Back to Journals » Veterinary Medicine: Research and Reports » Volume 12
Antigenic and Molecular Characterization of Virulent Newcastle Disease Viruses Circulating in Ethiopia Between 1976 and 2008
Authors Bari FD, Gelaye E , Tekola BG , Harder T, Beer M, Grund C
Received 17 December 2020
Accepted for publication 13 May 2021
Published 4 June 2021 Volume 2021:12 Pages 129—140
DOI https://doi.org/10.2147/VMRR.S297281
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Young Lyoo
Fufa D Bari,1,2 Esayas Gelaye,3 Berhe Gebreegziabher Tekola,3 Timm Harder,1 Martin Beer,1 Christian Grund1
1Institute of Infectology, Friedrich-Loeffler-Institut, Greifswald, Germany; 2Department of Microbiology, Immunology and Veterinary Public Health, College of Veterinary Medicine and Agriculture, Addis Ababa University, Bishoftu, Ethiopia; 3National Veterinary Institute, Bishoftu, Ethiopia
Correspondence: Fufa D Bari
Department of Microbiology, Immunology and Veterinary Public Health, College of Veterinary Medicine and Agriculture, Addis Ababa University, P O Box 34, Bishoftu, Ethiopia
Tel +251929190312
Fax +251114339933
Email [email protected]
Introduction: Newcastle disease virus (NDV) cultures held in the isolate collections in Ethiopia between 1976 and 2008 were not characterized using biological and molecular techniques. The already characterized NDV isolates belonged to genotype VI but the genetic nature of previously collected isolates, which could shade light on the history of introduction into the country and their evolutionary relationships, were not established.
Methods: A total of 14 NDVs (11 obtained from outbreak cases in chickens and three commercial vaccinal strains used in the country) were inoculated into specific pathogen free (SPF) embryonated chicken eggs (ECE). Allantoic fluids harvested from grown SPF ECE were tested by heamagglutination (HA) and heamagglutination inhibition (HI) tests. Partial F gene sequences were generated for all samples and molecular evolutionary relationships were reconstructed together with reference sequences freely available online. The pathogenicities of the isolates were assessed in vivo by determining their intracerebral pathogenicity index (ICPI) in day-old chicks and molecularly by determination of F gene cleavage sites.
Results: Of these, 12 viruses (two vaccines and 10 outbreaks) were successfully propagated as evidenced by a positive heamagglutination (HA) test. These 12 propagated viruses were further characterized by heamagglutination inhibition (HI) test, of which only three viruses reacted with monoclonal antibody (MAb 617/616) specific for pigeon paramyxovirus-1. In addition, all 14 viruses were characterized by partial fusion (F) gene sequencing and phylogenetic tree reconstruction. The Ethiopian NDV isolates clustered with genotype VI viruses, forming two clades (groups 1 and 2) that have ancestral relationships with Egypt-1990 and Sudan-1975 like viruses.
Discussion: The characterized genotype VI NDVs were genetically similar to currently circulating NDVs in Ethiopia. The isolates had cleavage sites consistent with mesogenic/velogenic NDV with a mean ICPI value of 1.76, indicating that the isolates were velogenic. Two and four highly virulent viruses were thermostable at 56°C for 2 hours and 1 hour, respectively. To reduce chicken mortality and production losses, proper control of the disease should be instituted using high quality and protective vaccines together with strong biosecurity measures.
Keywords: antigenic, NDV, fusion gene sequence, molecular characterization, pathogenicity, velogenic
Introduction
Newcastle disease (ND) is one of the most significant contagious poultry diseases globally. Epizootics of the disease result in significant economic losses associated with high mortality and high morbidity (each approaching 100% in severe cases). It causes more significant problems in developing countries where proper ND control measures are lacking. In Ethiopia, the disease has been endemic in domestic chickens since 1972 and it is ranked as the highest priority chicken disease.1 The causative agent of the disease is Newcastle disease virus (NDV or avian paramyxovirus 1 (APMV-1)), whcih belongs to species Avian orthoavulavirus 1, genus Orthoavulavirus, subfamily Avulavirinae, family Paramyxoviridae, and order Mononegavirales.2 Genetically, NDV encodes a non-segmented, single-stranded, negative-sense RNA genome containing six genes encoding for the nucleocapsid protein (NP), phosphoprotein (P), matrix protein (M), fusion protein (Fp), hemagglutinin-neuraminidase (HN) and large polymerase protein (L).3,4 NDV genes are arranged as 3ʹ NP-P-M-Fp-HN-L 5ʹ.
NDV isolates display a wide spectrum of virulence in chickens, ranging from in-apparent infection to 100% mortality. Based on their virulence in chickens, NDVs are classified as lentogenic (avirulent), mesogenic (intermediately virulent) and velogenic (highly virulent) (Office International Des Epizooties, OIE5). Velogenic strains are responsible for severe disease and cause significant mortality in susceptible flocks. These virulent strains have resulted in five major panzootics globally, each caused by different genotypes of NDV.6 NDV virulence and pathogenicity is experimentally assessed using a combination of an in vivo and in vitro testing. The most commonly used in vivo virulence assessment is the intracerberal pathogenicity index (ICPI, value greater or equal to 0.7) assessed in chicks 24–40 hours old while the only OIE accepted in vitro virulence assessment is the presence of multiple basic amino acids at the F gene cleavage site.5
NDV has a wide host range that includes domestic chickens, wild birds like pigeons, and other migratory birds.7 The propensity of wild birds to act as reservoirs of NDV can pose significant challenges to backyard chicken production as a result of frequent contact between the domestic and wild species. Frequent outbreaks of the disease occur annually in different parts of the country due to lack of vaccination coverage in backyard poultry production and/or due to a failure to elicit effective protection in vaccinated farms. Some limited molecular characterization of several recently isolated NDVs were reported,8–10 but there is a gap in knowledge related to the original source of introduction, evolution, and strain heterogeneity of NDV. In order to effectively control ND and ultimately improve the food and economic security of Ethiopia, there is a critical need to determine the NDV strain heterogeneity across historical isolates and to compare them to the commercially available vaccine isolates used for immunization. In this paper we describe the biological and molecular characterizations of 11 NDV isolates collected between 1976 and 2008 in order to determine the strain variability and compare it to contemporary vaccine strains.
Materials and Methods
History of Viruses
Fourteen NDVs (three vaccine strains and 11 outbreak isolates) originally isolated and stored at −70 °C in the National Veterinary Institute, Bishoftu, Ethiopia, were available for both biological and molecular characterization. Isolates were originally cultured between 1976 and 2008, including vaccinal strains, and thus represent strains circulating in the community over a prolonged period of time. The limitation of the study is that only two viruses collected before 1997 were available and included in the study. Details concerning the source of the viruses (farm or field) and year of collection are presented in Table 1. The vaccine strains were also obtained from the National Veterinary Institute. These isolates were collected from three different areas: Bishoftu, Dukem, and Wolaita (Figure 1).
 |
Table 1 Pathogenicity Determinations of NDV Isolates and Vaccinal Strains |
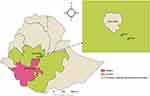 |
Figure 1 NDV outbreak areas in Ethiopia. The three green colours on the map show the locations from which the outbreak samples originated. The map was drawn using QGIS software. |
Propagation of the Viruses
The viruses were propagated in specific pathogen free (SPF) embryonating chicken eggs 9–10 days old by inoculating 0.2 milliliters (mL) of undiluted virus sample per egg into the allantoic sac. All inoculated SPF embryonated chicken eggs were candled twice per day for four consecutive days. All the eggs were kept at 4 °C for at least two hours after detecting death of embryo or at the end of the incubation period before harvesting allantoic fluid (AF). The collected AFs were centrifuged at 3500 revolutions per minute (rpm) for 10 minutes, supernatant harvested, pooled per virus, aliquoted and kept at −70 °C for subsequent use. The same batches of each aliquot were further checked for contamination with Mycoplasma species and other bacteria by culturing in appropriate media and by conducting standard biochemical tests for suspected bacterial organisms.
Hemagglutination (HA) Test
HA tests were performed essentially as described by OIE.5 The HA test positive AF with an HA titre of at least 1:32 and free from bacterial contamination were taken forward for further experiments. HA negative AF were blindly passaged for a second time in the embryonated chicken eggs, and if the second harvested AF was still negative for HA test, it was considered as negative for NDV.
Inactivation of Infectious AF of NDV and Hemagglutination Inhibition Test
The hemagglutination inhibition (HI) tests were performed using 4HA units of ß-propiolactone inactivated NDV, 15 APMV-1 (NDV) serotype specific hyperimmune sera and six APMV-1 specific monoclonal antibodies (mAb), namely, mAb −10, −27, −39, −51, -u85, and −617/616 according to Alexander et al.11 Of the six mAbs tested, mAb-39 and mAb-161/617 are known to be specific for pigeon paramyxovirus-1 (PPMV-1). The antibodies are titrated double-folded in 25 µL of 0.85% NaCl buffer to which 25 µL of 4HA diluted virus was added to all wells except to control wells and incubated at room temperature for 45 minutes. Finally, 25 µL of 1% washed chicken erythrocyte suspension was added to all wells, gently tapped and incubated at RT for 30 minutes. Beta-propiolactone inactivated NDV Ulster was used as a positive control in both HA and HI tests. Furthermore, two types of negative control were used for HI tests: reference sera without virus, and buffer without sera and virus. Reciprocal logarithm (log)2 titers of the highest dilution at which no agglutination was observed was taken as HI titer.
Thermostability Test
The thermostability test was performed as described by Grund et al.12 Briefly, virus samples were aliquoted in 1.5 mL eppindrof tubes and incubated in a water bath at 56 °C to be heated. The incubated samples were then taken out of the water bath at different time intervals of 5, 15, 30, 60, 90, and 120 minutes and immediately transferred onto ice. The viruses were then tested for their HA assay.
Pathogenicity Determination
Pathogenicity of the isolates was determined by intracerebral pathogenicity index testing (in vivo) and determination of F gene cleavage site (in vitro).
Intracerebral Pathogenicity Index (ICPI; in vivo) Determination
The ICPIs of the isolates were determined in animal facilities of biosafety level 2 (BSL-2) at the Friedrich-Loeffler-Institut in accordance with a legally approved protocol (LVL MV/TSD/7221.3–2.2-005/07). Following the standard OIE5 Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 0.05 mL of 1:10 diluted virus in a sterile isotonic saline solution (0.85%, pH 7.26) was inoculated intracerebrally in one day old specific pathogen-free chicks (Lohmann Animal Health, Cuxhaven, Germany). The chicks were clinically monitored for eight consecutive days for development of clinical signs and mortality. One chick was included in each group as a non-infected negative control that was used to demonstrate evidence of contact transmission of the disease.
Cleavage Site Determination
In vitro (molecular) pathogenicity determination was conducted based on the sequencing of the F gene of all 14 viruses and determination of their cleavage sites of the deduced amino acid sequences. A one-step reverse transcription polymerase chain reaction (RT-PCR) was run in 25µL volume according to and using primers of Kim et al.13 The PCR products were separated by gel electrophoresis on 1.5% agarose gel (biotechnology grade or electrophoresis grade). The PCR products were excised and eluted from the agarose gel using QIAquick® Gel Extraction Kit following the manufacturer's recommendations. The amount of PCR product elutes from the agarose gel were then estimated by Nanodrop spectrophotometry (PeQLab Biotechnology, GmbH, Germany) and subsequently used for sequencing.
The amplicon was sequenced in both directions using the amplification primers using the Big DyeTM Terminator Cycle Sequencing Kit and an ABI Prism® DNA Sequencer (3130 Genetic Analyzer, Applied Biosystems). The gene sequencing reaction product was cleaned by Sigma Spin Post-Reaction Clean-Up Columns according to the manufacturer’s instructions (Sigma). The nucleotide sequences generated using Sanger Sequencer (sequence analyser) (Applied Biosystems) were edited by Chromas Lite software (Technelysium Pty Ltd, Australia) and used for molecular characterization.
Data Analysis
Genetic distances were calculated using the maximum composite likelihood model with 1000 bootstrap replicates as implemented in Molecular Evolutionary Genetics Analysis (MEGA) Version 6.14 Amino acid distances of the deduced amino acid sequences of the partial fusion gene sequences were calculated with the Poison correction model implemented in the same program. For phylogenetic analysis, NDV reference sequences were retrieved from Genbank, including previously published sequences of both local and geographically diverse strains.8,10,15 Sequences were aligned using the Clustal W program that was embedded in MEGA Version 6 software.16 MEGA was also used for deducing amino acid sequences from their nucleotide sequence data. A phylogenetic tree was reconstructed by initially selecting the best-fit model that was subsequently used for phylogenetic tree reconstruction in MEGA Version 6. The phylogenetic tree was reconstructed using the Kimura 2-Parameter and Gamma distributed invariant sites (G+I) rates by MEGA Version 6. We used the Minimum Evolution Method with the Kimura 2-Parameter model to estimate the transversion/transition bias of nucleotide substitution patterns with an assumption of homogeneity among lineages using a bootstrap value of 1000 replicates. Genetic diversity among Ethiopian isolates and the reference strains was estimated by calculating genetic distance using the p-distance model found in MEGA Version 6.
Results
Of the 14 NDV isolates (3 vaccine strains and 11 field strains), only 12 (including 10 outbreak isolates and 2 vaccine strains) were successfully propagated in ECE and were used for subsequent experiments. All 14 viruses' F gene sequences were successfully generated from allantoic fluids of SPF ECE or from original samples of the 2 viruses which failed to grow in SPF ECE and were included in molecular characterizations. Only the 12 NDVs grown in SPF ECEs were further characterized using biological techniques (Table 1), as these techniques require live virus.
Antigenic or Biological Characterization
Based on HI testing, all 10 Ethiopian NDV outbreak isolates belonged to Avian orthoavulavirus-1 (PPMV) as they gave the highest log reaction titer of ≥9. Notably, no field isolates belonged to the same HI type as vaccine strains used in the country (LaSota and/or I-2). In order to further refine the identification of the isolates, testing was performed with 6 Avian orthoavulavirus-1 specific monoclonal antibodies (MAbs), including two pigeon-type paramyxovirus-1 (PPMV-1) specific MAbs (MAb 39 and MAb 161/617). None of the isolates reacted with MAb 39 and only 3 isolates reacted (log titer ≥9) with MAb 617/616. Since these latter two MAbs are known to be specific for PPMV-1 of NDV variants, it suggests that 3 of the 10 isolates are likely PPMV-1; however the limited sero reactivity observed in these isolates suggests that they could be variant populations circulating in chickens. The other 7 isolates that did not react to the pigeon-type MAbs were other APMV-1s.
Thermostability Test
Thermostability of the hemagglutinin of the NDV isolates varied grossly between isolates. Interestingly, two isolates (R655/08 and R656/08) were thermostable at 56 °C for 2 hrs, while four isolates were stable for 1 hr. Some of these viruses have ICPI values above average (1.800 > 1.764), indicating that they are highly pathogenic, and are circulating in chickens in both commercial farms and smallholdings. In contrast, one isolate was thermostable for 30 minutes and one was stable for only 5 minutes (Table 1), similar to the vaccine strains tested.
In Vivo and Molecular (in Vitro) Pathogenicity Determinations
Molecular pathogenicity determination of the 10 viable outbreak NDS isolates showed the presence of an F gene cleavage site sequence typical of virulent NDV. Two distinct cleavage patterns, 112RRRKR117F↓ or 112RRQKR117F↓, indicate that the cleavages sites of the precursor F0 (in its F1 and F2 subunits) were observed (Table 1) at the F0 proteolytic sites of the field isolates in domestic chickens. Both are indicators of the presence of virulent genes in these viruses (either mesogenic or velogenic pathotypes). In contrast to the field isolates, the three vaccine strains included in this investigation encoded cleavage sites consistent with lentogenic viruses (112GRQGR117L↓; Table 1).
The observation of virulent proteolytic cleavage sites on linear amino acid sequences, as observed in the 10 field isolates, does not differentiate between mesogenic and velogenic pathotypes of the NDV. To differentiate between the two, in vivo pathogenicity (ICPI) determinations was carried out in day-old chicks. The results demonstrated that all field isolates tested were velogenic, having an ICPI value of between 1.625 and 1.875 (mean 1.764 ± 0.08; mean ± standard deviation [SD]) (Table 1). These values are higher than the 0.7 required for an isolate to be classified as virulent. Negative control (sentinel) chicks from all experimental groups infected interacerebrally acquired an infection with the respective isolates administered to the groups and died within four days of association with the chicks.
Mapping of Amino Acid Substitutions in the Fusion Proteins of NDV Isolates
A total of 86 aas (aa) were deduced from each of the 258bp nucleotide F gene sequences and these were used for phylogenetic tree reconstruction. Historical field isolate sequences were aligned with 27 recently circulating and characterized isolates publicly available in sequence databases (Figure 2). Many nucleotide changes that resulted in amino acid substitutions were observed, and these changes mostly occured in and around F gene cleavage sites and/or nearby reported neutralization sites (Figure 2). The vaccinal strains had a deduced cleavage site of 112GRQGR117L↓ while the NDV isolated from chickens found in the field or farm had cleavage sites of 112RRQKR117F↓ and 112RRRKR117F↓. The 112RRQKR117F↓ and 112RRRKR117F↓ cleavage residues indicate that the NDV isolates correspond to mesogenic/velogenic pathotypes.
Genetic Distance
The Ethiopian NDV isolates have a within-group genetic distance of 10.16% and a between-group genetic distance of 25.1% (Table 2). These isolates are grouped for ease of presentation as published by authors: group1 (sequences characterized in this study from the central part of the country), group 2 from the central part of Ethiopia,9 and group 3 from Amhara region, Northern Ethiopia10 (Table 2).
 |
Table 2 Mean (± Standard Error) Genetic Distance Within (a) and Between (b) the Two Groups of Genotype VI ND Viruses Studied |
The highest genetic distance between the current characterized individual NDV isolates compared was 13.1% (Table 3), which occurred between R644/08 and the other four group viruses: R647/08, R1823/09, R653/08, and R655/08. Phylogenetic tree reconstruction (Figure 3) showed that the current NDVs studied were grouped into two subgroups having a genetic distance of 30% from the seven group viruses that could make them deserving of further subclassification as subgroups having bootstrap values of more than 60. When compared with previously published sequences, the highest genetic distance of 25.1% (± 0.039 [SE]; Table 2b) was observed between isolates obtained from the central part of the country and those obtained from the northern part (group 3, Amhara region).
 |
Table 3 Detailed Mean Genetic Distance (± Std Error Shown Above the Diagonal Line) Between Ethiopian NDV Outbreak Isolates Characterized in the Study* |
 |
Figure 3 Evolutionary relationships of NDVs isolated from Ethiopia. Their evolutionary history was inferred using the Minimum Evolution (ME) method implemented using MEGA Version 6. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (5000 replicates) are shown next to the branches. The evolutionary distances were computed using the Maximum Composite Likelihood method. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The ME tree was searched using the close-neighbor-interchange (CNI) algorithm at a search level of 1. The neighbor-joining algorithm was used to generate the initial tree. The analysis involved 93 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair. There were a total of 258 positions in the final dataset. Evolutionary analyses were conducted using MEGA Version 6.16 |
Molecular Evolutionary Relationship
The F gene sequence generated here extends from position 4544 to 4915; however, only 258 nt sequences that extend from 4658 to 4915 (with reference to NDV vaccinal strain sequence NDV LaSota JF950510) were used for molecular evolutionary relationship analysis. The use of relatively shortened sequences was to allow reasonable comparison with more recently isolated viruses in the country (from 2011 to 2014) for which the sequence data is available in GenBank. Model selection showed that Kimura 2-Parameter with Gamma distribution and invariant sites was selected as the best-fit model. Phylogenetically, all NDVs characterized belonged to Class II NDV, but were grouped into two different genotypes. The vaccinal strains were clustered with known Class II genotype II while the NDV isolated from chickens found in the field or farm clustered with Class II genotype VI (Figure 3). Phylogenetically, the viruses were grouped into two groups: group 1 and group 2 that contained seven and four NDVs, respectively (Figure 3). The oldest or the historical NDV isolate of 1976 (R1825/09, Table 1) in Ethiopia and all other field or farm isolates belonged to the Class II genotype VI (Figure 3). As depicted on the tree, Egypt-1990 isolate (GenBank accession number: DQ096604) and Sudan-1975 virus (GenBank accession number: AY151383) could be ancestors of group 1 and group 2 viruses, respectively.
Discussion
This study revealed that velogenic genotype VI NDVs, commonly known as pigeon paramyxovirus type 1, were circulating in Ethiopia from 1976 to 2007. Furthermore, these isolates are antigenically heterogeneous to the vaccine strains, indicating that decreased immune protection associated with poor cross-reactivity could have contributed to the continued outbreaks that occurred in the face of vaccination. The virulence of these isolates was confirmed by both in vivo (ICPI) chicken challenge experiment and in vitro by determination of their F gene cleavage site according to the criteria recommended by the OIE.5 In addition, the observation of NDV transmission from intracerebrally infected chicks to in-contact sentinel chicks within four days after association suggests that the viruses were highly contagious.
The F gene sequence based on molecular evolutionary relationship analysis (Figure 3) demonstrated that Ethiopian isolates clustered with genotype VI NDVs. Notably, the F gene amino acid sequence data and the cleavage site colocalize in monophylogenetic clades with the exception of one isolate. Previous studies using similar molecular approaches also reported the circulation of genotype VI NDVs in Ethiopia.9,10 Dimitrov et al17 updated the phylogenetic classification of NDV based on full F gene sequence analysis using three phylogenetic tree reconstruction methods. Accordingly, all seven Ethiopian isolates included in the Dimitrov et al17 analysis were grouped into genotype XXI-VI-1. Interestingly, Dimitrov et al17 also reported that the Egyptian isolates were grouped into genotype XXI, which is consistent with the currents analysis and report. However, unlike the Fentie et al10 report, we did not observe La Sota vaccine strain-related isolates using molecular evolutionary relationship analysis, or pathogenicity determinations by cleavage site determination of F gene sequence and in vivo ICPI determination. Instead, in our study, the genotype VI Ethiopian isolates were clustered into two groups: group 1 (Egypt-1990 like: accession number DQ096604) and group 2 (Sudan-1975 like: accession number AY151383) viruses. Phylogenetic analysis of each of these monophylogenetic clades demonstrated that the group 1 isolates share a common ancestor with Egypt-1990, while the group 2 isolates descend from Sudan-1975 viruses (Figure 3). The ancestral relationship with Egypt could be possible in that, previously, Ethiopia imported chickens and eggs from Egypt, and introduction of the viruses via such imports could explain the common ancestor. In contrast, to our knowledge such trade activities did not occur between Sudan and Ethiopia. In fact, the Ethiopian isolates were detected during the 1970s when these viruses were isolated in Sudan and also in Egypt (GenBank accession numbers AY151382-84 and DQ096604, respectively).
The viruses characterized in this study suggest that the ND outbreaks occurred as the result of four different outbreaks, ie every five or ten years (Table 1) due to either introduction from elsewhere or possibly from local evolution and maintenance over time following first introduction. These outbreaks were caused by two different class II genotype VI NDVs, as depicted in Figure 3. A closer look into the molecular evolution discloses that, in 1997, when multiple isolates were obtained (6 of 11 field isolates), four different outbreaks of NDV were observed in the country, of which three affected both commercial and village chickens found in the same town. But the other two epizootics affected only village chickens found in two different areas. The four group Ethiopian viruses (Sudan-1990 like) occurred in three different years: 1984, 1997 and 2000 (Figure 1). Phylogenetic tree analysis indicated that the same virus might have affected these epidemiologically different areas (Figure 3) of the country. Since then, no isolate belonging to this group was kept at NVI, Ethiopia. A major question is, how was the APMV-1 virus introduced to Ethiopia? This question was addressed via molecular evolutionary relationship analysis of the F gene sequence of the oldest isolate kept at NVI (1976 isolate), which belonged to APMV-1 of genotype VI that shares a common ancestor with Egypt-1990 like NDVs. This finding does not agree with a previous report by Aldous et al,14 who suggested it belonged to Australian vaccinal strains produced in Ethiopia. The two relatively recent field isolates of 2007 were obtained from commercial farms in Bishoftu town, which have better management practices, vaccinate their chickens routinely and have the best biosecurity measures (level 3, according to the FAO standard).18
The high frequency of mutations observed in and around cleavage sites (Figure 2) and residue positions closer to reported neutralization residues,19 as compared to La Sota reference vaccine strain sequence, could suggest the emergence of novel variant virulent NDV populations. A high mutation rate significantly contributes to genetic diversity and genetic distance among and between the virus populations. Among Ethiopian isolates, the observation of a 23.5% genetic distance between current isolates and those from Amhara region (group 3, shown in Table 2b) is due to clear genetic differences between the two virulent genotype VI isolates and Las Sota vaccine strain-related genotype II viruses. The high variability estimate in our study could be due to use of the short sequence of the hyper variable part of the NDV (F gene) sequence.
The two viruses showed high thermostability for 2 hrs at 56 °C with high ICPI (>1.770) and were isolated 13 years apart, suggesting that these viruses can overcome high environmental temperatures for prolonged periods. These high thermostable viruses, if circulated among chicken populations, can endanger production more easily as most chickens are susceptible to NDV infection under local management conditions. Control of ND relies on mass vaccination of susceptible chickens with the class I less pathogenic vaccine strains. In this study, we have come to understand the nature of NDVs circulating in the country and deduced their similarities with and differences from in-use vaccine strains and traced the source of the emergence of the disease. The antigenic differences observed between vaccine strains and field outbreak viruses was high, and the locally produced vaccine might provide incomplete protection. Hence, as Miller et al20 suggested, vaccines with at least a fusion protein sequence closer to the challenge strains would most probably provide better protection and this can likely be achieved by developing vaccines using the two thermostable NDVs observed in this study.
Conclusion
Virulent genotype VI NDVs, having ancestral relationships with Sudan-1975 and Egypt-1990 strains, were circulating both in commercial and backyard poultry production in the country. As these isolates were obtained from these two categories of farm, it is important to conduct continuous surveillance of NDV in chickens and pigeons in the area because the genotype identified belonged to the pigeon type and chickens and pigeons usually mix together under smallholder farmers' management conditions. The clustering of previously characterized NDV isolates from Amhara region with La Sota strains raises great concern regarding the efficacy and proper inactivation of the existing vaccine used in the country. Of the 11 isolates propagated in SPF ECE, two were thermostable at 56 °C for 2 hrs. The high thermostability of these isolates could contribute to the continuous outbreak of the disease in the country as they can easily overcome prevailing environmental temperatures and they are also good candidates for killing thermostable vaccines from local strains. Ethiopia is producing ND vaccines for local use and proper ND control measures need to be instituted via vaccination campaigns.
Ethical Approval
Samples were collected from dead chickens in Ethiopia as part of routine post-mortem examinations conducted by the National Veterinary Institute, Bishoftu. The animal experiments were conducted at the Friedrich-Loeffler-Institut, Germany, according to German21 and EU legislation22 regarding animal welfare and approved by the State Office of Agriculture, Food safety, and Fishery in Mecklenburg, Western Pomerania, Germany (LALLF M-V) (LVL MV/TSD/7221.3-2.2-005/07). All experimental procedures were carried out in approved biosafety level 2 facilities at the Friedrich-Loeffler-Institut, Germany, following all relevant national as well as international regulations and according to ethical principles and care for experimental animals.
Acknowledgments
Excellent technical assistance provided by NDV and Avian Influenza laboratory technicians and students at the Friedrich-Loeffler-Institut, Germany, was greatly appreciated. We also thank Dr. Samson Leta, an epidemiologist in the College of Veterinary Medicine and Agriculture, Addis Ababa University, Ethiopia, for help in sketching the map (Figure 1).
Funding
The research project was funded by Rothamsted International, UK via African Research Fellowship Award to the first author. The funder played no role in the design and conclusion of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sambo E, Bettridge J, Dessie T, et al. Participatory evaluation of chicken health and production constraints in Ethiopia. Prev Vet Med. 2015;118:117–127. doi:10.1016/j.prevetmed.2014.10.014
2. Amarasinghe GK, Ayllon MA, Bao Y, et al. Taxonomy of the order Mononegavirales: update 2019. Arch Virol. 2019;164:1967–1980. doi:10.1007/s00705-019-04247-4
3. Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology.
4. Toyoda T, Sakaguchi T, Lmai K, et al. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle virus. Virology. 1987;58:242–247. doi:10.1016/0042-6822(87)90261-3
5. Office International des Epizooties. Newcastle disease (infection with newcastle disease virus). OIE Terresterial Manual, Chapter 2.3.14, Office International des Epizooties (OIE); 2012. Available from: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.14_NEWCASTLE_DIS.pdfpage.
6. Miller PJ, Haddas R, Simanov L, et al. Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect Gen Evol. 2015;29:216–229. doi:10.1016/j.meegid.2014.10.032
7. Artois M, Manvell R, Fromont E, et al. Serosurvey for newcastle disease and avian influenza A virus antibodies in great cormorants from France. J Wildlife Dis. 2002;38:169–171. doi:10.7589/0090-3558-38.1.169
8. Mulisa DD, Keno MS, Alemu RB, et al. Characterisation of Newcastle disease virus and poultry handling practices in live poultry markets in Ethiopia. SpringPlus. 2014;3:1–6.
9. Damene D, Fusaro A, Sombo M, et al. Characterisation of Newcastle disease virus isolates obtained from outbreak cases in commercial chickens and wild pigeons in Ethiopia. SpringPlus. 2016;5:476. doi:10.1186/s440064-016-2114-8
10. Fentie T, Heidari A, Aiello R. Molecular characterization of Newcastle disease viruses isolated from rural chicken in northwest Ethiopia reveals the circulation of three distinct genotypes in the country. Trop Anim Hlth Prod. 2014;46(2):299–304. doi:10.1007/s11250-013-0487-z
11. Alexander DJ. Newcastle disease, other avian paramyxoviruses and pneumovirus infections. In: Saif JM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editors. Diseases of Poultry.
12. Grund CH, Werner O, Gelderblow HR, et al. Avian paramyxovirus serotype 1 isolates from the spinal cord of parrots display a very low virulence. J Vet Med B. 2002;49:445–451. doi:10.1046/j.1439-0450.2002.00596.x
13. Kim LM, King DJ, Currey PE, et al. Phylogenetic diversity among low virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to poultry-origin isolates. J Virol. 2007;81(22):12641–12653. doi:10.1128/JVI.00843-07
14. Aldous EW, Mynn JK, Banks J, Alexander DJ. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 2003;32(3):239–256. doi:10.1080/030794503100009783
15. Ujvari D, Wehmann E, Kaleta EF, et al. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. 2003;96:63–73. doi:10.1016/S0168-1702(03)00173-4
16. Tamura K, Dudley J, Nei M, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi:10.1093/molbev/mst197
17. Dimitrov KM, Abolnik C, Afonso CL, et al. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol. 2019;74:103917. doi:10.1016/j.meegid.2019.103917.
18. Food and Agricultural Organization. Improved animal health for poverty reduction and sustainable livelihoods. FAO (Food and Agricultural Organization) Animal Production and Health Paper; 2002: 44.
19. Miller PJ, Kim LM, Ip HS, et al. Evolutionary dynamics of Newcastle disease virus. Virology. 2009;391:64–74. doi:10.1016/j.virol.2009.05.033
20. Miller PJ, King DJ, Alfonso CL, Suarez DL. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine. 2007;25:7238–7246. doi:10.1016/j.vaccine.2007.07.017
21. Anon. Tierschutzgesetz, Bundesgesetzblatt 19th June 2020, vol I. 2020: 1328.
22. EC. Directive 2010/63/EU on the protection of animals used for scientific purposes. Available from: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32010L0063&from=EN.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

