral nerve demyelination. Glia. 2007;55(7):712–722. doi:10.1002/GLIA.20493
49. Cebrián R, Rodríguez-Cabezas ME, Martín-Escolano R, et al. Preclinical studies of toxicity and safety of the AS-48 bacteriocin. J Adv Res. 2019;20:129–139. doi:10.1016/J.JARE.2019.06.003
50. Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2(1):1–9. doi:10.1038/sigtrans.2017.23
51. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1(6):a001651–a001651. doi:10.1101/CSHPERSPECT.A001651
52. Kulms D, Schwarz T. NF‐κB and Cytokines. Vitam Horm. 2006;74:283–300. doi:10.1016/S0083-6729(06)74011-0
53. Hayes MP, Freeman SL, Donnelly RP. IFN-gamma priming of monocytes enhances LPS-induced TNF production by augmenting both transcription and MRNA stability. Cytokine. 1995;7(5):427–435. doi:10.1006/CYTO.1995.0058
54. Bolívar S, Anfossi R, Humeres C, et al. IFN-β plays both pro- and anti-inflammatory roles in the rat cardiac fibroblast through differential STAT protein activation. Front Pharmacol. 2018;9:1368. doi:10.3389/FPHAR.2018.01368/BIBTEX
55. Dinarello CA. Proinflammatory cytokines. Chest. 2000;118(2):503–508. doi:10.1378/CHEST.118.2.503
56. Aparicio-Siegmund S, Garbers C. The biology of interleukin-27 reveals unique pro- and anti-inflammatory functions in immunity. Cytokine Growth Factor Rev. 2015;26(5):579–586. doi:10.1016/J.CYTOGFR.2015.07.008
57. Meisel C, Vogt K, Platzer C, Randow F, Liebenthal C, Volk HD. Differential regulation of monocytic tumor necrosis factor-alpha and interleukin-10 expression. Eur J Immunol. 1996;26(7):1580–1586. doi:10.1002/EJI.1830260726
58. Hilda JN, Das S, Tripathy SP, Hanna LE. Role of neutrophils in tuberculosis: a bird’s eye view. Innate Immun. 2020;26(4):240–247. doi:10.1177/1753425919881176
59. Baggiolini M, Loetscher P, Moser B. Interleukin-8 and the chemokine family. Int J Immunopharmacol. 1995;17(2):103–108. doi:10.1016/0192-0561(94)00088-6
60. Tavano R, Segat D, Fedeli C, et al. Formyl-peptide receptor agonists and amorphous SiO2-NPs synergistically and selectively increase the inflammatory responses of human monocytes and PMNs. Nanobiomedicine. 2016;3:2. doi:10.5772/62251
61. Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72(5):847–855.
62. Kukulski F, Bahrami F, Yebdri FB, et al. NTPDase1 controls IL-8 production by human neutrophils. J Immunol. 2011;187(2):644–653. doi:10.4049/JIMMUNOL.1002680
63. Aranha CC, Gupta SM, Reddy KVR. Assessment of cervicovaginal cytokine levels following exposure to microbicide Nisin gel in rabbits. Cytokine. 2008;43(1):63–70. doi:10.1016/J.CYTO.2008.04.005
64. Małaczewska J, Kaczorek-łukowska E, Wójcik R, Rękawek W, Siwicki AK. In vitro immunomodulatory effect of nisin on porcine leucocytes. J Anim Physiol Anim Nutr. 2019;103(3):882–893. doi:10.1111/JPN.13085
65. Mouritzen MV, Andrea A, Qvist K, Poulsen SS, Jenssen H. Immunomodulatory potential of Nisin A with application in wound healing. Wound Repair Regen. 2019;27(6):650–660. doi:10.1111/WRR.12743
66. Kindrachuk J, Jenssen H, Elliott M, et al. Manipulation of innate immunity by a bacterial secreted peptide: lantibiotic nisin Z is selectively immunomodulatory. Innate Immun. 2013;19(3):315–327. doi:10.1177/1753425912461456
67. DeForge LE, Preston AM, Takeuchi E, Kenney J, Boxer LA, Remick DG. Regulation of interleukin 8 gene expression by oxidant stress. J Biol Chem. 1993;268(34):25568–25576. doi:10.1016/S0021-9258(19)74429-9
68. Lekstrom-Himes JA, Kuhns DB, Alvord WG, Gallin JI. Inhibition of human neutrophil IL-8 production by hydrogen peroxide and dysregulation in chronic granulomatous disease. J Immunol. 2005;174(1):411–417. doi:10.4049/JIMMUNOL.174.1.411
69. Tan C, Aziz M, Wang P. The vitals of NETs. J Leukoc Biol. 2021;110(4):797–808. doi:10.1002/JLB.3RU0620-375R
70. Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207(9):1853–1862. doi:10.1084/JEM.20100239
71. Steinberg BE, Grinstein S. Unconventional roles of the NADPH oxidase: signaling, ion homeostasis, and cell death. Sci STKE. 2007;2007(379):pe11. doi:10.1126/STKE.3792007PE11
72. Yang H, Biermann MH, Brauner JM, Liu Y, Zhao Y, Herrmann M. New insights into neutrophil extracellular traps: mechanisms of formation and role in inflammation. Front Immunol. 2016;7:302. doi:10.3389/FIMMU.2016.00302
73. Begde D, Bundale S, Pise MV, Rudra J, Nashikkar N, Upadhyay A. Immunomodulatory efficacy of nisin—a bacterial lantibiotic peptide. J Pept Sci. 2011;17(6):438–444. doi:10.1002/PSC.1341
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.