Back to Journals » Cancer Management and Research » Volume 13
AVL9 is Upregulated in and Could Be a Predictive Biomarker for Colorectal Cancer
Authors Li D, Zeng Y, Shen P , Lin X, Yang T, Chen B, Ma Z , Wang H
Received 13 January 2021
Accepted for publication 22 March 2021
Published 9 April 2021 Volume 2021:13 Pages 3123—3132
DOI https://doi.org/10.2147/CMAR.S301844
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Danfeng Li,1,* Yongming Zeng,1,* Peilin Shen,2,* Xiaosheng Lin,1 Tian Yang,1 Binlie Chen,1,3 Zhiyan Ma,1,3 Huaiming Wang1
1Department of Gastrointestinal Surgery, The First Affiliated Hospital of Shantou University Medical College, Shantou, 515041, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Shantou University Medical College, Shantou, 515041, People’s Republic of China; 3Medical College, Shantou University, Shantou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Huaiming Wang
Department of Gastrointestinal Surgery, The First Affiliated Hospital of Shantou University Medical College Email [email protected]
Purpose: This study aimed to explore the function and clinical significance of AVL9 in colorectal cancer (CRC).
Materials and Methods: The GEO, TCGA, and GEPIA databases were searched to evaluate the expression level of AVL9, while the SurvExpress online tool was used to explore its related clinical survival prognosis. The cBioPortal and LinkedOmics databases were used to identify AVL9 expression-related genes. Protein–protein interaction (PPI) networks were analyzed using Cytoscape 3.7.1 and DAVID6.8, which was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) signal pathway enrichment. The immunohistochemistry of AVL9 in CRC was detected using an online tool protein atlas. RNA isolation and reverse transcription–quantitative polymerase chain reaction (RT-qPCR) assays were used to detect AVL9 expression in tissue and plasma samples.
Results: Our study confirmed that AVL9 was highly expressed in CRC lesions versus the adjacent normal tissues (P < 0.001). High AVL9 expression was negatively associated with survival outcomes (P < 0.05). GO analysis showed that AVL9 expression-related genes were enriched in single organismal cell–cell adhesion, post-transcriptional regulation of gene expression, and negative regulation of the vascular endothelial growth factor receptor signaling pathway (P < 0.05). On a KEGG pathway analysis, these genes were mainly involved in progesterone-mediated oocyte maturation, axon guidance, the insulin signaling pathway, and the ubiquitin-mediated proteolysis signaling pathways (P < 0.05). In the PPI analysis, the KBTBD2, KIAA1147, EPDR1, and RNF216 genes interacted with AVL9, and GEPIA predicted that their expression levels were all positively correlated with AVL9. Furthermore, a clinicopathological parameter analysis found that high AVL9 expression was positively correlated with differentiation and TNM stage. RT-qPCR analysis further showed that plasma AVL9 expression was upregulated in CRC patients versus healthy controls.
Conclusion: AVL9 could serve as a potential biomarker and therapeutic target for CRC.
Keywords: AVL9, biomarker, colorectal cancer, prognosis
Introduction
Colorectal cancer (CRC) is a common malignant tumor worldwide.1 According to the latest statistics from the American Cancer Society, CRC is the second most common cause of cancer death in the United States. In the past 10 years, there has been a rapid decline in its incidence among those aged 65 years and older, with a downward trend of approximately 3.3% annually. However, the incidence in the age group of 50–64 years has increased by 1% each year, while the incidence of CRC patients under 50 years of age has increased by approximately 2% annually. The mortality rate showed the same trend in the above age groups. It is worth noting that the mortality rate of CRC in those age <50 years increased by 1.3% per year.2 In fact, 90% of CRC-related deaths can be prevented if patients are diagnosed at an early stage; therefore, early diagnosis is the fundamental way to improve patient prognosis.3 The basic reason for poor patient prognosis is that the understanding of the exact molecular mechanism of the early occurrence and development of CRC remains very limited. Therefore, a deeper understanding of the specific pathogenesis of CRC and clarification of the reasons for the increase in morbidity and mortality in young and middle-aged adults can improve the overall prognosis of CRC.
The migration-associated gene AVL9 is located on the human chromosome 7.14:32,495,489–32,588,726 (111,980 nt in length). Recent studies have shown that AVL9 plays an important role in cell polarity, cell migration, and cell cycle progression and acts as an oncogene.4,5 Zhang et al revealed that AVL9 expression is upregulated in clear cell renal carcinomas and promotes cell migration.6 Moreover, our previous study identified that AVL9 is a target gene of the linc00662/miR-497-5p axis that promotes the development of CRC.7 However, the function and clinical application value of AVL9 in CRC remain largely unexplored, and in the present study, we aimed to explore the roles of AVL9 in CRC and its potential relationship with clinicopathologic parameters and identify whether it may serve as a new target for the diagnosis and treatment of CRC.
Materials and Methods
Data Acquisition
The expression level of AVL9 in CRC was analyzed using GEPIA (http://gepia.cancer-pku.cn/), Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo), and The Cancer Genome Atlas (TCGA) databases (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) databases. Overall survival (OS) and disease-free survival (DFS) analyses were performed using the online database SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). The online tool protein atlas (https://www.proteinatlas.org/) was used to detect AVL9 expression in different human tumors.
Bioinformatics Analysis
AVL9 expression-related genes were obtained from LinkedOmics (http://www.linkedomics.org/login.php) and cBioPortal (http://www.cbioportal.org/). A Spearman correlation coefficient of >|0.5| was defined as the screening condition, while a Venn diagram was generated using the online webtool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to identify overlapping related genes based on the screening conditions. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway enrichment analyses of AVL9 expression-related genes were performed using the online biological information database DAVID6.8 (http://david.ncifcrf.gov/) (P<0.05). The protein–protein interaction (PPI) network was predicted using the STRING database (http://string-db.org), “Multiple proteins” was selected to enter expression-related genes and AVL9, “Homo sapiens” was selected for the species, and “Medium confidence (0.400)” was selected for the confidence of the strength of the interactions between proteins. Then, the networking results were exported in the “TSV” format and imported into Cytoscape 3.7.1.
Tissue Samples, Blood Samples, and Clinical Data Collection
Human CRC tissues and their paired adjacent noncancerous specimens were collected from 50 patients who had undergone CRC surgical resection without receiving any other treatment before surgery and were pathologically diagnosed with CRC between 2014 and 2016. After excision, the tissues were quickly frozen and stored at −80°C. Patients were monitored once every 3 months in the first 2 years after surgery and then every 6 months thereafter; at each follow-up, a detailed history was collected and a comprehensive physical examination performed; data of routine blood parameters, liver function, plasma carbohydrate antigen19-9, carcinoembryonic antigen, and abdominal ultrasound were all required. OS was calculated from the date of diagnosis until the date of death or the end of the follow-up period. Plasma samples of CRC patients and healthy controls were obtained from The First Affiliated Hospital of Shantou University Medical College. The Ethics Committee of Shantou University Medical College approved this study, and each participant and healthy controls signed an informed consent form regarding their specimen use for scientific research, and the research was performed in accordance with the World Medical Association Declaration of Helsinki.
Quantitative Real-Time Polymerase Chain Reaction
RNAiso Plus reagent (TaKaRa, Dalian, China) was used to extract total RNA from frozen tissues and plasma samples. According to the manufacturer’s instructions, 500 ng of total RNA was reverse transcribed to cDNA using the PrimeScript RT Master Mix (TaKaRa). The SYBR Premix Ex Taq II Kit (TaKaRa) was used to determine the expression level of AVL9 using the StepOnePlus system (Applied Biosystems, CA, USA). Fold changes were calculated using the 2−ΔΔCt method. The primer sequences for AVL9 were as follows: forward 5′-GTGAGGCACGTGACTGAGAA-3′; and reverse: 5′-TTGTTGCTGTTCCACACCCT-3′. The primer sequences for GAPDH were as follows: forward 5′-CGCTCTCTGCTCCTCCTGTTC-3′; reverse: 5′-ATCCGTTGACTCCGACCTTCAC-3′. Data were obtained from at least three independent experiments and are presented as mean ± standard deviation.
Statistics
All data are presented as mean ± standard deviation. The chi-square test was used to analyze the data. Statistical analyses were performed using IBM SPSS 24.0 (IBM, USA). An unpaired t-test was used to evaluate the differential expression of AVL9 between cancer tissues and adjacent normal tissues. The Log rank test and Cox proportional hazards regression were used to estimate the prognostic value of AVL9. Receiver operating characteristic (ROC) curve analysis was performed using the “pROC” package8 in R software (version 4.03), and significant differences were considered at values of P < 0.05.
Results
Upregulation of AVL9 Predicted Poor Prognosis in CRC Patients
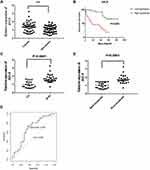
Based on our previous study and other published research, we first found that the relative expression level of AVL9 was significantly upregulated in CRC tissues in the GEPIA database. The data also indicated that AVL9 expression was upregulated in CRC versus normal tissues in the GEO microarray dataset GSE32323 and dramatically increased in CRC, findings that were consistent with the GEO and GEPIA data (Figure 1A–C). The online tool protein atlas was used to detect the expression of AVL9 in different human tumors, and the results showed that both AVL9 mRNA and protein were significantly expressed in CRC (Figure S1). Immunohistochemical analysis was performed to detect the AVL9 protein expression in 10 CRC tissues, and the results showed that 9 of 10 showed moderate staining and one of 10 showed strong staining. In the intensity analysis, we found that 2 of 10 were strong, while 8 of 10 were moderate. In the quantity analysis, we found that 7 of 10 were more than 75%, 2 of 10 were between 25% and 75%, and 1 of 10 was less than 25%; therefore, we can conclude that the AVL9 protein is also highly expressed in CRC (Figure S2). To further explore the relationship between AVL9 expression and survival outcomes, Kaplan-Meier curve analysis using the SurvExpress online tool showed that high expression of AVL9 was significantly associated with poor prognosis in terms of OS (Figure 2A, P = 0.0433; Figure 2B, P = 0.01585) and DFS (Figure 2C, P = 0.0002; Figure 2D, P = 0.0075).
Bioinformatics Analysis of AVL9 and the Genes Associated with Its Expression
To investigate the potential functions of AVL9 in CRC, the LinkedOmics and cBioportal online bioinformatics databases were used to analyze the AVL9 co-expression genes in CRC in TCGA dataset, we generated a Venn diagram using an online web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) to identify 160 genes that were common between the results of the two databases (Figure 3A), and GO function enrichment analysis of the DAVID6.8 database revealed that the relative expression genes were functionally concentrated in single organismal cell–cell adhesion, post-transcriptional regulation of gene expression, and negative regulation of vascular endothelial growth factor receptor signaling pathway (Figure 3B, P < 0.05). The KEGG pathway analysis showed that these genes were mainly involved in progesterone-mediated oocyte maturation, axon guidance, insulin signaling pathway, and ubiquitin-mediated proteolysis signaling pathways (Figure 3C, P < 0.05). PPI network analysis using Cytoscape 3.7.1 showed that the KBTBD2, KIAA1147, RNF216, EPDR1, and ANKIB1 genes were most closely related to AVL9 (Figure 4). Furthermore, according to the GEPIA data, we identified that the correlation between KBTBD2, KIAA1147, RNF216, EPDR1, and AVL9 were all positively correlated according to Pearson’s correlation coefficient (Figure S3A-D, P = 0).
AVL9 is Upregulated in Clinical Samples and Associated with the Clinical Characteristics of CRC
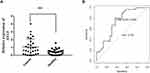
To validate the relationship between AVL9 expression and clinicopathological characteristics, we detected AVL9 in 50 paired CRC tissues and adjacent normal tissues. Reverse transcription–quantitative polymerase chain reaction (RT-qPCR) showed that AVL9 expression was higher in CRC tissues than in the adjacent normal tissues (Figure 5A, P < 0.001). Furthermore, patients with higher AVL9 expression had poorer OS times (Figure 5B, P < 0.0001) than those with lower AVL9 expression. In addition, AVL9 was more likely to be highly expressed in patients with TNM stage III or IV than in patients with TNM stage I or II (Figure 5C, P < 0.0001). Compared to patients with well-to-moderate, AVL9 expression in patients with poor-to-moderate was significantly increased (Figure 5C and D, P < 0.001). Moreover, the ROC curve analysis was used to analyze the predictive value of AVL9 expression in the tissues of patients with CRC. The cutoff value of 1.296 had a sensitivity of 72.9% and a specificity of 64%, while the area under the curve (AUC) was 0.683 (Figure 5E). To analyze the relationship between AVL9 expression and the clinical parameters of CRC patients, we divided them into high and low expression groups according to the medium expression value of AVL9 as the cutoff point (Table 1). Our data confirmed that AVL9 expression showed statistically significant differences according to differentiation (P = 0.01) and TNM stage (P = 0.028) but not according to sex (P = 0.729), age (P = 0.470), tumor size (P = 0.064), or N status (P = 0.204). Furthermore, multivariate survival analysis (Table 2) identified the AVL9 expression level as a covariate (hazard ratio [HR], 5.695; 95% confidence interval [CI], 1.860–17.442, P = 0.002). Collectively, upregulated expression of AVL9 may serve as a biomarker for CRC detection.
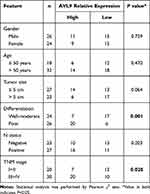 |
Table 1 AVL9 Expression and Clinicopathological Features in Patients with Colorectal Cancer |
 |
Table 2 Univariate and Multivariate Analyses of Clinical Pathological Factors of OS in 50 Patients with CRC |
AVL9 Was Upregulated in the Plasma of CRC Patients
We next confirmed the expression of AVL9 in 60 samples from CRC patient plasma and healthy control plasma based on the RT-qPCR assays, which confirmed that AVL9 expression in the plasma of CRC patients was remarkably upregulated versus that in the healthy controls (Figure 6A, P < 0.01). Subsequently, the ROC curve analysis was used to analyze the predictive value of AVL9 in the plasma of patients with CRC. The cutoff value was 0.602, with a sensitivity of 80.0% and specificity of 63.3%, and the AUC was 0.729 (Figure 6B). Taken together, these results indicate that AVL9 levels were elevated in CRC and might be useful as a non-invasive biomarker for detecting CRC in patients.
Discussion
CRC is one of the most common cancers and causes of cancer-related deaths worldwide. The high mortality rate of CRC patients is mainly due to the lack of biomarkers with high sensitivity and specificity. Many deaths can be avoided if CRC patients can be diagnosed and treated at an early stage. Therefore, determining effective biomarkers and prolonging the survival time of patients with CRC is of great clinical significance.9 AVL9, a migration-associated gene, was first described in budding yeast as an exocytosis gene,10 and recent studies have shown that it plays an important role in cell polarity, cell migration, and cell cycle progression.4,5 For instance, AVL9 expression is reportedly upregulated in clear cell renal carcinomas and promotes cell migration,6 and Liang et al11 revealed that AVL9 is a direct target of miR-203a-3p, which promotes the proliferation, migration, and invasion of non-small cell lung cancer. In our previous study, we found that AVL9 was a target gene of the linc00662/miR-497-5p axis, which promotes the occurrence and development of CRC.7 In present study, we aimed to increase our understanding of AVL9 expression using bioinformatics analysis. The GEO and TCGA databases were used to identify that AVL9 expression was upregulated in CRC tissues versus normal tissues, the survival time analysis showed that a higher level of AVL9 led to worse OS and DFS, these results show that AVL9 is a potential tumor-promoting gene of CRC.
To further study the mechanism of AVL9 expression upregulation in tumors, GO analyses showed that AVL9, together with its related genes, was functionally concentrated in tumor-related functions, such as single organismal cell–cell adhesion, suggesting that they may be involved in the migration of immune cells and tumor cells12 and post-transcriptional regulation of gene expression and negative regulation of the vascular endothelial growth factor receptor signaling pathway, which shed light on the mechanism of cancer in cell growth, differentiation, development, and apoptosis.13
KEGG pathway analyses also revealed that these genes were mainly involved in signaling pathways that promote tumor development. For instance, it has been well known that endocytosis plays a causative role in the cell homeostasis and proliferation control of cancer14 Similarly, previous studies reported that the insulin signaling pathway can promote tumor growth and invasion.15 Furthermore, it is well known that nerves and blood vessels have similar abilities to form branch networks, and there is strong evidence that the development of nerve and tumor vascular systems is regulated by axon-guided receptors and ligands.16 Ubiquitin-regulated protein degradation is necessary for useless proteins or proteins synthesized incorrectly in cells, and studies have shown that the dysregulation of ubiquitin activity can promote tumor progression.17 In summary, these findings suggest that AVL9, as well as its related genes, affects tumor progression, invasion, and metastasis. In the present study, the PPI network showed that AVL9 directly interacts with KBTBD2, KIAA1147, RNF216, and EPDR1; a further study found that all were positively correlated with AVL9 according to the Pearson’s correlation coefficient, and recent studies demonstrated that KBTBD2, KIAA1147, RNF216, and EPDR1 play an important role in the tumorigenesis and prediction of progression in cancer,18–21 that is, these genes may collaborate to promote the progression of CRC, but the specific mechanism among them requires further analysis in future studies.
To further verify the expression of AVL9 in CRC tissues and adjacent normal colorectal mucosa using RT-qPCR, we revealed that AVL9 expression was significantly increased in CRC tissues and closely associated with TNM staging and degree of differentiation. Furthermore, our data also indicated that the AVL9 low-expression group had a longer survival time than the high-expression group. In CRC diagnosis, owing to advantages such as convenience, minimally invasive, cheap, and repeatability, liquid biopsy provides a vital function versus colonoscopy examinations; however, the most commonly used diagnostic markers, namely, carcinoembryonic antigen, carbohydrate antigen19-9, and carbohydrate antigen 72–4, exhibit low specificity and sensitivity, particularly in early-stage CRC.22 Therefore, the main priority is to identify novel biomarkers to reliably detect early CRC and relapse in patients post-surgery, and emerging research has demonstrated that circulating tumor cells, ctDNA, miRNAs, and lncRNAs can improve CRC screening and diagnosis and predict recurrence and metastasis.23 In this study, to explore the potential diagnostic role of AVL9 in CRC, we first detected its expression in the plasma and found that it was significantly increased in CRC patients versus healthy controls; ROC curve analysis demonstrated AVL9 detection in the tissues and plasma exhibited good sensitivity and specificity as a diagnostic marker for CRC, as the AUC of AVL9 expression in the plasma was 0.729 with a cutoff value of 0.602. However, due to the small number of patients, this study can explain only the detection of AVL9, which plays a role in the early diagnosis of CRC, more large-scale studies and multicenter trials are still needed to confirm its advantages. Finally, AVL9 is expected to become a new biomarker of liquid biopsy for enabling the early diagnosis of CRC; assessment of progress, treatment effects, and prognosis as well as the evaluation of chemotherapy-acquired drug resistance in the near future.
Conclusion
We demonstrated here that the high expression and overexpression of AVL9 in CRC indicate poor prognosis. Detection of AVL9 in the tissues and plasma showed good efficacy, which revealed that it could act as a diagnostic marker for CRC.
Acknowledgments
This work was financially supported by the Shantou Science and Technology Bureau (No. 2006241552607), “Dengfeng Project” for the construction of high-level hospitals in Guangdong Province –the First Affiliated Hospital of Shantou University Medical College Supporting Funding (NO.202003-17).
Disclosure
The authors declare no conflicts of interest.
References
1. Cheng L, Eng C, Nieman LZ, et al. Trends in colorectal cancer incidence by anatomic site and disease stage in the United States from 1976 to 2005. Am J Clin Oncol. 2011;34(6):573–580. doi:10.1097/COC.0b013e3181fe41ed
2. Smith RA, von Eschenbach AC, Wender R, et al. American Cancer Society guidelines for the early detection of cancer: update of early detection guidelines for prostate, colorectal, and endometrial cancers. Also: update 2001-testing for early lung cancer detection. CA Cancer J Clin. 2001;51(1):
3. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi:10.3322/caac.21601
4. Linford A, Yoshimura S, Nunes Bastos R, et al. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012;22(5):952–966. doi:10.1016/j.devcel.2012.04.010
5. Li Y, Xu J, Xiong H, et al. Cancer driver candidate genes AVL9, DENND5A and NUPL1 contribute to MDCK cystogenesis. Oncoscience. 2014;1(12):854–865. doi:10.1016/j.devcel.2012.04.010
6. Zhang W, Wang J, Chai R, et al. Hypoxia-regulated lncRNA CRPAT4 promotes cell migration via regulating AVL9 in clear cell renal cell carcinomas. Onco Targets Ther. 2018;11:4537–4545. doi:10.2147/OTT.S169155
7. Wang H, Yu M, Hu W, et al. Linc00662 promotes tumorigenesis and progression by regulating miR-497-5p/AVL9 axis in colorectal cancer. Front Genet. 2020;10:1385. doi:10.3389/fgene.2019.01385
8. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12(1):77. doi:10.1186/1471-2105-12-77
9. Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–241. doi:10.3322/caac.21149
10. Harsay E, Schekman R. Avl9p, a member of a novel protein superfamily, functions in the late secretory pathway. Mol Biol Cell. 2007;18(4):1203–1219. doi:10.1091/mbc.e06-11-1035
11. Liang J, Sun T, Wang G, et al. Clinical significance and functions of miR-203a-3p/AVL9 axis in human non-small-cell lung cancer. Per Med. 2020;17(4):271–282. doi:10.2217/pme-2019-0108
12. Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi:10.1038/nrm2720
13. Corbett AH. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol. 2018;52:96–104. doi:10.1016/j.ceb.2018.02.011
14. Polo S, Pece S, Di Fiore PP. Endocytosis and cancer. Curr Opin Cell Biol. 2004;16(2):156–161. doi:10.1016/j.ceb.2004.02.003
15. Varewijck AJ, Janssen JA. Insulin and its analogues and their affinities for the IGF1 receptor. Endocr RElat Cancer. 2012;19(5):F63–F75. doi:10.1530/ERC-12-0026
16. Klagsbrun M, Eichmann A. A role for axon guidance receptors and ligands in blood vessel development and tumor angiogenesis. Cytokine Growth Factor Rev. 2005;16(4–5):535–548. doi:10.1016/j.cytogfr.2005.05.002
17. Khan OM, Carvalho J, Spencer-Dene B, et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128(4):1326–1337. doi:10.1172/JCI97325
18. Dai K, Huang Y, Chen Z, Sun X, Yang L, Jiang Y. Kbtbd2 inhibits the cytotoxic activity of immortalized NK cells through down-regulating mTOR signaling in a mouse hepatocellular carcinoma model. Eur J Immunol. 2018;48(4):683–695. doi:10.1002/eji.201747281
19. Taskesen E, Mishra A, van der Sluis S, et al; International FTD-Genomics Consortium. Susceptible genes and disease mechanisms identified in frontotemporal dementia and frontotemporal dementia with amyotrophic lateral sclerosis by DNA-methylation and GWAS. Sci Rep. 2017;7(1):8899. doi:10.1038/s41598-017-09320-z
20. Wang H, Wang Y, Qian L, et al. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1-dependent autophagy. Oncotarget. 2016;7(32):51174–51183. doi:10.18632/oncotarget.9433
21. Gimeno-Valiente F, Riffo-Campos ÁL, Ayala G, et al. EPDR1 up-regulation in human colorectal cancer is related to staging and favours cell proliferation and invasiveness. Sci Rep. 2020;10(1):3723. doi:10.1038/s41598-020-60476-7
22. Carpelan-Holmström M, Louhimo J, Stenman UH, et al. CEA, CA 19-9 and CA 72-4 improve the diagnostic accuracy in gastrointestinal cancers. Anticancer Res. 2002;22(4):2311–2316.
23. Vacante M, Ciuni R, Basile F, Biondi A. The liquid biopsy in the management of colorectal cancer: an overview. Biomedicines. 2020;8(9):308. doi:10.1159/000494411
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.