Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
Autophagy Induced by BCL2-Related ceRNA Network Participates in the Occurrence of COPD
Authors Shi ZE, Zhang MY, Liu JY , Zhang WD, Hu DM, Wang QX, Ji XL, Jiang YY, Qu YQ
Received 11 December 2021
Accepted for publication 28 March 2022
Published 8 April 2022 Volume 2022:17 Pages 791—808
DOI https://doi.org/10.2147/COPD.S347733
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Zhuang-E Shi,1 Meng-Yu Zhang,1 Jian-Yu Liu,1 Wen-Di Zhang,1 Dong-Mei Hu,1 Qing-Xiang Wang,1 Xiu-Li Ji,2 Yuan-Yuan Jiang,3 Yi-Qing Qu3
1Department of Pulmonary and Critical Care Medicine, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Shandong Key Laboratory of Infectious Respiratory Diseases, Jinan, People’s Republic of China; 2Department of Pulmonary Disease, Jinan Traditional Chinese Medicine Hospital, Jinan, People’s Republic of China; 3Department of Pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University, Shandong Key Laboratory of Infectious Respiratory Diseases, Jinan, People’s Republic of China
Correspondence: Yi-Qing Qu, Department of pulmonary and Critical Care Medicine, Qilu Hospital of Shandong University, Shandong Key Laboratory of Infectious Respiratory Diseases, Jinan, People’s Republic of China, Tel +86 531 8216 9335, Fax +86 531 8296 7544, Email [email protected]
Purpose: Chronic obstructive pulmonary disease (COPD) is a predominant cause of mortality worldwide. Autophagy, which depends on a lysosomal degradation pathway, plays an essential role in the occurrence of COPD. The aim of our study was to identify the potential function of autophagy and construct a BCL2-related competing endogenous RNA (ceRNA) network that induces autophagy in COPD.
Methods: Blood sample data from GSE31568, GSE24709, and GSE61741 were collected from the Gene Expression Omnibus (GEO) database. Differentially expressed miRNAs in COPD and controls were identified via GEO2R. Transcription factors were obtained from FunRich. DIANA, miRDB, miRTarBase, and TargetScan were used to predict target genes of miRNAs. Autophagy genes were collected from the Human Autophagy Database (HADb). The GSE151052 dataset was used to identify autophagy-related differentially expressed genes in tissues. Functional enrichment and protein–protein interaction (PPI) network analyses were conducted via Metascape and the STRING network. Spearman correlation analysis was used to analyze the relationship between autophagy-related differentially expressed genes and lung function. The BCL2-related ceRNA network was modeled by Cytoscape.
Results: We obtained 41 differentially expressed miRNAs and 10 significantly different transcription factors. We identified 19 autophagy-related differentially expressed genes that were significantly different (P< 0.05) in tissue samples. The most significant enrichment in Metascape was an autophagy item, which further confirmed autophagy participation in the occurrence of COPD. PPI network analysis found four genes (BCL2, BECN1, MAPK8, and ITPR1), among which BCL2 was correlated with both FEV1/FVC and FEV1 prediction. Finally, the BCL2-related ceRNA network was constructed to clarify the interaction of RNAs and occurrence of autophagy, including 18 miRNAs and 65 lncRNAs.
Conclusion: We identified 19 autophagy-related differentially expressed genes that participated in COPD; among them, BCL2 was correlated with lung function, and a BCL2-related ceRNA network was constructed, which further revealed the potential mechanism of autophagy involvement in COPD.
Keywords: COPD, autophagy, bioinformatics, lung function, BCL2, ceRNA
Introduction
Chronic obstructive pulmonary disease (COPD) is major cause of morbidity and mortality in chronic diseases worldwide, and is one of the most significant causes of death in all disease at most countries.1 The increasing morbidity of COPD imposes a huge economic and social burden.2–5 Lung function is one of the diagnostic criteria of COPD that can also reflect the severity of COPD.1
According to published studies, numerous biological functions take part in the occurrence of COPD, for instance, apoptosis, cell proliferation, and autophagy.6–8 However, the mechanism of autophagy is not clear in COPD. Autophagy depends on a lysosomal degradation pathway, and under certain circumstances, this pathway is considered a non-apoptotic cell death pathway and is necessary for cell differentiation, homeostasis, survive, development, which also takes part in the inflammation, aging, cancer and so on.9,10 It is related to lung,11 liver,12 and heart disease.13 In addition, autophagy plays a vital role in the process of infection via participating in host-pathogen interactions.14,15 Studies have also demonstrated that autophagy participates in the occurrence and development of idiopathic pulmonary fibrosis (IPF),16 pulmonary arterial hypertension (PAH),17 and acute lung injury (ALI).18 Autophagy plays a significant role in cellular behavior in COPD.19–21 However, there has been little research on autophagy participation in the occurrence of COPD.22 In this study, we identified the role of autophagy in COPD via bioinformatics analysis and further research.
Studies indicate that autophagy-related genes have number of biological functions23 that take part in phagocytosis by LC3-associated phagocytosis (LAP),24,25 secretion, and exocytosis.26 BCL2 is one of the autophagy-related genes. Qin et al found that nur77 bound to BCL2 and decreased the affinity of Beclin-1 for BCL2 to promote autophagy in COPD.27 Recent evidence suggests that the upregulation of BCL2 is related to the “abnormal” inflammation in COPD, and takes part in auto-maintenance.28 BCL2 plays an important role in maintaining the internal environment of airway epithelial cells. Cigarette smoke (CS) blocks epithelial cells in chronic airway inflammation by inhibiting Bik expression.22 Research has found that BCL2 could sustain hyperplastic mucus cells, and CS could enhance the expression of BCL2 and help promote the survival of mucus cells.10 Noncoding RNAs, which cannot encode proteins, are classified into three groups according their nucleotide lengths: less than 50 nt, 50–500 nt, and more than 500 nt and are significant in life processes.21 MiRNA and lncRNA are noncoding RNAs, and miRNAs participate in the occurrence and development of most cancers, for instance malignant lung tumors,29 liver carcinoma,30 breast cancer,31 and gastric carcinoma,32 among others. In addition, studies have indicated that miRNAs have an important function in COPD33 along with lncRNAs.34 lncRNAs interact with miRNAs and regulate the expression of genes. Consequently, lncRNAs can act as competing endogenous RNAs (ceRNA) to compete with their target miRNAs and regulate gene expression. CeRNA also takes part in the development of COPD. Zhao reported that lncRNA RP11-86H7.1 promoted inflammatory reactions, thus promoting the development of COPD.35
The reason there are still few effective treatments is that COPD pathogenesis remains unclear. In order to further understand this biological process, we constructed a BCL2-related ceRNA network to clarify the processes that induce autophagy in COPD. Meanwhile, it is vital as well to find potential biomarkers that can be used to guide treatment for COPD.
Methods
Data Sources
We used “COPD,” “noncoding RNA profiles by array,” and “Homo sapiens” as key words and collected three miRNA datasets of blood samples, GSE31568,36 GSE24709, and GSE61741,37 from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov) from the GPL9040 platform. The Human Autophagy Database (HADb) network was developed in the Laboratory of Experimental Cancer Research headed by Dr. Guy Berchem, and integrates human genes and proteins that directly or indirectly take part in autophagy, and was up-to-date as described in literature. Autophagy genes were obtained from the “clustering” module of HADb (http://www.autophagy.lu),38–40 which contained 231 autophagy genes. Then a mRNA dataset that belongs to a lung tissue sample, GSE151052, was acquired from the GEO database GPL17556 platform via searching the key words “COPD,” “expression profiling by array,” “Homo sapiens,” and “tissue.” The lung function datasets GSE7122041 also came from the GEO database by searching “COPD,” “expression profiling by high throughput sequencing,” “Homo sapiens,” and “tissue,” which belong to the GPL11532 platform. The detail of the five datasets was shown in Table 1. The flow charts of this study were presented in Figures 1 and 2.
 |
Table 1 The Detail of Datasets |
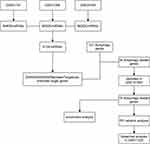 |
Figure 1 The detailed process of identifying autophagy participated in the occurrence of COPD. |
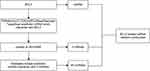 |
Figure 2 A flow diagram of BCL2-related ceRNA network. |
Differential Expression Analysis
GEO2R is a GEO web analysis tool to identify differentially expressed genes via experimental conditions between various groups in a GEO series. This tool was applied to analyze the differentially expressed genes in the GSE31568, GSE24709, and GSE61741 datasets. The screening criteria were adj.p<0.05 and log (fold change [FC])>1. The NetworkAnalyst network (https://www.networkanalyst.ca)42–46 was used to analyze 38 differentially expressed autophagy-related genes in GSE151052. NetworkAnalyst is a comprehensive gene expression profiling and visual analytics network. We used the “gene expression table” module, input the expression data, and chose the “limma” statistical method; adj.p<0.05 was the screening criterion.
Prediction of Transcription Factors and Target Genes for Differentially Expressed miRNAs (DEmiRNAs)
Transcription factors of 41 DEmiRNAs were obtained from FunRich (version 3.1.3),47 which was used mainly for functional enrichment analysis and interaction network analysis of genes and proteins. This stand-alone software tool was designed to handle multiple gene and protein datasets irrespective of the organism. The function of “miRNA enrichment” in FunRich was used in our study to obtain the transcription factors of 41 DEmiRNAs. The top 10 transcription factors were identified in our study, and P < 0.05 was considered to be statistically significant. Then the target genes of the 41 DEmiRNAs were predicted via DIANA microT-CDS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index),48,49 miRDB (http://mirdb.org),50,51 miRTarBase (http://mirtarbase.cuhk.edu.cn),52 and TargetScan (http://www.targetscan.org).53 These tools use differential algorithms to predict target genes. The overlap of the four databases was used in further analysis.
Functional Enrichment Analysis and Protein–Protein Interaction (PPI) Network Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis can enrich many single genes and determine the common function and pathway for those genes. GO can classify the genes, proteins, ncRNAs and complexes, and then describe their biological roles.54,55 KEGG can link the genetic information and acquire further information via computerization. Consequently, GO and KEGG methods were used to do a functional enrichment and pathway analysis. Moreover, we constructed a PPI network to help us identify the interactions within proteins, so as to furthermore research the molecular mechanism of diseases. As a consequence, a PPI network was constructed of autophagy-related differentially expressed genes in GSE151052. Metascape (https://metascape.org),56 a gene annotation and analysis resource, which was involved in gene enrichment analysis and PPI analysis. When autophagy-related differentially expressed genes were input into the network, a document containing the result of enrichment and PPI can be exported. String (https://www.string-db.org)57 predicted protein-protein interactions with a direct and indirect association via computational prediction and integration with other databases. Metascape and the String network were used for enrichment and protein-protein interaction (PPI) network analysis in our study.
Construction of the BCL2-Related ceRNA Network
In our study, the ceRNA network of BCL2 was constructed with miRNA and lncRNA to reveal the interaction between RNAs and to induce autophagy in COPD. MiRNAs were obtained by using DIANA microT-CDS, miRTarBase, starBase (http://starbase.sysu.edu.cn),58 TargetScan and the intersection with DEmiRNAs. LncRNAs were found via starBase and DIANA Lncbase (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex),59 respectively. Finally, miRNAs and lncRNAs were input using Cytoscape (version 3.8.2)60 as a tool, and the BCL2-related ceRNA was structured via setting up nodes and edges in Cytoscape.
Statistical Analysis
The 41 DEmiRNAs were first collected in GEO databases and statistical analysis was performed using the GEO2R tool on the GEO database, and the statistically significant standards were adj.p<0.05 and logFC>1. Then acquired autophagy-related differentially expressed miRNAs in GEO151052 were analyzed by “limma” via the NetworkAnalyst network. The expression information of BCL2, BECN1, MAPK8, ITPR1 as well as the lung function data of COPD patients in GSE71220 were collected. Spearman analysis was applied to analyze the correlation between gene expression and lung function using SPSS 24.0.
Results
Differentially Expressed miRNAs in COPD
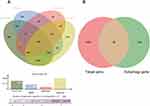
We first obtained GSE31568, GSE24709, and GSE61741 datasets from blood samples. Those datasets came from the GPL9040 platform. GSE31568 contained 70 control samples and 24 COPD samples, GSE24709 contained 19 control samples and 24 COPD samples, and GSE61741 contained 94 control samples and 47 COPD samples. More details are shown in Table 1. Then we obtained the differentially expressed miRNAs by using the GEO2R tool. There were 453 upregulated DEmiRNAs and 410 downregulated DEmiRNAs in GSE31568, 464 upregulated DEmiRNAs and 399 downregulated DEmiRNAs in GSE24709, 435 upregulated DEmiRNAs and 413 downregulated DEmiRNAs in GSE61741. Details are shown in Figure 3A–C); a total of 863, 863, and 848 DEmiRNAs were obtained. According to the screening criteria adj.p<0.05 and logFC>1, we obtained 113, 186, and 110 DEmiRNAs, respectively. Finally, 41 overlapping DEmiRNAs were obtained (Figure 3D), including 30 upregulated DEmiRNAs and 11 downregulated DEmiRNAs as shown in Supplementary Table 1. In addition, we also predicted the transcription factors of 41 DEmiRNAs via the FunRich tool and identified the top 10 transcription factors with the most significant differences, EGR1, SP1, POU2F1, SP4, NKX6-1, RORA, MEF2A, SOX1, ZFP161, and YY1. More details are presented in Table 2 and Figure 4.
 |
Table 2 The Top10 Trancription Factor for DEMs |
 |
Figure 4 The column chart of transcription factor for DEmiRNAs, the top10 was showed. |
Screening Autophagy-Related Genes According to the Predicted Target Genes of DEmiRNAs
To explore the relationship between COPD and autophagy and to clarify whether autophagy participates in the occurrence of COPD, the target genes of DEmiRNAs were predicted and autophagy genes were collected. A total of 11,447 target genes were predicted by the DIANA microT-CDS database, 7368 target genes were predicted by MiRDB database, 3255 target genes were predicted by the miRTarBase database, and 15,035 target genes were predicted via the TargetScan database, the overlap were 1596 target molecules, as shown in Figure 5A. In addition, 231 autophagy genes were downloaded from HADb as shown in Supplementary Table 2. The overlap between 231 autophagy genes and 1596 target genes was 38 autophagy-related genes. The results were shown in Figure 5B and the autophagy-related gene list was shown in Supplementary Table 3.
Validation That Autophagy-Related Genes Were Differentially Expressed in Tissue Samples in COPD Patients
In order to acquire data on the differential expression of autophagy-related genes in COPD and reduce the sample differences, we used tissue samples for validation. GSE151052, a lung tissue sample dataset, was downloaded from GEO, and we acquired the expression data on the 38 autophagy-related genes in GSE151052. Then we identified 19 autophagy-related differentially expressed genes that were significantly different (P < 0.05) via the NetworkAnalyst network: EIF4EBP1, DNAJB9, RAB1A, CHMP2B, BECN1, FKBP1A, PTEN, EDEM1, ITGA6, SAR1A, WIPI2, MAPK8, ITPR1, UVRAG, CDKN1B, TP53INP2, SIRT1, VEGFA, and BCL2. The box plots were made via the NetworkAnalyst network and the heatmap was displayed via http://www.bioinformatics.com.cn, a free online platform for data analysis and visualization. More details were presented in Figures 6 and 7.
 |
Figure 7 The hot map of 19 autophagy-related remarkable difference genes (p<0.05). |
Functional Enrichment and PPI Network Analysis of the 19 Differentially Expressed Autophagy-Related Genes
In this study, we used GO and KEGG enrichment analysis to clarify the potential biological function of these genes via the Metascape network. The results showed that the 19 genes were mainly involved in the autophagy process in COPD. The top five enrichment items were ko04140 (autophagy), GO:0009991 (response to extracellular stimulus), GO:0001666 (response to hypoxia), CORUM:6472 (Bcl-2-Beclin1-UVRAG-PI (3) KC3 complex), and GO:0010506 (regulation of autophagy). They related to autophagy almost. The first item obviously confirmed our theory that autophagy plays an important role in the occurrence of COPD. In addition, the article 5 explained genes took part in the regulation of autophagy in COPD. So, KEGG and GO enrichment analysis showed that autophagy-related differentially expressed genes play significant role in autophagy and regulation of autophagy in the occurrence of COPD. The enriched ontology cluster results were presented in Figure 8A, The enrichment factor dot plot by http://www.bioinformatics.com.cn is shown in Figure 8B The enrichment network by cluster ID and the enrichment network by P-value are displayed in Figure 8C and D.
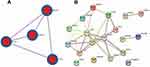
PPI network analysis was used to determine the interaction between 19 autophagy-related differentially expressed genes by Metascape and the STRING network. We found that there were close interactions among 12 genes, for instance, BCL2, BECN1, MAPK8, ITPR1, and so on. In addition, we also found ITGA6, CHMP2B, and TP53INP2 were independent of each other at 19 autophagy-related differentially expressed genes. The network is shown in Figure 9B. In addition, we obtained the relationship of BCL2, BECN1, MAPK8 and ITPR1 genes from the PPI network analysis via Metascape. It was shown that BCL2 with BECN1, MAPK8, and ITPR1 were densely connected, as well as with BECN1, but MAPK8 and ITPR1 had no obvious connection. The network was shown in Figure 9A.
Associations of BCL2, BECN1, MAPK8, and ITPR1 Expression Levels with Lung Function
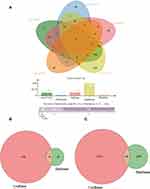
Lung function is not only used to diagnose COPD but also as a tool to evaluate its severity.1 In order to explore the effect of autophagy in COPD, we analyzed the relationship between BCL2, BECN1, MAPK8, and ITPR1 gene expression and lung function in COPD. We obtained the GSE71220 dataset from the GEO database and acquired the expression data of BCL2, BECN1, MAPK8, ITPR1 genes and FEV1/FVC and FEV1 predictive data. Then by Spearman relationship analysis we found that BCL2 and FEV1/FVC, BCL2 and FEV1 predict, ITPR1 and FEV1 predict all had significant correlation in COPD via SPSS 24.0. Spearman analysis results suggested that the expression level of BCL2 was related to FEV1/FVC (P = 0.008) and FEV1 predict (P < 0.001). The expression level of ITPR1 was related to FEV1 predict (P = 0.001) in COPD, while the expression levels of BECN1 and MAPK8 were not related to FEV1/FVC (P = 0.31, P = 0.151) and FEV1 predict (P = 0.472, P = 0.331). The expression level of ITPR1 was not related to FEV1/FVC (P = 0.185). Detailed information was presented in Tables 3 and 4.
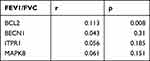 |
Table 3 The SPEARMAN Relationship Between FEV1/FVC and BCL2, BECN1, ITPR1, MAPK8 in COPD |
 |
Table 4 The SPEARMAN Relationship Between FEV1 Predict and BCL2, BECN1, ITPR1, MAPK8 in COPD |
Construction of the BCL2-Related ceRNA Network
We found there was a strong association between expression of BCL2 and FEV1/FEV and FEV1 predict in COPD, while ITPR1 was associated only with FEV1 predict. In addition, BCL2 is one of the most important oncogenes in apoptosis research, and the relationship between BCL2 and COPD is an active research topic. In order to explain the interaction between RNAs when autophagy is induced in COPD, we chose the BCL2 gene and structured a BCL2-related ceRNA network. We obtained 284, 68, 319, and 1255 predicted miRNAs via the DIANA (microT-CDS), miRTarBase, starBase, and TargetScan databases. The overlap of the four database-predicted results were 18 miRNAs: hsa-miR-448, hsa-miR-449a, hsa-miR-96-5p, hsa-miR-20a-5p, hsa-miR-153-3p, hsa-miR-30b-5p, hsa-miR-15b-5p, hsa-miR-365a-3p, hsa-miR-125a-5p, hsa-miR-497-5p,hsa-miR-182-5p, hsa-miR-15a-5p,hsa-miR-503-5p, hsa-miR-125b-5p, hsa-miR-195-5p, hsa-miR-34a-5p, hsa-miR-21-5p, and hsa-miR-16-5p. Finally, we found hsa-miR-153-3p and hsa-miR-497-5p not only were included in those 18 miRNAs but also belong to the DEmiRNAs. The intersection graph is shown in Figure 10A. As a result, hsa-miR-153-3p and hsa-miR-497-5p were used for further prediction. A total of 341 lncRNAs were obtained via the starBase database, and 1958 lncRNAs were obtained as shown in Figure 10B and C). Eventually, the BCL2-related ceRNA network was constructed by Cytoscape and consisted of 65 lncRNAs and 2 miRNAs, as well as BCL2, which clarified the relationship between RNAs when autophagy is induced in COPD. Details are shown in Figure 11.
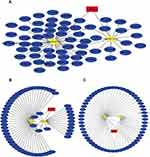 |
Figure 11 The ceRNA network of BCL2-miRNAs-LncRNAs. (A–C) The BCL2-related ceRNA was constructed via cytoscape. The red represents the BCL2, the yellow is miRNAs and the blue is lncRNAs. |
Discussion
COPD is the third leading cause of disease-related death worldwide.61 Researchers have found that COPD is an age-associated disease,62 and with the increase in aging populations, the morbidity of age-related diseases such as COPD is increasing. However, because there are no effective therapies, mortality and disability in COPD are increasing,63–66 along with its financial and social burdens. As a consequence, more research on the pathogenesis of COPD to guide treatment is needed.
Increasing evidence shows that autophagy may take part in the occurrence of COPD,67 but there are few studies of this phenomenon. An increasing number of studies have suggested that miRNAs and lncRNAs play a role and act as biomarkers to diagnose and evaluate the prognosis.68,69 They also have found a relationship with autophagy in cancer. Fu found that miR-9, miR-31, miR-34c, miR-196, and miR-221/222 are involved in autophagy.70 Autophagy and proliferation could be suppressed by overexpression of miR-502 in colon cancer cells,71–74 but its value in COPD is not clear. In this study, we first studied the DEmiRNAs and determined that autophagy plays a role in in the occurrence of COPD; furthermore, we obtained the autophagy-related gene BCL2 and constructed a BCL2-related ceRNA to clarify the mechanism of autophagy in COPD.
We obtained 1596 predicted genes via four predictive databases, 231 autophagy genes via HADb. We found 38 autophagy-related genes in blood samples of COPD. For further confirmation of the role of autophagy in COPD, we further undertook a differential expression analysis for 38 autophagy-related genes in lung tissue samples and found a total of 19 autophagy-related genes that were differentially expressed. Blood samples and tissue displayed a consistently differential expression trend, indicating that COPD is a systemic disease with not only pulmonary features but also extrapulmonary features.75,76 For instance, cigarette smoking is the predominant etiology of COPD.1 Cigarette smoke is rich in many compounds, such as high concentrations of acrolein and other carcinogenic organic compounds. Studies have reported an inextricable link between acrolein and oxidant stress,77 and multiple lines of evidence indicate plasma acrolein and lung tissue acrolein are increased synchronously, which further supports COPD as a systematic disease.78–80 Yasuo found that the longer the period of smoking, the higher acrolein in the plasma, inducing oxidant stress and small airway obstruction and resulting in the progression of COPD.80 Our study could identify COPD is a systemic disease from the gene perspective.
Then we found the major enrichment item was autophagy, showing that autophagy takes part in the occurrence of COPD. Several studies have suggested that autophagy in part of the pathogenesis in the occurrence of COPD. CS is the leading risk factor in COPD. Some studies have shown that autophagy and selective autophagy create a harmful reaction in epithelial cells in COPD,81–85 promoting airway inflammation and mucus hyperproduction,86,87 clarifying the role of autophagy in occurrence of COPD. We also constructed a PPI network in order to understand the mechanism of autophagy-related differentially expressed genes and found a strong interaction among BCL2, BECN1, MAPK8, and ITPR1. In this study, we researched the relationship between BCL2, BECN1, MAPK8, and ITPR1 gene expression and lung function in COPD to find a biomarker to help further research on the influence of autophagy in COPD. We found BCL2 and FEV1/FVC, BCL2 and FEV1 predict, ITPR1 and FEV1 predict in COPD and had a significant correlation via Spearman relationship analysis. BCL2 encodes an integral outer mitochondrial membrane protein that blocks the apoptotic death of some cells such as lymphocytes, and as a consequence, BCL2 is associated with autophagy.88,89 Studies have shown that BCL2 is correlated with lung function and may take part in the occurrence and progression of COPD.90 BCL2 is a mitochondrial outer membrane permeabilization protein that mediates cell apoptosis, which induces the occurrence and progression of COPD.91,92 As a result, autophagy is associated with the occurrence of COPD through the differential expression of BCL2 in mediating cell apoptosis.
In recent years, the ceRNA network has been widely studied and it has been shown that ceRNA affects the development of numerous cancers, for example, lung cancer,93,94 breast cancer,95 and gastric cancer.32 It was also studied in COPD,35 but it was deficient. Last, we choose the autophagy-related gene BCL2 for further research. A BCL2-related ceRNA network was constructed to clarify the mechanism of autophagy and find biomarkers to guide treatment in COPD. The ceRNA network may also clarify the mechanism of RNAs in inducing autophagy and participating in the occurrence and development of COPD.
There are several limitations in our research. First, we used bioinformatics analysis throughout the whole research process and did not verify experimental studies. Second, we did not demonstrate the process of RNAs interaction in ceRNA network, and it needs to be further researched and experimentally validated. Third, we acquired autophagy-related genes in blood samples and in order to decrease the sample differences, we choose tissue samples to verify differential gene expression, which could increase errors.
Conclusion
In conclusion, 19 autophagy-related differentially expressed genes were found and were identified as participating in the occurrence of COPD. There was a correlation between BCL2 and lung function, and a BCL2-related ceRNA network was constructed to explain the interaction between RNAs that induce autophagy in COPD.
Abbreviations
COPD, chronic obstructive pulmonary disease; ceRNA, competing endogenous RNAs; GEO, Gene Expression Omnibus; HADb, Human Autophagy Database; PPI, protein protein interaction network; IPF, idiopathic pulmonary fibrosis; PAH, pulmonary arterial hypertension; ALI, acute lung injury; LAP, LC3-associated phagocytosis; GO, Gene Ontology, KEGG, Kyoto Encyclopedia of Genes and Genomes; CS, cigarette smoke.
Data Sharing Statement
MiRNAs, genes and lung function data were downloaded from GEO database (https://www.ncbi.nlm.nih.gov). Autophagy genes were obtained from Human Autophagy Database (HADb; http://www.autophagy.lu).
Ethics Statement
Ethical approval was obtained from the Research Ethics Committee of Qilu Hospital of Shandong University as per the Helsinki declaration and its later amendments.
Consent for Publication
All authors participating in this research consent to the publication.
Funding
This study was supported by the National Natural Science Foundation of China (grant no: 81800039) and Jinan clinical research center for prevention and control project of major respiratory diseases (grant no: 201912011).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Global Initiative for Chronic Obstructive Lung Disease. The global strategy for diagnosis, management and prevention of COPD (updated 2020), the pocket guide (updated 2020) and the complete list of references examined by the committee is available on the GOLD website. Available from: www.goldcopd.org.
2. European Respiratory Society on behalf of the Forum of International Respiratory Societies (FIRS). The global impact of respiratory disease, second edition; 2017. Available from: https://www.who.int/gard/publications/The_Global_Impact_of_Respiratory_Disease.pdf.
3. Guarascio AJ, Ray SM, Finch CK, Self TH. The clinical and economic burden of chronic obstructive pulmonary disease in the USA. Clinicoecon Outcomes Res. 2013;5:235–245. doi:10.2147/CEOR.S34321
4. Murray CJL, Barber RM, Foreman KJ, et al. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: quantifying the epidemiological transition. Lancet. 2015;386(10009):2145–2191. doi:10.1016/S0140-6736(15)61340-X
5. Murray CJ, Atkinson C, Bhalla K, et al. The state of US health, 1990–2010: burden of diseases, injuries, and risk factors. JAMA. 2013;310(6):591–608. doi:10.1001/jama.2013.13805
6. Zheng M, Hong W, Gao M, et al. Long noncoding RNA COPDA1 promotes airway smooth muscle cell proliferation in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2019;61(5):584–596. doi:10.1165/rcmb.2018-0269OC
7. Zong D, Li J, Cai S, et al. Notch1 regulates endothelial apoptosis via the ERK pathway in chronic obstructive pulmonary disease. Am J Physiol Cell Physiol. 2018;315(3):C330–C340. doi:10.1152/ajpcell.00182.2017
8. Gouzi F, Blaquiere M, Catteau M, et al. Oxidative stress regulates autophagy in cultured muscle cells of patients with chronic obstructive pulmonary disease. J Cell Physiol. 2018;233(12):9629–9639. doi:10.1002/jcp.26868
9. Mebratu YA, Schwalm K, Smith KR, Schuyler M, Tesfaigzi Y. Cigarette smoke suppresses Bik to cause epithelial cell hyperplasia and mucous cell metaplasia. Am J Respir Crit Care Med. 2011;183(11):1531–1538. doi:10.1164/rccm.201011-1930OC
10. Hussain SS, George S, Singh S, et al. A small molecule BH3-mimetic suppresses cigarette smoke-induced mucous expression in airway epithelial cells. Sci Rep. 2018;8(1):13796. doi:10.1038/s41598-018-32114-w
11. Racanelli AC, Kikkers SA, Choi AMK, Cloonan SM. Autophagy and inflammation in chronic respiratory disease. Autophagy. 2018;14(2):221–232. doi:10.1080/15548627.2017.1389823
12. Ueno T, Komatsu M. Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol. 2017;14(3):170–184. doi:10.1038/nrgastro.2016.185
13. Shirakabe A, Ikeda Y, Sciarretta S, Zablocki DK, Sadoshima J. Aging and autophagy in the heart. Circ Res. 2016;118(10):1563–1576. doi:10.1161/CIRCRESAHA.116.307474
14. Liao SX, Sun PP, Gu YH, Rao XM, Zhang LY, Ou-Yang Y. Autophagy and pulmonary disease. Ther Adv Respir Dis. 2019;13:1753466619890538. doi:10.1177/1753466619890538
15. Tominello TR, Oliveira ERA, Hussain SS, et al. Emerging roles of autophagy and inflammasome in ehrlichiosis. Front Immunol. 2019;10:1011. doi:10.3389/fimmu.2019.01011
16. Ghavami S, Yeganeh B, Zeki AA, et al. Autophagy and the unfolded protein response promote profibrotic effects of TGF-beta1 in human lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2018;314(3):L493–L504. doi:10.1152/ajplung.00372.2017
17. Farber HW, Loscalzo J. Pulmonary arterial hypertension. N Engl J Med. 2004;351(16):1655–1665. doi:10.1056/NEJMra035488
18. Gao Y, Wang N, Li RH, Xiao YZ. The role of autophagy and the chemokine (C-X-C Motif) ligand 16 during acute lung injury in mice. Med Sci Monit. 2018;24:2404–2412. doi:10.12659/MSM.906016
19. Fujii S, Hara H, Araya J, et al. Insufficient autophagy promotes bronchial epithelial cell senescence in chronic obstructive pulmonary disease. Oncoimmunology. 2012;1(5):630–641. doi:10.4161/onci.20297
20. Ryter SW, Lee SJ, Choi AM. Autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. Expert Rev Respir Med. 2010;4(5):573–584. doi:10.1586/ers.10.61
21. Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–844. doi:10.1038/nrg2683
22. Vij N, Chandramani-Shivalingappa P, Van Westphal C, Hole R, Bodas M. Cigarette smoke-induced autophagy impairment accelerates lung aging, COPD-emphysema exacerbations and pathogenesis. Am J Physiol Cell Physiol. 2018;314(1):C73–C87. doi:10.1152/ajpcell.00110.2016
23. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi:10.1016/j.cell.2018.09.048
24. Cadwell K, Debnath J. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol. 2018;217(3):813–822. doi:10.1083/jcb.201706157
25. Heckmann BL, Boada-Romero E, Cunha LD, Magne J, Green DR. LC3-associated phagocytosis and inflammation. J Mol Biol. 2017;429(23):3561–3576. doi:10.1016/j.jmb.2017.08.012
26. DeSelm CJ, Miller BC, Zou W, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21(5):966–974. doi:10.1016/j.devcel.2011.08.016
27. Qin H, Gao F, Wang Y, et al. Nur77 promotes cigarette smoke induced autophagic cell death by increasing the dissociation of Bcl2 from Beclin-1. Int J Mol Med. 2019;44(1):25–36. doi:10.3892/ijmm.2019.4184
28. Siganaki M, Koutsopoulos AV, Neofytou E, et al. Deregulation of apoptosis mediators’ p53 and bcl2 in lung tissue of COPD patients. Respir Res. 2010;11(1):46. doi:10.1186/1465-9921-11-46
29. Jin X, Guan Y, Zhang Z, Wang H. Microarray data analysis on gene and miRNA expression to identify biomarkers in non-small cell lung cancer. BMC Cancer. 2020;20(1):329. doi:10.1186/s12885-020-06829-x
30. Zhang X, Xu X, Ge G, et al. miR498 inhibits the growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep. 2019;41(3):1638–1648. doi:10.3892/or.2018.6948
31. Rodriguez-Martinez A, de Miguel-perez D, Ortega FG, et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 2019;21(1):21. doi:10.1186/s13058-019-1109-0
32. Zhang K, Zhang L, Mi Y, et al. A ceRNA network and a potential regulatory axis in gastric cancer with different degrees of immune cell infiltration. Cancer Sci. 2020;111(11):4041–4050. doi:10.1111/cas.14634
33. Zhu M, Ye M, Wang J, Ye L, Jin M. Construction of potential miRNA-mRNA regulatory network in COPD plasma by bioinformatics analysis. Int J Chron Obstruct Pulmon Dis. 2020;15:2135–2145. doi:10.2147/COPD.S255262
34. Qi X, Chen H, Fu B, et al. LncRNAs NR-026690 and ENST00000447867 are upregulated in CD4(+) T cells in patients with acute exacerbation of COPD. Int J Chron Obstruct Pulmon Dis. 2019;14:699–711. doi:10.2147/COPD.S191815
35. Zhao J, Pu J, Hao B, et al. LncRNA RP11-86H7.1 promotes airway inflammation induced by TRAPM2.5 by acting as a ceRNA of miRNA-9-5p to regulate NFKB1 in HBECS. Sci Rep. 2020;10(1):11587. doi:10.1038/s41598-020-68327-1
36. Keller A, Leidinger P, Bauer A, et al. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011;8(10):841–843. doi:10.1038/nmeth.1682
37. Keller A, Leidinger P, Vogel B, et al. miRNAs can be generally associated with human pathologies as exemplified for miR-144. BMC Med. 2014;12(1):224. doi:10.1186/s12916-014-0224-0
38. Goodwin JM, Dowdle WE, DeJesus R, et al. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Rep. 2017;20(10):2341–2356. doi:10.1016/j.celrep.2017.08.034
39. Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;16(7):487–511. doi:10.1038/nrd.2017.22
40. Abdul Rahim SA, Dirkse A, Oudin A, et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br J Cancer. 2017;117(6):813–825. doi:10.1038/bjc.2017.263
41. Obeidat M, Ding X, Fishbane N, et al. The effect of different case definitions of current smoking on the discovery of smoking-related blood gene expression signatures in chronic obstructive pulmonary disease. Nicotine Tob Res. 2016;18(9):1903–1909. doi:10.1093/ntr/ntw129
42. Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. doi:10.1093/nar/gkz240
43. Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc. 2015;10(6):823–844. doi:10.1038/nprot.2015.052
44. Xia J, Benner MJ, Hancock RE. NetworkAnalyst–integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–174. doi:10.1093/nar/gku443
45. Xia J, Lyle NH, Mayer ML, Pena OM, Hancock RE. INVEX–a web-based tool for integrative visualization of expression data. Bioinformatics. 2013;29(24):3232–3234. doi:10.1093/bioinformatics/btt562
46. Xia J, Fjell CD, Mayer ML, Pena OM, Wishart DS, Hancock RE. INMEX–a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res. 2013;41:W63–70. doi:10.1093/nar/gkt338
47. Fonseka P, Pathan M, Chitti SV, Kang T, Mathivanan S. FunRich enables enrichment analysis of OMICs datasets. J Mol Biol. 2021;433(11):166747. doi:10.1016/j.jmb.2020.166747
48. Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi:10.1093/nar/gkt393
49. Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28(6):771–776. doi:10.1093/bioinformatics/bts043
50. Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. doi:10.1186/s13059-019-1629-z
51. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi:10.1093/nar/gkz757
52. Huang HY, Lin YC, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48(D1):D148–D154. doi:10.1093/nar/gkz896
53. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
54. Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056.
55. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi:10.1038/75556
56. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
57. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi:10.1093/nar/gkaa1074
58. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi:10.1093/nar/gkt1248
59. Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi:10.1093/nar/gkv1270
60. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
61. Barnes PJ. Targeting cytokines to treat asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2018;18(7):454–466. doi:10.1038/s41577-018-0006-6
62. Easter M, Bollenbecker S, Barnes JW, Krick S. Targeting aging pathways in chronic obstructive pulmonary disease. Int J Mol Sci. 2020;21(18):18. doi:10.3390/ijms21186924
63. Guilleminault L, Rolland Y, Didier A. [Characteristics of non-pharmacological interventions in the elderly with COPD. Smoking cessation, pulmonary rehabilitation, nutritional management and patient education]. Rev Mal Respir. 2018;35(6):626–641. French. doi:10.1016/j.rmr.2017.12.004
64. Kennedy BK, Berger SL, Brunet A, et al. Geroscience: linking aging to chronic disease. Cell. 2014;159(4):709–713. doi:10.1016/j.cell.2014.10.039
65. Vaz Fragoso CA, Gill TM. Defining chronic obstructive pulmonary disease in an aging population. J Am Geriatr Soc. 2010;58(11):2224–2226. doi:10.1111/j.1532-5415.2010.03128.x
66. Dall TM, Gallo PD, Chakrabarti R, West T, Semilla AP, Storm MV. An aging population and growing disease burden will require a large and specialized health care workforce by 2025. Health Aff. 2013;32(11):2013–2020. doi:10.1377/hlthaff.2013.0714
67. Chen S, Wang Y, Zhang H, et al. The antioxidant mitoq protects against CSE-induced endothelial barrier injury and inflammation by inhibiting ROS and autophagy in human umbilical vein endothelial cells. Int J Biol Sci. 2019;15(7):1440–1451. doi:10.7150/ijbs.30193
68. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4(1):199–227. doi:10.1146/annurev.pathol.4.110807.092222
69. Ma Y, Zhang J, Wen L, Lin A. Membrane-lipid associated lncRNA: a new regulator in cancer signaling. Cancer Lett. 2018;419:27–29. doi:10.1016/j.canlet.2018.01.008
70. Fu LL, Wen X, Bao JK, Liu B. MicroRNA-modulated autophagic signaling networks in cancer. Int J Biochem Cell Biol. 2012;44(5):733–736. doi:10.1016/j.biocel.2012.02.004
71. Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy-regulating microRNAs and Cancer. Front Oncol. 2017;7:65. doi:10.3389/fonc.2017.00065
72. Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2013;32(12):1570–1579. doi:10.1038/onc.2012.167
73. Mullany LE, Herrick JS, Wolff RK, Stevens JR, Samowitz W, Slattery ML. MicroRNA-transcription factor interactions and their combined effect on target gene expression in colon cancer cases. Genes Chromosomes Cancer. 2018;57(4):192–202. doi:10.1002/gcc.22520
74. Uddin MN, Li M, Wang X. Identification of transcriptional markers and microRNA-mRNA regulatory networks in colon cancer by integrative analysis of mRNA and microRNA expression profiles in colon tumor stroma. Cells. 2019;8(9):9. doi:10.3390/cells8091054
75. Singer JP, Katz PP, Iribarren C, et al. Both pulmonary and extra-pulmonary factors predict the development of disability in chronic obstructive pulmonary disease. Respiration. 2013;85(5):375–383. doi:10.1159/000338110
76. Eisner MD, Blanc PD, Yelin EH, et al. COPD as a systemic disease: impact on physical functional limitations. Am J Med. 2008;121(9):789–796. doi:10.1016/j.amjmed.2008.04.030
77. Stevens JF, Maier CS. Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol Nutr Food Res. 2008;52(1):7–25. doi:10.1002/mnfr.200700412
78. Sievi NA, Senn O, Brack T, et al. Impact of comorbidities on physical activity in COPD. Respirology. 2015;20(3):413–418. doi:10.1111/resp.12456
79. Agusti A, Soriano JB. COPD as a systemic disease. COPD. 2008;5(2):133–138. doi:10.1080/15412550801941349
80. Yasuo M, Droma Y, Kitaguchi Y, et al. The relationship between acrolein and oxidative stress in COPD: in systemic plasma and in local lung tissue. Int J Chron Obstruct Pulmon Dis. 2019;14:1527–1537. doi:10.2147/COPD.S208633
81. Lam HC, Cloonan SM, Bhashyam AR, et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest. 2013;123(12):5212–5230. doi:10.1172/JCI69636
82. An CH, Wang XM, Lam HC, et al. TLR4 deficiency promotes autophagy during cigarette smoke-induced pulmonary emphysema. Am J Physiol Lung Cell Mol Physiol. 2012;303(9):L748–L757. doi:10.1152/ajplung.00102.2012
83. Chen ZH, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 2008;3(10):e3316. doi:10.1371/journal.pone.0003316
84. Chen ZH, Lam HC, Jin Y, et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A. 2010;107(44):18880–18885. doi:10.1073/pnas.1005574107
85. Mizumura K, Cloonan SM, Nakahira K, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. 2014;124(9):3987–4003. doi:10.1172/JCI74985
86. Li D, Hu J, Wang T, et al. Silymarin attenuates cigarette smoke extract-induced inflammation via simultaneous inhibition of autophagy and ERK/p38 MAPK pathway in human bronchial epithelial cells. Sci Rep. 2016;6(1):37751. doi:10.1038/srep37751
87. Zhou JS, Zhao Y, Zhou HB, et al. Autophagy plays an essential role in cigarette smoke-induced expression of MUC5AC in airway epithelium. Am J Physiol Lung Cell Mol Physiol. 2016;310(11):L1042–L1052. doi:10.1152/ajplung.00418.2015
88. Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2(9):647–656. doi:10.1038/nrc883
89. Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: bcl-2 proteins and apoptosis: an update. Arch Toxicol. 2015;89(3):289–317. doi:10.1007/s00204-014-1448-7
90. Sata M, Takabatake N, Inoue S, et al. Intronic single-nucleotide polymorphisms in Bcl-2 are associated with chronic obstructive pulmonary disease severity. Respirology. 2007;12(1):34–41. doi:10.1111/j.1440-1843.2006.00959.x
91. Weaver CV, Liu SP. Differentially expressed pro- and anti-apoptogenic genes in response to benzene exposure: immunohistochemical localization of p53, Bag, Bad, Bax, Bcl-2, and Bcl-w in lung epithelia. Exp Toxicol Pathol. 2008;59(5):265–272. doi:10.1016/j.etp.2007.02.012
92. Zeng H, Kong X, Peng H, et al. Apoptosis and Bcl-2 family proteins, taken to chronic obstructive pulmonary disease. Eur Rev Med Pharmacol Sci. 2012;16(6):711–727.
93. Li R, Yang YE, Jin J, et al. Identification of lncRNA biomarkers in lung squamous cell carcinoma using comprehensive analysis of lncRNA mediated ceRNA network. Artif Cells Nanomed Biotechnol. 2019;47(1):3246–3258. doi:10.1080/21691401.2019.1647225
94. Huo C, Zhang MY, Li R, et al. Comprehensive analysis of TPX2-related ceRNA network as prognostic biomarkers in lung adenocarcinoma. Int J Med Sci. 2020;17(16):2427–2439. doi:10.7150/ijms.49053
95. Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234(6):9105–9117. doi:10.1002/jcp.27587
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.