Back to Journals » Infection and Drug Resistance » Volume 16
ApoE Mimetic Peptide COG1410 Exhibits Strong Additive Interaction with Antibiotics Against Mycobacterium smegmatis
Authors Zhao YY, Wang C, Wang WX, Han LM, Zhang C, Yu JY, Chen W , Hu CM
Received 17 January 2023
Accepted for publication 22 March 2023
Published 28 March 2023 Volume 2023:16 Pages 1801—1812
DOI https://doi.org/10.2147/IDR.S403232
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yan-Yan Zhao,1,* Chun Wang,1,* Wei-Xiao Wang,2 Li-Mei Han,1 Caiyun Zhang,2 Jiao-Yang Yu,3 Wei Chen,2 Chun-Mei Hu1,4
1Department of Tuberculosis, the Second Hospital of Nanjing, Nanjing University of Chinese Medicine, Nanjing, 210003, People’s Republic of China; 2Clinical Research Center, the Second Hospital of Nanjing, Nanjing University of Chinese Medicine, Nanjing, 210003, People’s Republic of China; 3Key Laboratory of Resources Biology and Biotechnology in Western China, Ministry of Education, College of Life Sciences, Northwest University, Xi’an, 710069, People’s Republic of China; 4The Clinical Infectious Disease Center of Nanjing, Nanjing, 210003, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Chen; Chun-Mei Hu, Email [email protected]; [email protected]
Background: Drug-resistant tuberculosis (TB) is an emerging threat to public health worldwide. Antimicrobial peptide (AMP) is a promising solution to solve the antimicrobial resistance crisis. The apolipoprotein E mimetic peptide COG1410 has been confirmed to simultaneously have neuroprotective, anti-inflammatory, and antibacterial activity. However, whether it is effective to inhibit growth of mycobacteria has not been investigated yet.
Methods: The peptide COG1410 was synthesized with conventional solid-phase peptide synthesis and qualified by HPLC and mass spectrometry. Micro-dilution method was used to determine the minimal inhibitory concentration. A time-kill assay was used to determine the bactericidal dynamics of antimicrobial peptide and relative antibiotics. Static biofilm formation was conducted in 24-well plate and the biofilm was separated from planktonic cells and collected. The mechanism of action of COG1410 was explored by TEM observation and ATP leak assay. The localization of COG1410 was observed by confocal laser scan microscopy. The drug–drug interaction was determined by a checkerboard assay.
Results: COG1410 was a potent bactericidal agent against M. smegmatis in vitro and within the macrophages with MIC 16 μg/mL, but invalid against M. abscess and M. tuberculosis. A time-kill assay showed that COG1410 killed M. smegmatis as potent as clarithromycin, but faster than LL-37, another short synthetic cationic peptide. 1× MIC COG1410 almost reduced 90% biofilm formation of M. smegmatis. Additionally, COG1410 was able to penetrate the cell membrane of macrophage and inhibit intracellular M. smegmatis growth. TEM observation and ATP leak assay found that COG1410 disrupted cell membrane and caused release of cell contents. Confocal fluorescence microscopy showed that FITC-COG1410 aggregated around cell membrane instead of entering the cytoplasm. Although COG1410 had relative high cytotoxicity, it exhibited strong additive interaction with regular anti-TB antibiotics, which reduced the working concentration of COG1410 and expanding safety window. After 30 passages, there was no induced drug resistance for COG1410.
Conclusion: COG1410 was a novel and potent AMP against M. smegmatis by disrupting the integrity of cell membrane.
Keywords: antimicrobial peptide, COG1410, Mycobacterium smegmatis, additive interaction
Introduction
Tuberculosis (TB) is still a great challenge for public health worldwide, especially in lower-middle-income countries. Mycobacterium tuberculosis (MTB) is a causative agent that possesses a thick, waxy, hydrophobic cell envelope, as well as drug degrading and modifying enzymes, causing intrinsic resistance to many antibiotics. Chromosomal mutations also confer drug resistance via modifying or overexpressing the drug target, or preventing prodrug activation.1 In particular, drug resistance of MTB is in the continuous evolution process, which depends on bacterial fitness, strain’s genetic background and its capacity to adapt to the surrounding environment.2 According to WHO TB report in 2020, 6.3% and 1.2% of patients with bacteriologically confirmed pulmonary TB (2.1 million) were diagnosed as MDR-TB and pre-XDR-TB or XDR-TB, respectively.3 Approximately one in three of patients who developed MDR-TB were enrolled on treatment and the treatment success rate ranged only from 56% to 69% among WHO regions. Treatment of drug-resistant TB is actually both complex and expensive.4 What is worse, only a few new TB drugs including bedaquiline and linezolid have entered clinical practice in the past 50 years.5 Novel anti-TB drugs are urgently needed, to enhance bioavailability and efficacy against drug-resistant TB.
Antimicrobial peptides (AMPs) possess broad-spectrum antibacterial and immunomodulatory activities against infectious bacteria, viruses, and fungi, which are a hot topic of interest.6 Because of its special bactericidal mechanisms, AMPs are considered as one of the most effective therapeutic agents against TB.7 The mechanism of action of AMPs is either through direct disruption of the normal mycobacterial cell wall or entering the cell and binding to the intercellular targets.8,9 For example, LL-37 is a short synthetic cationic AMP with an α-helical structure derived endogenously from the human cathelicidin hCAP18.10 Recent studies have demonstrated that LL-37 bound to the cell wall of MTB, causing disintegration and rapid rupture within minutes.11 On the other hand, AMPs are able to induce autophagy for bacterial death and clearance, such as Iztli peptide 1, which was reported to induce autophagy in elimination of MTB.12 Additionally, the major target of AMPs is the bacterial cell membrane, which makes it complicated for bacteria to evade AMPs’ killing, while maintain cell membrane’s functional and structural integrity.13 AMPs are considered as a promising solution against current intractable antimicrobial resistance of MTB.
ApoE is one of the primary apolipoproteins involved in lipid metabolism in the central nervous system, which is a reservoir of cryptic bioactive peptides.14,15 Some mimetic peptides show potent antimicrobial and immunomodulatory activity.16–19 COG1410 is a synthetic peptide originated from ApoE, which is confirmed to have neuroprotective and anti-inflammation activities.20,21 In our previous study, we showed that COG1410 displayed strong bacterial killing against a few bacterial pathogens, high stability in human plasma, and a low propensity for resistance development.22 However, whether it is capable of inhibiting mycobacteria has not been studied in detail. In this study, we investigated its antimicrobial efficiency against mycobacteria.
Materials and Methods
Peptide
COG1410 is a synthetic mimetic peptide, derived from human apolipoprotein E (ApoE), a large multifunctional glycoprotein involved in the metabolism of fats in the body of mammals. COG1410 consists of 12 amino acids originated from 138 to 149 of ApoE N terminal domain, with aminoisobutyric acid (Aib) substitutions at positions 140 and 145.23 The COG1410 peptide was synthesized with conventional solid-phase peptide synthesis at a purity of 95% and qualified by HPLC and mass spectrometry in Polypeptide Labs (San Diego, CA). In all experiments, peptides were dissolved in sterile milli-Q water immediately before use.
Bacteria and Cells
Mycobacteria were cultured at 37°C in Middlebrook 7H9 broth (BD Bioscience) supplemented with 10% Middlebrook oleic acid dextrose and catalase enrichment (OADC), 2% (w/v) glycerol, and 0.08% (v/v) Tween 80, or Middlebrook 7H10 agar supplemented with 10% OADC. These mycobacteria used in this study were M. smegmatis mc2155, M. abscess 19977, the clinically isolated multiple drug-resistant (MDR) M. avium strain 8–70, the clinically isolated MDR M. intracellular strain 8–18, and M. tuberculosis H37Rv. To prepare the log-phase culture, M. smegmatis and M. abscess were incubated for 24 h, M. avium and M. intracellular were incubated for 7 d, M. tuberculosis was incubated for 14 d. Mouse macrophage cell line RAW264.7 (Shenzhen Yibaishun Science and Technology Ltd.) was cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (FBS) and 1% penicillin–streptomycin solution, at 37°C with 5% CO2.
Minimal Inhibitory Concentration Measurements
The log-phase cultures of mycobacteria were prepared as above, diluted and dispensed into 96-well plates, with final McFarland (MCF) unit 0.05 (105 CFU/mL), 100 µL for each well and 4 wells for each sample. Different concentrations of COG1410 and antibiotics were prepared by twofold gradient dilution and added into the wells with equal volume. Negative control was a drug-free medium. The plates were sealed with parafilm and incubated at 37°C for 48 h for M. smegmatis and M. abscess, 7 d for M. avium and M. intracellular, and 14 d for M. tuberculosis. The OD value was measured spectrophotometrically at 600 nm. The MIC assay was performed in triplicate.
Time-Kill Assay
The log-phase culture of M. smegmatis mc2155 was prepared as above. Different concentrations of COG1410 were added and incubated for 48 h. The OD600 values were monitored every one hour using a microplate reader. No peptide was used as negative control. The experiments were repeated twice.
The killing kinetics of COG1410 against mycobacteria were evaluated based on colony formation unit (CFU) reduction. Different concentrations of COG1410 and antibiotics (1× MIC) were added and incubated at 37°C. The CFU was checked at different time points. LL-37 was used as a positive control. Three independent experiments were performed.
Biofilm Assay
The biofilm assay was performed as described previously.24 The log-phase culture of M. smegmatis mc2155 was prepared as above and transferred to the 24-well plate with final McFarland (MCF) unit 0.05 in 2 mL Middlebrook 7H9 broth. Gradient COG1410 was diluted and added into the 24-well plate, four wells for each treatment. No AMP was used as negative control. The plates were incubated at 37°C for 3 days. The planktonic culture under the biofilm was extracted using a syringe. The remaining biofilm settled to the bottom and was suspended in 1 mL PBS. After a full vortex, the CFU within the planktonic and the biofilm suspension was measured by serial dilution and plating on 7H10 agar plates. Two independent experiments were performed.
Intracellular Activity in Infected Macrophages
The intracellular killing assay was performed as described previously with the following modifications.25 1 mL 5×105 RAW264.7 cells were inoculated in 1 mL DMEM with 10% FBS in a 24-well plate and incubated in the 5% CO2 incubator at 37°C for 20 h. 1 mL M. smegmatis mc2155 (5 × 106) was transferred into each well and co-incubated with the macrophage cells for 4 h. Then, the adherent cells were washed with the culture medium and PBS to remove extracellular bacteria. Macrophages were treated with 16 µg/mL COG1410. Samples were taken from different time points, washed 3 times with PBS and lysed with 1% Triton-X100. The intracellular survival of M. smegmatis was determined by plating serially diluted culture on Middlebrook7H10 agar with 10% OADC plates, and the number of colonies is counted after 3 days. Three independent experiments were performed.
ATP Leak Assay
ATP leak assay was performed using the Enhanced ATP Assay Kit of Beyotime (S0027) according to the manufacturer’s manual. Briefly, the log-phase culture of M. smegmatis was prepared above, washed and suspended in PBS. The suspension was exposed to 16 µg/mL COG1410 or 0.5 µg/mL of ethambutol and incubated at 37°C for 2 h. The supernatant was collected and used for determination of ATP levels. In total, 100 µL supernatant was mixed with 100 µL working solution, and the chemiluminescence was measured using a plate reader. Untreated samples were used as negative control. Three independent experiments were performed.
Transmission Electron Microscopy
The log-phase culture of M. smegmatis was prepared as above, then washed three times with 1× PBS and resuspended in PBS. The bacteria were incubated with 1× MIC COG1410 at 37°C for 2 h. Untreated sample was used as a negative control. The samples for TEM were prepared according to the steps described previously.22 M. smegmatis cells were fixed in 4% paraformaldehyde and 2.5% glutaraldehyde and then fixed in 1% buffered osmium tetroxide. After dehydration, cells were embedded in epoxy resin. Ultrathin sections of samples were made and placed on copper grids, stained with uranyl acetate and lead citrate, and applied to TEM for image collection.
Confocal Laser Scanning Microscopy
To explore the localization of COG1410, the log-phase cultures of mc2155, S. aureus and E. faecium were prepared, washed, and suspended in PBS. The cultures were incubated with 16 µg/mL FITC-labelled COG1410 for 1 h at 37°C. For each strain, 5 µL of culture was spotted on a clean slide coated with a thin layer of 1% agarose. Fluorescence was observed by Olympus FV3000 confocal laser scanning microscope. For FITC, the excitation and emission were 488 nm and 525 nm, respectively.
Cytotoxicity
Cytotoxicity of COG1410 was determined by CCK-8 assay according to the manufacturer’s instructions (Yeasen Biotech). 104 RAW 264.7 macrophage cells or human hepatic L02 cells were dispensed into the 96-well plate, incubated at 37°C and 5% CO2 for 24 h. Subsequently, COG1410 was added to the medium at different concentrations and incubated for another 24 h. Then, 10 µL of CCK-8 solution was added to each well and incubated for 4 h. The absorbance was measured spectrophotometrically at OD450 nm and compared with untreated cells and blank control.
Drug-Resistance Development Assay
M. smegmatis culture was incubated with sub-lethal concentration (1/32 MIC) of rifampicin or COG1410 for the 1 day. 20 µL culture was taken daily and added to 2 mL fresh medium containing the corresponding antibiotics. The concentrations of antibiotics were doubled every 10 passages, and the cultures were collected every 5 passages for the whole 30 passages. The collected bacteria were tested for susceptibility by micro-dilution methods. The MIC change fold was used to assess the rate of developed resistance of M. smegmatis to COG1410 and the antibiotic.
Combination with Antibiotics
Antimicrobial interactions were determined by the checkerboard assay. In a 96-well plate, COG1410 was diluted serially along the ordinate, while the antibiotics were diluted along the abscissa. Then, a log-phase culture of M. smegmatis was added, approximately 105 CFU/mL each well. The plates were then incubated at 37°C and OD600 was measured after 48 h. The fractional inhibitory concentration index (FICI) was calculated for each combination using this equation: FICI = FIC(A) + FIC(B), where FIC(A) = MIC of drug A in combination/MIC of drug A alone, and FIC(B) = MIC of drug B in combination/MIC of drug B alone. FICI of ≤0.5 was interpreted as synergy, 0.5 < FICI ≤ 1.0 as an additive, 1.0 < FICI ≤4.0 as indifferent, and FICI > 4.0 as antagonism.
Results
COG1410 Displays Potent Bactericidal Activity Against M. smegmatis
The antimicrobial activity of COG1410 against mycobacteria was determined by measuring its minimum inhibitory concentrations (MICs). It was invalid against M. tuberculosis, M. abscess, M. avium, and M. intracellular, with MIC over 128 µg/mL (Table S1). To our interest, COG1410 displayed potent bactericidal activity against M. smegmatis, with an MIC value of 16 µg/mL (Figure 1A). A time-kill assay showed that COG1410 did not kill M. tuberculosis (Figure S1) but killed M. smegmatis faster than LL-37, another short synthetic cationic peptide killing M. smegmatis.26 1× MIC COG1410 exhibited significant bactericidal activity within 5 min and killed more than 70% M. smegmatis cells within 10 min, while 1× MIC LL-37 reduced less than 45% after 2 h (Figure 1B). Compared to clinical anti-mycobacterium antibiotics, COG1410 was similar to clarithromycin (CLR), but stronger than azithromycin (AZM) and rifampicin (RFP) (Figure 1C). However, it is worthy to note that COG1410 could not eliminate M. smegmatis cells, even after 48 h, which was similar to the tested anti-mycobacterium antibiotics, causing about 7% bacteria remained (Figure 1D).
COG1410 Inhibits M. smegmatis Biofilm Formation
Biofilm is a common pathogenic factor of mycobacteria.27 The biofilm of M. smegmatis was mainly on the air–medium interface. Therefore, we utilized syringe to exact the planktonic culture and let the biofilm settle down to the bottom. Colony forming units (CFUs) within the biofilms in the presence or absence of COG1410 were measured, as well as the planktonic cultures. To our interest, 1/2 MIC COG1410 was effective on inhibiting the biofilm formation of M. smegmatis, and 1× MIC COG1410 almost reduced 90% biofilm compared to untreated control (Figure 2A). The inhibiting efficiency increased along with more AMP. Similarly, the planktonic culture under the biofilm was significantly inhibited by COG1410 as well (Figure 2B). These data showed that COG1410 was a good biofilm inhibitor of M. smegmatis.
COG1410 Reduces Bacterial Load Within Macrophages
Mycobacteria can survive and proliferate in macrophages.28 To investigate whether COG1410 could play the antimicrobial effect within macrophages, we first established an intracellular infection cell model with M. smegmatis. After co-incubation of 4 h, the extracellular bacteria were removed by washing and the intercellular bacteria were counting through cell lysis. As shown in Figure 3A, the extracellular bacteria were 2-log less than its intracellular counterpart, which indicated that M. smegmatis successfully enter the cytoplasm of macrophages. Then, we decided to measure the intracellular bactericidal activity of COG1410. We found that COG1410 was able to reduce bacterial load within the macrophage cells compared to the untreated control. At 6 h post of infection, approximately 40% of bacteria were inhibited by 1 MIC COG1410. Over time, M. smegmatis proliferated in the cells, but were significantly slower in the presence of COG1410 (Figure 3B). These data showed that COG1410 could pass through cell membrane to reach the intracellular pathogen.
COG1410 Disrupts Cell Membrane of M. smegmatis
To explore the mechanism of action of COG1410 against M. smegmatis, we observed the bacterial morphology treated with COG1410 via transmission electron microscopy. The untreated cells were bacillus full of black cell contents, while COG1410 treated cells became transparent with incomplete membrane (Figure 4). These images showed that COG1410 treatment impaired cell membrane and released cell contents, causing cell death.
 |
Figure 4 COG1410 treatment disrupted cell membrane of M. smegmatis. (A) M. smegmatis was exposed to 1× MIC COG1410 for 2 h. (B) The cells in PBS were negative control. Scale bar: 500 nm. |
To further confirm this mechanism, the leakage of ATP was detected. As early as 10 min after treatment, a significant ATP leak was observed. As time went on, the leak increased (Figure 5). Ethambutol (EMB) was chosen as a negative control, which is a first-line drug for TB and NTM infection. The mechanism of action of EMB is to inhibit arabinosyl transferase, which is involved in mycobacterial cell wall constitution.29 Sharply different from COG1410, EMB never induced ATP leak. Therefore, the mechanism of action of COG1410 was different from that of EMB, mainly disrupting membrane integrity.
COG1410 is Localized Around Cell Membrane of M. smegmatis
To investigate whether COG1410 binds directly to the bacterial membrane or enters the cytoplasm, we treated M. smegmatis with 16 µg/mL FITC-labelled COG1410 for 1 h and observed the localization of FITC-1410 by confocal fluorescence microscopy. We found that FITC-1410 aggregated around cell membrane of M. smegmatis. Also, we noticed ovoid or spherical cells of M. smegmatis, which might be associated with morphology change under stress.30 In contrast, FITC-1410 entered the cytoplasm of Staphylococcus aureus and Enterococcus faecium (Figure 6). Based on our previous study, COG1410 was active against E. faecium and M. smegmatis, but not S. aureus.22 Therefore, we infer that the major target of COG1410 against M. smegmatis is the cell membrane and whether it enters the cytoplasm does not matter.
 |
Figure 6 COG1410 was localized around the cell membrane. The localization of FITC-labelled COG1410 was observed in M. smegmatis, S. aureus, and E. faecium by CLSM. Scale bar: 4 µm. |
COG1410 Has Relative High Cytotoxicity
One of the limiting factors for the use of AMPs as therapeutic agents is their cytotoxicity to mammalian cells. To evaluate the cytotoxic effect of COG1410, we tested its toxicity on mouse macrophage RAW264.7 cells and human hepatic L02 cells by CCK-8 assay. We found that COG1410 had no significant cytotoxic effect on the two types of cells below 32 µg/mL (Figure 7). Further structural modification is needed to reduce the cytotoxicity of COG1410.
There is No Induced Drug-Resistance for COG1410
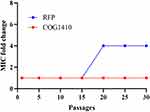
The development of drug resistance in mycobacteria is one of the major challenges. AMPs have different targets of action from conventional antibiotics, and they are less likely to induce resistance in bacteria. In order to determine possible resistance development, we exposed M. smegmatis to sublethal doses of COG1410 and rifampicin (RFP) and monitored the susceptibility after serial passages. We found that M. smegmatis easily developed resistance against RFP, of which MIC increased 4 folds after 20 passages. However, there was no induced drug-resistance for COG1410 even after 30 passages (Figure 8), which suggested that COG1410 was potent to inhibit growth of M. smegmatis.
COG1410 Displays Strong Additive Interaction with Anti-TB Antibiotics
Interaction between COG1410 and conventional antibiotics used to treat TB or NTM infection was evaluated by checkerboard assay. The FICI index of each combination against M. smegmatis was summarized in Table 1. All of the tested antibiotics had FICI between 0.5 and 1 with COG1410 except Ciprofloxacin, which indicated that COG1410 had additive interaction with these antibiotics. Especially, COG1410 enhanced 16 folds of antimicrobial activity of Ethambutol, Rifampicin, Azithromycin, and Cefoxitin. With 1/2 MIC amikacin, 1/16 MIC COG1410 completely inhibited bacterial growth of M. smegmatis within 48 h. Similarly, combined with 8 µg/mL COG1410, only 1/8 MIC Linezolid or 1/16 MIC Cefoxitin was needed to achieve the inhibitory effect (Figure 9). Therefore, the additive interaction between COG1410 and the antibiotics reduced the working concentration of COG1410 and expanded the safety window.
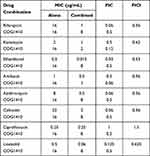 |
Table 1 Antimicrobial Activity of Combination of COG1410 with Antibiotics Against M. smegmatis |
COG1410 did not exhibit similar activity with antibiotics against M. abscess and M. tuberculosis (Table S2), which suggested that COG1410 was not capable of creating a hole in the cell membranes of these two pathogenic mycobacteria.
Discussion
In our previous study, we found that COG1410 was a potent antimicrobial peptide against pan-drug resistant A. baumannii through disruption of the integrity of cell membrane and stimulation of intracellular ROS production. Strong synergistic interaction between COG1410 and polymyxin B reduced working concentration of COG1410, broadening its application landscape.22 Here, we investigated the bacteriostatic activity of COG1410 against mycobacteria. COG1410 took effect faster than LL-37 and killed more than 70% M. smegmatis cells within 10 min. Besides planktonic cells, COG1410 could significantly inhibit biofilm formation. Remarkably, COG1410 exhibited intracellular activity, inhibiting M. smegmatis’s growth within the macrophage. Confocal fluorescence microscopy observation showed that COG1410 mainly aggregated around the cell wall of M. smegmatis. TEM observation and ATP leak assay revealed that COG1410 disrupted integrity of bacterial cell membrane. After 30 passages, there were no drug-resistant colonies against COG1410. Remarkably, COG1410 was confirmed to have additive interaction with all the tested clinical anti-TB antibiotics. COG1410 neither killed M. abscess and M. tuberculosis directly nor exhibited additive activity with antibiotics against these two pathogenic mycobacteria. In our preliminary screening, we reported that COG1410 inhibited M. abscess and M. tuberculosis, which was due to early observation and hasty conclusion. The inhibitory effect against mycobacteria must be very cautious since they grow much slower than common pathogens like E. coli.
M. smegmatis is usually accepted as a surrogate to screen anti-TB compounds. For example, an in vivo-mimic silkworm infection model with M. smegmatis was established for screening of therapeutically effective anti-MTB antibiotics.31 However, in our study, COG1410 was active against M. smegmatis but not against other NTM species and MTB. It has been reported that Candida albicans and Staphylococcus aureus are the best surrogates to predict anti-MTB activity of AMPs, better than M. smegmatis.32 Consistently, our previous study also found that COG1410 did not inhibit growth of S. aureus.22 M. smegmatis belongs to rapidly-growing mycobacterium, while MTB is slowly-growing, which are different in terms of cell diameter, length, cytoplasmic volume, and total ribosome number.33 Additionally, the porins in the cell wall between M. smegmatis and M. tuberculosis are different, which determine the permeation properties of hydrophilic molecules. The latter has a lower number of porins, and the porins have lower channel-forming activity.34 These differences may influence the efficacy of COG1410 against M. tuberculosis. As well, M. smegmatis is confirmed as a good model for studying the general biological properties of mycobacteria, but not suitable for pathogenicity and virulence.33,35 Therefore, from our experience, in vitro screening model of M. smegmatis is not suitable for anti-MTB drugs.
It is known that the natural multilayered cell wall rich in lipids restricts antibiotic permeability within mycobacteria. Application of AMPs is able to address this challenge by forming membrane pores or disrupting the integrity of cell membrane and facilitating the entry of chemical antibiotics, which might be a model of antimicrobial synergy between AMPs and antibiotics.36,37 At present, abundant natural or synthetic AMPs against mycobacteria have been identified.38 Most of them displayed synergistic interaction with antibiotics. For example, AS-48, a bacteriocin produced by Enterococcus faecalis, was active against MTB and other nontuberculous mycobacterial species (NTMs), showing synergistic combination with ethambutol.39 The synthetic AMPs HHC-8 and MM-10 showed strong synergism with rifampicin against M. smegmatis with FICs 0.09, as well as MTB.40 Some of the AMPs showed additive interaction with anti-mycobacterial antibiotics. For example, NZX, a fungal peptide, had additive effect with isoniazid and ethambutol both in vitro and in a murine TB-model.41 It is worth noting that no AMPs like COG1410 showed additive interaction with so many anti-mycobacteria antibiotics. 8 µg/mL COG1410 (1/2 MIC) enhanced the efficacy of rifampicin, ethambutol, azithromycin, cefoxitin, and linezolid by 16, 33, 16, 16, and 8 folds, respectively. Especially, 1 µg/mL COG1410 and 0.5 µg/mL amikacin were able to inhibit growth of M. smegmatis. As a whole, COG1410 is a good adjunct for clinical antibiotics.
To date, many AMPs have been isolated from various natural sources. AntiTbPdb is a database, which collected around 1010 experimentally verified anti-mycobacterial peptides.42 Some peptides with proven anti-TB activity, better pharmacokinetic properties, and minimal or no toxicity are promising candidates to tackle drug-resistant MTB and enter preclinical trials.43 There are also encouraging examples from re-design and modification of natural peptides, which enhance anti-TB activity. For example, Nisin A is a prototype I antibiotic and used as a safe food additive. Through amino acid replacement, nisin V (M21V) had significantly higher anti-TB activity.44 Since it possesses neuroprotective, anti-inflammatory, and antibacterial effects, COG1410 is a promising lead AMP, which is worthy of further improvement with structure modification.
Conclusions
In this study, we confirmed that COG1410 was a novel and potent bactericidal AMP against M. smegmatis. COG1410 aggregated around cell membrane and finally disrupted its integrity, causing release of cell contents. COG1410 was very stable in human plasma, which benefits from substitution of two unnatural amino acids. Drug resistance was not easily developed. Further modification of its structure was necessary to enhance anti-TB activity and reduce cytotoxicity.
Acknowledgments
We would like to thank Dr. George F. Li in Cognosci, Inc., United States, for providing COG1410 peptide. This study was supported by Natural Science Foundation of Jiangsu province (BK20221172) and National Natural Science Foundation of China (82272340), the Six Talent Peaks Project in Jiangsu Province (WSN-160) and “333 talent project” of Jiangsu province. Funders had no role in study design, data collection or analysis, preparation of the manuscript or the decision to publish it.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. 2017;41(3):354–373. doi:10.1093/femsre/fux011
2. Allué-Guardia A, García JI, Torrelles JB. Evolution of drug-resistant mycobacterium tuberculosis strains and their adaptation to the human lung environment. Front Microbiol. 2021;12:612675. doi:10.3389/fmicb.2021.612675
3. World Health Organization. Global Tuberculosis Report 2021. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO; 2021.
4. Tadolini M, Garcia-Prats AJ, D’Ambrosio L, et al. Compassionate use of new drugs in children and adolescents with multidrug-resistant and extensively drug-resistant tuberculosis: early experiences and challenges. Eur Respir J. 2016;48(3):938–943. doi:10.1183/13993003.00705-2016
5. Chakraborty S, Rhee KY. Tuberculosis drug development: history and evolution of the mechanism-based paradigm. Cold Spring Harb Perspect Med. 2015;5(8):a021147. doi:10.1101/cshperspect.a021147
6. Bardan A, Nizet V, Gallo RL. Antimicrobial peptides and the skin. Expert Opin Biol Ther. 2004;4(4):543–549. doi:10.1517/14712598.4.4.543
7. Nakatsuji T, Gallo RL. Antimicrobial peptides: old molecules with new ideas. J Invest Dermatol. 2012;132(3 Pt 2):887–895. doi:10.1038/jid.2011.387
8. Tomasinsig L, Zanetti M. The cathelicidins--structure, function and evolution. Curr Protein Pept Sci. 2005;6(1):23–34. doi:10.2174/1389203053027520
9. AlMatar M, Makky EA, Yakıcı G, Var I, Kayar B, Köksal F. Antimicrobial peptides as an alternative to anti-tuberculosis drugs. Pharmacol Res. 2018;128:288–305. doi:10.1016/j.phrs.2017.10.011
10. Dürr UH, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758(9):1408–1425. doi:10.1016/j.bbamem.2006.03.030
11. Deshpande D, Grieshober M, Wondany F, et al. Super-resolution microscopy reveals a direct interaction of intracellular mycobacterium tuberculosis with the antimicrobial peptide LL-37. Int J Mol Sci. 2020;21(18):6741. doi:10.3390/ijms21186741
12. Peláez Coyotl EA, Barrios Palacios J, Muciño G, et al. Antimicrobial peptide against mycobacterium tuberculosis that activates autophagy is an effective treatment for tuberculosis. Pharmaceutics. 2020;12(11):1071. doi:10.3390/pharmaceutics12111071
13. Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30(3):131–141. doi:10.1016/j.it.2008.12.003
14. Pitas RE, Boyles JK, Lee SH, Hui D, Weisgraber KH. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B, E(LDL) receptors in the brain. J Biol Chem. 1987;262(29):14352–14360. doi:10.1016/S0021-9258(18)47945-8
15. Zanfardino A, Bosso A, Gallo G, et al. Human apolipoprotein E as a reservoir of cryptic bioactive peptides: the case of ApoE 133–167. J Peptide Sci. 2018;24(7):e3095. doi:10.1002/psc.3095
16. Wang CQ, Yang CS, Yang Y, Pan F, He LY, Wang AM. An apolipoprotein E mimetic peptide with activities against multidrug-resistant bacteria and immunomodulatory effects. J Peptide Sci. 2013;19(12):745–750. doi:10.1002/psc.2570
17. Pane K, Sgambati V, Zanfardino A, et al. A new cryptic cationic antimicrobial peptide from human apolipoprotein E with antibacterial activity and immunomodulatory effects on human cells. FEBS J. 2016;283(11):2115–2131. doi:10.1111/febs.13725
18. Puthia M, Marzinek JK, Petruk G, Ertürk Bergdahl G, Bond PJ, Petrlova J. Antibacterial and anti-inflammatory effects of apolipoprotein E. Biomedicines. 2022;10(6). doi:10.3390/biomedicines10061430
19. Petruk G, Elvén M, Hartman E, et al. The role of full-length apoE in clearance of Gram-negative bacteria and their endotoxins. J Lipid Res. 2021;62:100086. doi:10.1016/j.jlr.2021.100086
20. Chiu LS, Anderton RS, Cross JL, et al. Assessment of R18, COG1410, and APP96-110 in excitotoxicity and traumatic brain injury. Transl Neurosci. 2017;8:147–157. doi:10.1515/tnsci-2017-0021
21. Cao F, Jiang Y, Wu Y, et al. Apolipoprotein E-mimetic COG1410 reduces acute vasogenic edema following traumatic brain injury. J Neurotrauma. 2016;33(2):175–182. doi:10.1089/neu.2015.3887
22. Wang B, Zhang FW, Wang WX, et al. Apolipoprotein E mimetic peptide COG1410 combats pandrug-resistant Acinetobacter baumannii. Front Microbiol. 2022;13:934765. doi:10.3389/fmicb.2022.934765
23. Laskowitz DT, McKenna SE, Song P, et al. COG1410, a novel apolipoprotein E-based peptide, improves functional recovery in a murine model of traumatic brain injury. J Neurotrauma. 2007;24(7):1093–1107. doi:10.1089/neu.2006.0192
24. Cruz J, Flórez J, Torres R, et al. Antimicrobial activity of a new synthetic peptide loaded in polylactic acid or poly(lactic-co-glycolic) acid nanoparticles against Pseudomonas aeruginosa, Escherichia coli O157:H7 and methicillin resistant Staphylococcus aureus (MRSA). Nanotechnology. 2017;28(13):135102. doi:10.1088/1361-6528/aa5f63
25. Nur Ain Mohd A, Nur Izzati R, Fatin Amira A, et al. Immunomodulation of murine macrophages RAW264.7 infected with mycobacterium smegmatis. Asian J Med Biomed. 2020;4(SI1):40–46.
26. Gupta K, Singh S, van Hoek ML. Short, synthetic cationic peptides have antibacterial activity against Mycobacterium smegmatis by forming pores in membrane and synergizing with antibiotics. Antibiotics. 2015;4(3):358–378. doi:10.3390/antibiotics4030358
27. Esteban J, García-Coca M. Mycobacterium Biofilms. Front Microbiol. 2018;8. doi:10.3389/fmicb.2017.02651
28. Bhat KH, Yaseen I. Mycobacterium tuberculosis: macrophage takeover and modulation of innate effector responses. In: Mycobacterium-Research and Development. IntechOpen; 2018.
29. Zhu C, Liu Y, Hu L, Yang M, He Z-G. Molecular mechanism of the synergistic activity of ethambutol and isoniazid against Mycobacterium tuberculosis. J Biol Chem. 2018;293(43):16741–16750. doi:10.1074/jbc.RA118.002693
30. Anuchin AM, Mulyukin AL, Suzina NE, Duda VI, El-Registan GI, Kaprelyants AS. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiology. 2009;155(Pt 4):1071–1079. doi:10.1099/mic.0.023028-0
31. Yagi A, Uchida R, Hamamoto H, Sekimizu K, Kimura K-I, Tomoda H. Anti-Mycobacterium activity of microbial peptides in a silkworm infection model with Mycobacterium smegmatis. J Antibiot. 2017;70(5):685–690. doi:10.1038/ja.2017.23
32. Ramón-García S, Mikut R, Ng C, et al. Targeting Mycobacterium tuberculosis and other microbial pathogens using improved synthetic antibacterial peptides. Antimicrob Agents Chemother. 2013;57(5):2295–2303. doi:10.1128/AAC.00175-13
33. Yamada H, Chikamatsu K, Aono A, et al. Smegmatis be used as a real alternative for M. tuberculosis? Eur Respir J. 2016;48(suppl 60):PA2789.
34. Kartmann B, Stengler S, Niederweis M. Porins in the cell wall of Mycobacterium tuberculosis. J Bacteriol. 1999;181(20):6543–6546. doi:10.1128/JB.181.20.6543-6546.1999
35. Jas T, J R, Rajan A, Shankar V. J R, Rajan A, Shankar V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J Infect Public Health. 2020;13(9):1255–1264. doi:10.1016/j.jiph.2020.06.023
36. Duong L, Gross SP, Siryaporn A. Developing Antimicrobial Synergy With AMPs. Front Med Technol. 2021;3:640981. doi:10.3389/fmedt.2021.640981
37. Wu CL, Hsueh JY, Yip BS, Chih YH, Peng KL, Cheng JW. Antimicrobial peptides display strong synergy with vancomycin against vancomycin-resistant E. faecium, S. aureus, and wild-type E. coli. Int J Mol Sci. 2020;21(13):4578.
38. Abedinzadeh M, Gaeini M, Sardari S. Natural antimicrobial peptides against Mycobacterium tuberculosis. J Antimicrob Chemother. 2015;70(5):1285–1289. doi:10.1093/jac/dku570
39. Aguilar-Pérez C, Gracia B, Rodrigues L, et al. Synergy between circular bacteriocin AS-48 and Ethambutol against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2018;62(9):e00359–00318. doi:10.1128/AAC.00359-18
40. Sharma A, Gaur A, Kumar V, et al. Antimicrobial activity of synthetic antimicrobial peptides loaded in poly-Ɛ-caprolactone nanoparticles against mycobacteria and their functional synergy with rifampicin. Int J Pharm. 2021;608:121097. doi:10.1016/j.ijpharm.2021.121097
41. Rao KU, Henderson DI, Krishnan N, et al. A broad spectrum anti-bacterial peptide with an adjunct potential for tuberculosis chemotherapy. Sci Rep. 2021;11(1):4201. doi:10.1038/s41598-021-83755-3
42. Usmani SS, Kumar R, Kumar V, Singh S, Raghava GPS. AntiTbPdb: a knowledgebase of anti-tubercular peptides. Database. 2018;2018. doi:10.1093/database/bay025
43. Oliveira GS, Costa RP, Gomes P, Gomes MS, Silva T, Teixeira C. Antimicrobial peptides as potential anti-tubercular leads: a concise review. Pharmaceuticals. 2021;14(4):323. doi:10.3390/ph14040323
44. Carroll J, Field D, O’Connor PM, et al. Gene encoded antimicrobial peptides, a template for the design of novel anti-mycobacterial drugs. Bioeng Bugs. 2010;1(6):408–412. doi:10.4161/bbug.1.6.13642
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.