Back to Journals » Infection and Drug Resistance » Volume 16
Antimicrobial Resistance Patterns, Sequence Types, Virulence and Carbapenemase Genes of Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates from a Tertiary Care Teaching Hospital in Zunyi, China
Authors Shen M, Chen X, He J, Xiong L, Tian R, Yang G, Zha H, Wu K
Received 23 November 2022
Accepted for publication 10 January 2023
Published 29 January 2023 Volume 2023:16 Pages 637—649
DOI https://doi.org/10.2147/IDR.S398304
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Meijing Shen,* Xianghao Chen,* Jingyue He, Lin Xiong, Rengui Tian, Guangwu Yang, He Zha, Kaifeng Wu
Department of Laboratory Medicine, the First People’s Hospital of Zunyi (The Third Affiliated Hospital of Zunyi Medical University), Zunyi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Kaifeng Wu; He Zha, Department of Laboratory Medicine, the First People’s Hospital of Zunyi (The Third Affiliated Hospital of Zunyi Medical University), Zunyi, People’s Republic of China, Email [email protected]; [email protected]
Purpose: Carbapenem-resistant Klebsiella pneumoniae (CRKP) has seriously threatened public health worldwide. This study aimed to investigate the antimicrobial resistance patterns, sequence types (STs), virulence and carbapenemase genes of CRKP isolates from patients in Zunyi, China.
Methods: CRKP isolates were collected from the First People’s Hospital of Zunyi between January 2018 and December 2020. Antimicrobial susceptibility was determined using a VITEK® 2 analyzer and confirmed using either the broth dilution method, Kirby–Bauer method, or E-test assays. Carbapenemase production was examined using a modified carbapenem inactivation method. STs of the studied isolates were determined by multilocus sequence typing, and the presence of carbapenemase and virulence genes was examined using polymerase chain reaction assays.
Results: In total, 94 CRKP isolates were collected. All studied isolates produced carbapenemase, and the most common carbapenemase gene was New Delhi metallo-β-lactamase (NDM; 72.3%), followed by Klebsiella pneumoniae carbapenemase (KPC; 24.5%), and Verona integron-encoded metallo-β-lactamase (VIM; 3.2%). Of the studied isolates, 74.3% exhibited multidrug-resistant (MDR) phenotype, and 25.7% were either pandrug-resistant (PDR) or extensively drug-resistant (XDR) phenotypes. The most prevalent sequence type was ST2407 (37.2%), followed by ST76 (21.3%) and ST11 (11.7%). The NDM gene was present in 97.1% of ST2407 isolates and 90.0% of ST76 isolates, whereas the KPC gene was present in 90.9% of ST11 isolates. The majority of the isolates carried wabG, uge, and fimH virulence genes, with prevalence rates of 94.7%, 92.6%, and 94.7%, respectively.
Conclusion: This study describes NDM-producing ST2407 and ST76, as well as KPC-producing ST11, as the major clonal types of CRKP isolates in Zunyi, China. All CRKP isolates were resistant to multiple types of antibiotics, and the majority of isolates carried carbapenemase and virulence genes. Clonal spread of NDM-producing CRKP ST2407 and ST76, and KPC-producing CRKP ST11 should be strictly monitored.
Keywords: Klebsiella pneumoniae, antimicrobial resistance patterns, virulence genes, sequence types, carbapenemase genes
Introduction
Klebsiella pneumoniae (Kp; K. pneumoniae) is an important hospital and community-acquired pathogen that primarily causes pneumonia, urinary tract infections, bloodstream-associated infections, meningitis, and pyogenic liver abscess.1–3 Carbapenem antibiotics represent the most important treatment for Enterobacteriaceae; however, with the widespread use of such antibiotics, carbapenem-resistant K. pneumoniae (CRKP) is increasingly prevalent worldwide.4,5
The effect of carbapenems can be abrogated by mechanisms including hydrolysis by carbapenemases, loss of outer membrane proteins and over-expression of efflux pumps.6 In K. pneumoniae, resistance to carbapenems is mainly mediated by carbapenemases, which can hydrolyze carbapenem antibiotics. The most important carbapenemases are K. pneumoniae carbapenemase (KPC), metallo-β-lactamases, such as the New Delhi metallo-β-lactamase (NDM), Imipenemase metallo-β-lactamase (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and Oxacillin hydrolyzing enzymes-48 (OXA-48).7–9 Since the carbapenemase phenotype is closely related to the use of antimicrobial agents against Kp infection, it is important to continue detecting the genes that are responsible for carbapenem resistance in CRKP isolates.
Multidrug-resistant (MDR) bacterial pathogens have increased worldwide and are considered a public health threat.10 Several recent investigations have reported the emergence of MDR bacterial pathogens from different origins that increase the necessity of the proper use of antibiotics.11–15 Therefore, the routine application of antimicrobial susceptibility testing to detect the antibiotic of choice, as well as the screening of emerging MDR strains, is warranted.
The presence of virulence genes in K. pneumoniae is commonly associated with poor outcomes, including prolonged disease course and high mortality, in infected patients. It has been reported that virulence genes are associated with the pathogenicity of Klebsiella pneumoniae.16 Indeed, virulence determinants encoded by magA, rmpA, kfu, wabG, uge, fimh, etc. have been found to play important roles in the pathogenesis of Kp infection.17 For example, regulator of mucoid phenotype A (rmpA) is capable of activating the production of capsular polysaccharide.18 Mucoviscosity-associated gene A (magA) was recognized as a capsule polymerase, which is associated with the occurrence of liver abscess during Kp infection.19 Type 1 fimbrial adhesin (fimH) is an important mediator in attachment of Kp to the epithelial cells and hence facilitates colonization and invasion of the host.20 wabG and uge genes which encode products contribute to the production of both capsular polysaccharide and lipopolysaccharide.21 Klebsiella ferric iron uptake (kfu) is implicated in the uptake of iron from the host, which is associated with virulent hypermucoviscosity phenotype.22 Shi et al described the high prevalence of rmpA, kfu, fimH, wabG, iroN, iutA, and entB virulence genes in Kp isolates.23 Although Yang et al also reported the high prevalence of uge and fimH virulence genes,24 the prevalence of magA and rmpA virulence genes was lower than that reported by Shi et al.23 Thus, the frequency of virulence genes in CRKP isolates remains to be determined.
Previous studies have investigated the epidemiology and antimicrobial resistance patterns of CRKP isolates in some cities in China;6,25,26 however, no detailed reports of antimicrobial resistance patterns and STs have been described in Zunyi, Guizhou Province, China. The present study aimed to investigate the antimicrobial resistance patterns, STs, and presence of virulence genes (magA, rmpA, wabG, uge, fimH, and kfu) and carbapenemase genes (KPC, NDM, OXA-48, IMP, and VIM) in CRKP isolates from patients at the First People’s Hospital of Zunyi (the Third Affiliated Hospital of Zunyi Medical University) in Zunyi, Guizhou Province, China, between January 2018 and December 2020.
Materials and Methods
Isolation and Identification of CRKP
This study was conducted at the First People’s Hospital of Zunyi (Third Affiliated Hospital of Zunyi Medical University), a university-affiliated teaching hospital in Zunyi, Guizhou Province, China. K. pneumoniae strains were isolated using standard microbial assays and identified using Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF-MS) (Bio-Mérieux, Marcyl’ètoile, France). Between January 2018 and December 2020, 3387 K. pneumoniae isolates were identified and collected.
According to the Clinical and Laboratory Standards Institute (CLSI) M100-S31 criteria,27 CRKP was defined when K. pneumoniae displayed a minimal inhibition concentration (MIC) for imipenem ≥4 μg/mL, meropenem ≥4 μg/mL, or ertapenem ≥2 μg/mL. Based on these inclusion criteria, a total of 94 non-repeated clinical CRKP isolates were enrolled.
String Test
As previously described,28 all isolates were cultured at 37°C on blood agar plates (Autobio, Zhengzhou, China). The colony was gently touched and lifted using an inoculation loop. A positive test result was defined as a viscous string > 5 mm in length, which was observed visually.
DNA Preparation of CRKP Isolates
Pure bacterial colonies were added to 800 μL distilled water in a 1.5 mL EP tube, mixed vigorously, boiled for 30 min to release genomic DNA and plasmids, and centrifuged at 12,000 rpm for 10 min. Afterwards, the supernatant was collected and used as template DNA.
Antimicrobial Susceptibility Testing
Bacterial susceptibility to cephalosporins (cefazolin, cefuroxime, ceftriaxone, ceftazidime, and cefepime), monoamides (aztreonam), cephalomycins (cefotetan), carbapenems (imipenem, ertapenem, and meropenem), β-lactam combination agents (piperacillin/tazobactam, ampicillin/sulbactam, cefoperazone/sulbactam, and ceftazidime/avibactam), quinolones (ciprofloxacin and levofloxacin), aminoglycosides (gentamicin, tobramycin, and amikacin), glycylcyclines (tigecycline), and sulfonamides (trimethoprim/sulfamethoxazole) was analyzed in this study. The broth dilution method was used to determine the minimum inhibitory concentration (MIC) of tigecycline (Yangtze River Pharmaceutical-Group Co., Ltd, Taizhou, China), and the MICs were interpreted using FDA (http://www.fda.gov) MIC breakpoint standards. The Kirby–Bauer method was used to determine the susceptibility to cefuroxime (Oxoid, Hampshire, UK), cefoperazone/sulbactam (Oxoid, Hampshire, UK), ceftazidime/avibactam (Pfizer Co., Ltd, NewYork, USA).27 E-test assays were used to determine the MIC of imipenem (BIO-KONT, Wenzhou, China), meropenem (BIO-KONT, Wenzhou, China) and ertapenem (BIO-KONT, Wenzhou, China). The MIC values for other antibiotics were determined on a VITEK®2 Compact system (bioMérieux, Marcy l’ètoile, France) using the supporting antimicrobial susceptibility GN13 (bioMérieux, Marcy l’ètoile, France). Escherichia coli ATCC® 25922 was used for quality control in the antimicrobial susceptibility test. The antimicrobial susceptibility results were interpreted according to CLSI M100-S31.27
As previously mentioned,29 CRKP isolates were classified as multidrug-resistant (MDR: non-susceptibility to more than one agent in three or more classes of antibiotics), extensively drug-resistant (XDR: non-susceptibility to one or more agents in all but one or two classes), or pandrug-resistant (PDR: non-susceptibility to representative agents of all classes).
Detection of Carbapenemase Genes and Phenotypes
Polymerase chain reaction (PCR) testing was carried out to examine the presence of carbapenemase genes using a previously described method.30 The carbapenemase genes KPC, NDM, OXA-48, IMP, and VIM were amplified with a T100 PCR Amplifier (Bio-Rad, Hercules, CA, USA), and the products were analyzed by gel electrophoresis and ultraviolet imaging. Primers were synthesized by Shenzhen Huada Gene Technology Company (Shenzhen, China) and primer sequences are displayed in Supplementary Table 1. The PCR amplification parameters were pre-denaturation at 95 °C for 3 min, 95 °C for 15s, 55 °C for 30s, 72 °C for 1 min, 30 cycles, and extension at 72° C for 5 min.
According to the (CLSI) M100-S3127 and the protocol described by Tsai et al,31 the modified carbapenem inactivation method (mCIM) and EDTA-carbapenem inactivation method (eCIM) were performed to determine carbapenemase phenotypes.
Detection of Virulence Genes
The virulence genes magA, rmpA, wabG, uge, fimH, and kfu were analyzed according to available information.32 Briefly, these virulence genes were amplified by PCR on a T100 PCR Amplifier (Bio-Rad, Hercules, CA, USA), and the products were analyzed by gel electrophoresis and ultraviolet imaging. Primers were synthesized by the Shenzhen Huada Gene Technology Company (Shenzhen, China). The sequences of the primers are listed in Supplementary Table 2. The PCR amplification was performed under the conditions of pre-denaturation at 95 °C for 3 min, 95 °C for 15s, 55 °C for 30s, 72 °C for 1 min, 35 cycles, and extension at 72 °C for 5 min.
Multilocus Sequence Typing (MLST)
MLST analysis was performed as previously described.33 Briefly, according to the typing standards of the Institute Pasteur MLST and Whole Genome MLST database (http://bigsdb.pasteur.fr/klebsiella/), seven housekeeping genes of K. pneumoniae (gapA, infB, mdh, pgi, phoE, tonB, rpoB) were amplified by PCR. The primers sequences are listed in Supplementary Table 3. The PCR amplification parameters were pre-denaturation at 94 °C for 2 min, 35 cycles of 94 °C for 20s, 55 °C for 30s, 72 °C for 30s, and extension at 72 °C for 5 min. The amplified products were sent to Shanghai Bioengineering Co., Ltd for Sanger sequencing. The sequences of these housekeeping genes were uploaded to the MLST database for allele codes and STs of the isolates.
Data Processing
The WHONET software (version 5.6; WHO, Geneva, Switzerland) was used to calculate the antimicrobial susceptibility rates of the CRKP isolates. Statistical analysis software (SPSS software version 26.0; IBM, Armonk, NK, USA) was used to analyze the significance of the differences between groups. The χ2 test was used, and a p value less than 0.05 (p <0.05) was considered as statistically significant.
Results
Phenotypic Characteristics of the Recovered CRKP Isolates
The proportion of CRKP out of the total Kp clinical isolates was 1.4% (17/1214) in 2018, 2.3% (22/948) in 2019, and 4.5% (55/1225) in 2020, showing an increasing yearly trend. Based on their phenotypic characteristics, the CRKP isolates were divided into four typical phenotypes (Supplementary Figure 1). After an overnight growth on the blood plate, 74 (78.7%) isolates formed white, moist, and convex colonies; 10 (10.6%) isolates formed grey, moist, convex, mucous colonies, and colony fusion could be easily seen; 8 (8.5%) isolates formed grey, convex, and smooth colonies; and 2 (2.1%) isolates formed grey, moist, convex, and mucous colonies with a positive string test.
General Information on the CRKP Clinical Isolates
Table 1 displays that 75.5% (71/94) of the isolates were collected from children (≤14 years old), and 24.5% were collected from adults. Sputum (68.1%, 64/94), urine (13.8%, 13/94), and blood (5.3%, 5/94) were the main specimen types. CRKP isolates were mainly collected from pediatric and intensive care units (ICU).
 |
Table 1 Basic Information on CRKP Isolates in Zunyi, China |
In vitro Antimicrobial Susceptibility
To have an understanding on the antimicrobial resistance patterns, the antibiotic susceptibilities of all the strains were determined (Table 2). All isolates were resistant to cefazolin, cefuroxime, ceftriaxone, ceftazidime, imipenem, meropenem, ertapenem, ampicillin/sulbactam, piperacillin/tazobactam, and cefoperazone/sulbactam. Most CRKP isolates were resistant to cefepime (97.9%), aztreonam (71.3%), and cefotetan (75.5%). In comparison, CRKP isolates were less resistant to ciprofloxacin (23.4%), levofloxacin (17.0%), gentamicin (22.3%), tobramycin (16.0%), amikacin (14.9%), and trimethoprim/sulfamethoxazole (24.5%). The results showed that 21.3% of the isolates were resistant to tigecycline, and 76.6% were resistant to ceftazidime/avibactam.
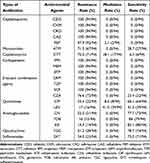 |
Table 2 Antimicrobial Susceptibility of 94 CRKP Isolates |
The occurrence of MDR, XDR, and PDR in the recovered isolates was further analyzed. As shown in Table 3, 21.4% of the isolates were the XDR phenotype, and 4.3% of the isolates were the PDR phenotype. The antibiotic resistance genes carried by MDR and XDR isolates were statistically significant (Supplementary Table 4).
 |
Table 3 The Occurrence of MDR, XDR and PDR Among the CRKP Isolates |
Carbapenemase Phenotypes of the CRKP Isolates
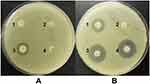
To determine the carbapenemase phenotypes of the studied isolates, the modified carbapenem inactivation and EDTA-carbapenem inactivation assays were performed. The mCIM results demonstrated that all CRKP isolates produced carbapenemases. A representative image is shown in Figure 1A. The eCIM results revealed that 71 isolates (75.5%) produced metallo-β-lactamase, whereas 23 isolates (24.5%) produced serine carbapenemase. A representative image is shown in Figure 1B.
Correlation Between Carbapenemase Phenotypes and Carbapenemase Genes
To determine the relationship between the carbapenemase genes and carbapenemase phenotypes, we examined the presence of carbapenemase genes in all the CRKP isolates. The results showed that NDM was detected in 68 isolates (72.3%, 68/94), with a coincidence rate of 98.5% between genotypes and phenotypes. KPC was detected in 23 isolates (24.5%, 23/94), and the coincidence rate between genotypes and phenotypes was 95.7%. VIM was detected in three isolates (3.2%), which completely coincided with the phenotype of the isolates (Figure 2). OXA-48 and IMP were not detected in any of these isolates.
Prevalence of Virulence Genes
To have an understanding on the virulence determinants of the isolates, wabG, fimH, uge, kfu, rmpA and magA virulence genes were detected using PCR assays. Among the 94 CRKP isolates, 89 (94.7%), 89 (94.7%), 87 (92.6%), and 13 (13.8%) isolates carried the wabG, fimH, uge, and kfu virulence genes, respectively. rmpA and magA were not detected in the isolates. The results are shown in Figure 3.
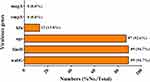 |
Figure 3 Prevalence of virulence genes in the CRKP isolates. |
Prevalence of Carbapenemase Genes in Children and Adults
To determine the difference in the prevalence of carbapenemase genes between children and adults, the ratio of carbapenemase genes between the two populations was compared. Among the 68 NDM-positive isolates, 63 isolates (92.6%, 63/68) were from children, and 5 strains (7.4%, 5/68) were from adults. The KPC genes were present in 5 child isolates (21.7%, 5/23) and 18 adult isolates (78.3%, 18/23). The VIM gene was detected only in children. The prevalence of NDM-producing CRKP in children was significantly higher than that in adults. (P<0.001) (Figure 4).
Sequence Types of the CRKP Isolates
Regarding the STs of the isolates, 94 CRKP isolates were subjected to MLST analysis. The 94 CRKP isolates belonged to 19 STs (Figure 5). Thirty-five isolates (37.2%, 35/94) belonged to ST2407, and most isolates (94.3%, 33/35) were from children. ST76 accounted for 21.3% (20/94) of the isolates (from children), whereas ST11 accounted for 11.7% (11/94) of the isolates (from adults).
 |
Figure 5 Sequence types of the CRKP isolates. Abbreviation: ST, sequence type. |
Carbapenemase Gene Characteristics and Antimicrobial Susceptibilities of the Top Three STs
As demonstrated in Figure 6, the majority of ST2407 clones (34/35, 97.1%) and ST76 clones (18/20, 90.0%) carried the NDM gene, whereas the majority of ST11 clones (10/11, 90.9%) carried the KPC gene. Figure 7 displays the antimicrobial susceptibilities of the top three STs. ST2407, ST76, and ST11 clones were completely resistant to cefazolin, cefuroxime, ceftriaxone, ceftazidime, cefotetan, cefepime, ampicillin/sulbactam, piperacillin/tazobactam, cefoperazone/sulbactam, imipenem, meropenem, and ertapenem. Most ST2407 clones were sensitive to tobramycin, ciprofloxacin, amikacin, gentamicin, and trimethoprim/sulfamethoxazole, whereas only 20.0% of ST2407 clones were sensitive to levofloxacin. The ST76 clones were completely sensitive to tobramycin, ciprofloxacin, amikacin, gentamicin, and levofloxacin, while 85.0% of the isolates were sensitive to trimethoprim/sulfamethoxazole. Only 18.2% of ST11 isolates were sensitive to gentamicin, tobramycin, and amikacin; 9.1% of the isolates were sensitive to ciprofloxacin and levofloxacin; and 36.4% of the isolates were sensitive to trimethoprim/sulfamethoxazole.
 |
Figure 7 Antimicrobial susceptibility of the top three STs. *P <0.05, ***P <0.001. Abbreviation: ST, sequence type. |
Discussion
This study described that all CRKP isolates were completely resistant to cephalosporins, but more than 70% isolates maintained sensitivity to antimicrobial agents, including gentamicin, tigecycline, tobramycin, and amikacin. All CRKP isolates carried carbapenemase genes, most frequently the NDM gene. The studied isolates were found to have 19 different STs and ST2407, ST76, and ST11 were the leading STs, suggesting their diversity in the CRKP Zunyi isolates. This study strengthened our knowledge on the molecular epidemiology and antimicrobial susceptibility of CRKP isolates in China, which will aid in the clinical treatment and control of CRKP infection.
The prevalence of CRKP in Zunyi increased with year from 2018 to 2020, but the proportion was at relatively low level of no more than 5%. According to the 2021 China Antimicrobial Resistance Surveillance (http://www.carss.cn/) report, the proportion of CRKP increased from 3.0% in 2005 to 27.1% in 2021, and a particularly high CRKP proportion (more than 50%) was found in Zhejiang Province and other regions.34 Compared to the situation in other countries and most cities in China,34–36 the management of infections due to CRKP remains under control in this region.
The studied CRKP isolates were highly resistant to some of commonly used antibiotics, especially β-lactam antibiotics with resistance rates to most antibiotics of more than 97.0%. Ceftazidime/avibactam has been mainly used to treat severe infections caused by carbapenem-resistant Enterobacteriaceae, which produce KPC or OXA-48-like carbapenemases and are ineffective against metallo-β-lactamase producers.37,38 As previously reported,39–41 the present study revealed that the majority of the KPC-producing isolates (95.7%) were sensitive to ceftazidime/avibactam, while metallo-β-lactamase isolates were completely resistant to ceftazidime/avibactam.
The CLSI recommends applying the modified carbapenem inactivation method and EDTA-carbapenem inactivation method as a reliable way to detect carbapenem resistance phenotypes in Enterobacteriaceae.27 Consistent with the results of Tsai et al,31 our results depicted that the presence of carbapenemase genes of NDM, VIM, and KPC correlated well with the phenotypes of the isolates, indicating the potential of detecting the carbapenemase resistance phenotype to predict the carbapenemase gene from CRKP isolates.
Resistance to carbapenems in Kp is associated with various mechanisms. In China, it has been reported that the presence of KPC is responsible for carbapenem resistance in adult and pediatric isolates.25,42 The present work demonstrated that the NDM and KPC genes detected in CRKP were mainly isolated in children and adults, respectively. Although OXA-48 is a prevalent carbapenemase type associated with Kp in some European countries, such as France and Turkey,43,44 it is seldom detected in Kp isolates in China,45,46 and the present study further support the observation that OXA-48 is not currently the primary carbapenemase types associated with Kp in China.
In the United States and Europe, ST258 is the dominant sequence that hosts KPC enzymes.43 In most cities of China, it was the ST11, a clone closely related to ST258, which was responsible for most K. pneumoniae infections.47–50 In this study, neither ST258 nor ST11 was the most prevalent clone in Zunyi. This study revealed that ST2407, isolated mainly from children, was the most prevalent CRKP isolate in this region. For clones circulating in children, a study in Shenzhen found that ST20 was the dominant clone, which carries the KPC enzymes.51 In Jiangsu Province, and Wuhan, China, the main antimicrobial resistance gene in pediatric patients carried by CRKP was KPC2, and the prevalent strains were ST11 with high virulence and antimicrobial resistance.6,25 Another work in Shenzhen demonstrated that ST307 was the most dominant genotype, and NDM represented the main resistance mechanism.52 On the other hand, most of the isolates belonged to ST2407, which produces NDM enzymes, as revealed in this work. We highlight the diversity of CRKP STs circulating worldwide and the importance of monitoring ST2407 in K. pneumoniae infection.
An important finding of the present work is the high presence of wabG, uge, and fimH, but the extremely low presence of magA and rmpA among the strains under study. In this study, it was found that more than 90% CRKP Zunyi isolates carried the uge, fimH, and wabG virulence genes. In agreement with the results reported by Yang et al,24 the prevalence of magA and rmpA genes was not as high as that reported by Shi et al who described a relatively high prevalence of magA and rmpA virulence genes in both hypervirulent and moderate-virulent Kp isolates.23 Association between the virulence level of Kp and the presence of these virulence-associated genes has been investigated; however, except for magA and allS, the presence of other genes showed no connection with bacterial virulence.23 Although these virulence genes play important roles in the pathogenesis of Kp infection, the absence of some virulence genes, such as magA and rmpA, may suggest the existence of redundant molecules implicated in the pathogenesis of Kp infection. In addition, the discrepancy in the presence of virulence genes suggests that it may not be a good choice to evaluate the virulence of the Kp isolates merely by examining the presence of virulence-associated genes.
In agreement with other studies,53 we found that ST11 demonstrated a higher rate of resistance to levofloxacin, tobramycin, ciprofloxacin, amikacin, gentamicin, and trimethoprim/sulfamethoxazole. Other studies have also highlighted a higher risk of ST11 than non-ST11 in patients in the ICU.54,55 Thus, although ST11 was not the most prevalent sequence in Zunyi, the emergence of ST11 warrants careful monitoring. Su et al revealed that ST76 carrying the KPC gene was the main CRKP sequence type in the intensive care and neurosurgery units in the hospital.56 Similar results were reported in the eastern region of Heilongjiang Province, China. Contrary to their results, the present study showed that ST76 circulating in Zunyi mainly hosts NDM enzymes. This suggests the existence of different antimicrobial resistance mechanisms for certain STs.
This study had some limitations. First, the number of isolates collected was relatively small; hence, further isolates are needed to gain a comprehensive insight into the prevalence of CRKP isolates. Second, the subtypes of carbapenemase genes in NDM, VIM, and KPC were not determined.
Conclusion
The present work depicts the prevalence of NDM-producing CRKP ST2407 and ST76, as well as KPC-producing ST11, as the major clonal types in Zunyi, Guizhou Province, China. All CRKP isolates were resistant to multiple types of antibiotics, and the majority of isolates carried carbapenemase and virulence genes. Clonal spread of NDM-producing CRKP ST2407 and ST76, and KPC-producing CRKP ST11 should be strictly monitored.
Abbreviations
CRKP, carbapenem-resistant Klebsiella pneumoniae; Kp, Klebsiella pneumoniae; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; VIM, Verona integron-encoded metallo-β-lactamase; IMP, Imipenemase metallo-β-lactamase; OXA-48, Oxacillin hydrolyzing enzymes-48; mCIM, modified carbapenem inactivation method; eCIM, EDTA-carbapenem inactivation method; MLST, multilocus sequence typing. MDR, multidrug-resistant; XDR, extensively drug-resistant; PDR, pandrug-resistant; CZO, cefazolin; CXM, cefuroxime; CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; CTT, cefotetan; IPM, imipenem; MEM, meropenem; ETP, ertapenem; SAM, ampicillin/sulbactam; TZP, piperacillin/tazobactam; SCF, cefoperazone/sulbactam; CZA, ceftazidime/avibactam; CIP, ciprofloxacin; LEV, levofloxacin; CN, gentamicin; TOB, tobramycin; AK, amikacin; TGC, tigecycline; SXT, trimethoprim/sulfamethoxazole.
Ethics Statement
This study was approved by the Ethics Committee of the First People’s Hospital of Zunyi (No. 2020-068). The need for written informed consent was waived because the samples were routinely collected and patients’ anonymous information was provided by the microbiology hospital laboratory. This study completely followed the principles outlined in the Declaration of Helsinki.
Funding
This project was supported by the Guizhou High-level (BAI) Innovative Talents Project (QIANKehe Platform & Talents-GCC[2022]042-1); National Natural Science Foundation of China (No.81760475); Innovation Group Project provided by the Guizhou Provincial Department of Education (QianJiaoheKYzi [2021]019); Key Discipline Project of Clinical Laboratory Diagnostics funded by Guizhou Provincial Health Commission (QianWeijianhan[2021]160); Key Discipline Project of Clinical Laboratory Diagnostics funded by Zunyi Municipal Health Bureau (2022-1444); and Zunyi United Science and Technology Fund Project (Zunyi Kehe HZzi [2019]147, [2019]173 & [2022] 97).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–359. doi:10.1038/s41579-019-0315-1
2. Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi:10.1186/s12941-017-0191-3
3. Wang G, Zhao G, Chao X, Xie L, Wang H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int J Environ Res Public Health. 2020;17(17). doi:10.3390/ijerph17176278
4. Zhang R, Chan EW, Zhou H, Chen S. Prevalence and genetic characteristics of carbapenem-resistant Enterobacteriaceae strains in China. Lancet Infect Dis. 2017;17(3):256–257. doi:10.1016/S1473-3099(17)30072-5
5. Zhang D, Hu S, Sun J, et al. Antibiotic consumption versus the prevalence of carbapenem-resistant Gram-negative bacteria at a tertiary hospital in China from 2011 to 2017. J Infect Public Health. 2019;12(2):195–199. doi:10.1016/j.jiph.2018.10.003
6. Tao G, Tan H, Ma J, Chen Q. Resistance Phenotype and Molecular Epidemiology of Carbapenem-Resistant Klebsiella pneumoniae Isolated from Nanjing Children’s Hospital in Jiangsu Province, China. Infect Drug Resist. 2022;15:5435–5447. doi:10.2147/IDR.S377068
7. Vamsi SK, Moorthy RS, Hemiliamma MN, Chandra Reddy RB, Chanderakant DJ, Sirikonda S. Phenotypic and genotypic detection of carbapenemase production among gram negative bacteria isolated from hospital acquired infections. Saudi Med J. 2022;43(3):236–243. doi:10.15537/smj.2022.43.3.20210809
8. Li J, Huang Z, Tang M, et al. Clonal Dissemination of Multiple Carbapenemase Genes in Carbapenem-Resistant Enterobacterales Mediated by Multiple Plasmids in China. Infect Drug Resist. 2021;14:3287–3295. doi:10.2147/IDR.S327273
9. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi:10.1128/CMR.11.4.589
10. Cerceo E, Deitelzweig SB, Sherman BM, Amin AN. Multidrug-Resistant Gram-Negative Bacterial Infections in the Hospital Setting: overview, Implications for Clinical Practice, and Emerging Treatment Options. Microb Drug Resist. 2016;22(5):412–431. doi:10.1089/mdr.2015.0220
11. Algammal AM, Ibrahim RA, Alfifi KJ, et al. A First Report of Molecular Typing, Virulence Traits, and Phenotypic and Genotypic Resistance Patterns of Newly Emerging XDR and MDR Aeromonas veronii in Mugil seheli. Pathogens. 2022;11(11):1262. doi:10.3390/pathogens11111262
12. Algammal AM, Hashem HR, Al-Otaibi AS, et al. Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 2021;21(1):237. doi:10.1186/s12866-021-02287-y
13. Algammal AM, Abo Hashem ME, Alfifi KJ, et al. Sequence Analysis, Antibiogram Profile, Virulence and Antibiotic Resistance Genes of XDR and MDR Gallibacterium anatis Isolated from Layer Chickens in Egypt. Infect Drug Resist. 2022;15:4321–4334. doi:10.2147/IDR.S377797
14. Makharita RR, El-Kholy I, Hetta HF, et al. Antibiogram and Genetic Characterization of Carbapenem-Resistant Gram-Negative Pathogens Incriminated in Healthcare-Associated Infections. Infect Drug Resist. 2020;13:3991–4002. doi:10.2147/IDR.S276975
15. Kareem SM, Al-Kadmy IMS, Kazaal SS, et al. Detection of gyrA and parC Mutations and Prevalence of Plasmid-Mediated Quinolone Resistance Genes in Klebsiella pneumoniae. Infect Drug Resist. 2021;14:555–563. doi:10.2147/IDR.S275852
16. Vargas JM, Moreno Mochi MP, Nunez JM, et al. Virulence factors and clinical patterns of multiple-clone hypermucoviscous KPC-2 producing K. pneumoniae. Heliyon. 2019;5(6):e01829. doi:10.1016/j.heliyon.2019.e01829
17. Aljanaby A. Role of rmpA, wabG, uge, Ycfm, fimh1, EntB, Ybt-irp2 and kfu genes in pathogenicity of Klebsiella pneumoniae: an overview. Int J Chemtech Res. 2017;10:391.
18. Struve C, Roe CC, Stegger M, et al. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio. 2015;6(4):e00630. doi:10.1128/mBio.00630-15
19. Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705. doi:10.1084/jem.20030857
20. Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun. 2009;77(11):5016–5024. doi:10.1128/IAI.00585-09
21. Cortes G, Borrell N, de Astorza B, Gomez C, Sauleda J, Alberti S. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun. 2002;70(5):2583–2590. doi:10.1128/IAI.70.5.2583-2590.2002
22. Bachman MA, Oyler JE, Burns SH, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. 2011;79(8):3309–3316. doi:10.1128/IAI.05114-11
23. Shi Q, Lan P, Huang D, et al. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. 2018;18(1):94. doi:10.1186/s12866-018-1236-2
24. Yang K, Liu S, Li H, et al. NDM-1-Positive K. pneumoniae at a Teaching Hospital in Southwestern China: clinical Characteristics, Antimicrobial Resistance, Molecular Characterization, Biofilm Assay, and Virulence. Can J Infect Dis Med Microbiol. 2020;2020:9091360. doi:10.1155/2020/9091360
25. Jin Z, Wang Z, Gong L, et al. Molecular epidemiological characteristics of carbapenem-resistant Klebsiella pneumoniae among children in China. AMB Express. 2022;12(1):89. doi:10.1186/s13568-022-01437-3
26. Le T, Wang L, Zeng C, Fu L, Liu Z, Hu J. Clinical and microbiological characteristics of nosocomial, healthcare-associated, and community-acquired Klebsiella pneumoniae infections in Guangzhou, China. Antimicrob Resist Infect Control. 2021;10(1):41. doi:10.1186/s13756-021-00910-1
27. CLSI. Performance Standards for Antimicrobial Susceptibility Testing: 31th Informational Supplement. Clinical and Laboratory Standards Institute (CLSI); 2021.
28. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–112. doi:10.1016/j.ijid.2015.06.023
29. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
30. Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi:10.18388/abp.2015_1148
31. Tsai YM, Wang S, Chiu HC, Kao CY, Wen LL. Combination of modified carbapenem inactivation method (mCIM) and EDTA-CIM (eCIM) for phenotypic detection of carbapenemase-producing Enterobacteriaceae. BMC Microbiol. 2020;20(1):315. doi:10.1186/s12866-020-02010-3
32. Remya PA, Shanthi M, Sekar U. Characterisation of virulence genes associated with pathogenicity in Klebsiella pneumoniae. Indian J Med Microbiol. 2019;37(2):210–218. doi:10.4103/ijmm.IJMM_19_157
33. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
34. Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008-2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi:10.1080/22221751.2020.1799721
35. Lan P, Jiang Y, Zhou J, Yu Y. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. doi:10.1016/j.jgar.2021.02.020
36. Haji SH, Aka STH, Ali FA. Prevalence and characterisation of carbapenemase encoding genes in multidrug-resistant Gram-negative bacilli. PLoS One. 2021;16(11):e0259005. doi:10.1371/journal.pone.0259005
37. Nordmann P, Poirel L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin Infect Dis. 2019;69(Suppl 7):S521–S528. doi:10.1093/cid/ciz824
38. van Duin D, Bonomo RA. Ceftazidime/Avibactam and Ceftolozane/Tazobactam: second-generation beta-Lactam/beta-Lactamase Inhibitor Combinations. Clin Infect Dis. 2016;63(2):234–241. doi:10.1093/cid/ciw243
39. Shirley M. Ceftazidime-Avibactam: a Review in the Treatment of Serious Gram-Negative Bacterial Infections. Drugs. 2018;78(6):675–692. doi:10.1007/s40265-018-0902-x
40. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):e121–e124. doi:10.1016/j.cmi.2019.08.020
41. Soriano A, Carmeli Y, Omrani AS, Moore LSP, Tawadrous M, Irani P. Ceftazidime-Avibactam for the Treatment of Serious Gram-Negative Infections with Limited Treatment Options: a Systematic Literature Review. Infect Dis Ther. 2021;10(4):1989–2034. doi:10.1007/s40121-021-00507-6
42. Chen D, Li H, Zhao Y, et al. Characterization of carbapenem-resistant Klebsiella pneumoniae in a tertiary hospital in Fuzhou, China. J Appl Microbiol. 2020;129(5):1220–1226. doi:10.1111/jam.14700
43. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi:10.1016/S1473-3099(13)70190-7
44. Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: the Impact and Evolution of a Global Menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
45. Yu F, Wang S, Lv J, et al. Coexistence of OXA-48-Producing Klebsiella pneumoniae and Escherichia coli in a Hospitalized Patient Who Returned from Europe to China. Antimicrob Agents Chemother. 2017;61(4). doi:10.1128/AAC.02580-16
46. Han R, Shi Q, Wu S, et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
47. Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi:10.1016/j.jgar.2020.09.004
48. Tan D, Zhang Y, Cheng M, et al. Characterization of Klebsiella pneumoniae ST11 Isolates and Their Interactions with Lytic Phages. Viruses. 2019;11(11):1080. doi:10.3390/v11111080
49. Yu X, Zhang W, Zhao Z, et al. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae isolates with focus on antimicrobial resistance. BMC Genomics. 2019;20(1):822. doi:10.1186/s12864-019-6225-9
50. Zeng L, Deng Q, Zeng T, Liu Y, Zhang J, Cao X. Prevalence of Carbapenem-Resistant Klebsiella pneumoniae Infection in Southern China: clinical Characteristics, Antimicrobial Resistance, Virulence, and Geographic Distribution. Microb Drug Resist. 2020;26(5):483–491. doi:10.1089/mdr.2018.0401
51. Patil S, Chen X, Wen F. Exploring the phenotype and genotype of multi-drug resistant Klebsiella pneumoniae harbouring blaCTX-M group extended-spectrum beta-lactamases recovered from paediatric clinical cases in Shenzhen, China. Ann Clin Microbiol Antimicrob. 2019;18(1):32. doi:10.1186/s12941-019-0331-z
52. Patil S, Chen H, Guo C, et al. Emergence of Klebsiella pneumoniae ST307 Co-Producing CTX-M with SHV and KPC from Paediatric Patients at Shenzhen Children’s Hospital, China. Infect Drug Resist. 2021;14:3581–3588. doi:10.2147/IDR.S324018
53. Wang Q, Wang X, Wang J, et al. Phenotypic and Genotypic Characterization of Carbapenem-resistant Enterobacteriaceae: data From a Longitudinal Large-scale CRE Study in China (2012-2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
54. Zhang Y, Jin L, Ouyang P, et al. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother. 2020;75(2):327–336. doi:10.1093/jac/dkz446
55. Liu Y, Zhang X, Cai L, Zong Z. Enhanced survival of ST-11 carbapenem-resistant Klebsiella pneumoniae in the intensive care unit. Infect Control Hosp Epidemiol. 2020;41(6):740–742. doi:10.1017/ice.2020.68
56. Su S, Li C, Zhao Y, et al. Outbreak of KPC-2-Producing Klebsiella pneumoniae ST76 Isolates in an Intensive Care Unit and Neurosurgery Unit. Microb Drug Resist. 2020;26(9):1009–1018. doi:10.1089/mdr.2019.0363
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.