Back to Journals » International Journal of General Medicine » Volume 15
An Integrative Transcriptomic Analysis Reveals EGFR Exon-19 E746-A750 Fragment Deletion Regulated miRNA, circRNA, mRNA and lncRNA Networks in Lung Carcinoma
Authors Chen L, Li S , Shi W, Wu Y
Received 21 April 2022
Accepted for publication 27 June 2022
Published 5 July 2022 Volume 2022:15 Pages 6031—6042
DOI https://doi.org/10.2147/IJGM.S370247
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Ling Chen,* Shenyi Li,* Weifeng Shi, Yibo Wu
The Affiliated Hospital of Jiangnan University, Wuxi, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yibo Wu; Weifeng Shi, The Affiliated Hospital of Jiangnan University, Wuxi, Jiangsu Province, People’s Republic of China, Tel +86-510-68089762 ; +86-510-68089762, Fax +86-510-68089762, Email [email protected]; [email protected]
Introduction: Competing endogenous RNA (ceRNA) appears to be an important post-transcriptional manner that regulates gene expression through a miRNA-mediated mechanism. Mutations in exon-19 of EGFR were frequently observed in lung cancer genes, which were associated with EGFR activity and EGFR-targeted therapies.
Methods: We explored the transcriptome regulated by mutation in EGFR exon-19 E746-A750 fragment via using a network modeling strategy. We applied transcriptome sequencing to detect the deletion process of EGFR exon-19 E746-A750 fragment. Bio-informatics analyses were used to predict the gene target pairs and explain their potential roles in tumorigenesis and progression of lung cancer.
Results: We conducted an explorative lncRNA/miRNA/circRNA and mRNA expression study with two groups of lung adenocarcinoma tissues, including EGFR exon-19 E746-A750 deletion group and EGFR exon-19 wild-type group. Meanwhile, we screen out the hub genes related to the EGFR-19-D patient. Significant pathways and biological functions potentially regulated by the deregulated 128 non-coding genes were enriched.
Conclusion: Our work provides an important theoretical, experimental and clinical foundation for further research on more effective targets for the diagnosis, therapy and prognosis of lung cancer.
Keywords: microRNAs, miRNAs, mRNAs, long non-coding RNAs, lncRNAs, gene regulating network, lung cancer
Introduction
Lung adenocarcinoma is one of the most common subtypes of non-small cell lung cancer (NSCLC) and is the leading cause of cancer-related death worldwide.1 As lung cancer was prone to early metastasis, and the prognosis of lung cancer patients was generally poor, so the 5-year survival rate used to be less than 80%.2 Although the treatment has been improved over the past decades, the survival rate remains low.3
Epidermal growth factor (EGFR) is immobilized on the cell surface and binds to its specific receptor to activate cells. Somatic mutations in the EGFR gene cause them to lose their ability to bind to receptors, which activates and causes cell to divide. Some studies have shown that mutations in the EGFR signaling pathway are potential therapeutic targets. Inhibition of EGFR signaling pathway activation can improve the prognosis of cancer patients with EGFR mutations.4 There are seven types EGFR exon-19 mutation in NSCLC. Among the somatic mutations of EGFR tyrosinase domain in NSCLC, the proportion of EGFR intra-exon 19 E746-A750 (EGFR-19-D) fragment deletion is the most common.5
MicroRNAs (miRNAs) is a non-coding single-stranded RNA molecule with about 22 nucleotides encoded by an endogenous gene that inducing degradation by binding specifically to the 3’-untranslated region (UTR) mRNA sequence,6 or inhibiting of mRNA translation.7 Long non-coding RNAs (lncRNAs) belong to a class of non-coding ribonucleic acids with a molecular weight greater than 200 nucleotides, which regulate the expression of epigenetic genes in the transcriptional and transcriptional levels of various physiological and pathological processes.8 In addition, the circular RNA (circRNA) molecule has a closed ring structure, which is composed of 5’-3ʹpolarity and a poly A tail.9 Without affection by RNA exonuclease, the expression is more stable and not easy to degrade.10
The new regulatory mechanism between non-coding RNA and coding RNA in Senna hypothesis contributes to the understanding of cancer initiation and development.11 It is reported that lncRNAs, as competitive endogenous RNA (ceRNA) or microRNA sponge, inhibit the expression of miRNA response elements (MREs) by competing with common miRNAs in tumorigenic process,12 such as imprinting, X inactivation and development. Previous studies mainly focused on the interaction network between lncRNA and RNA regulation.13 Some miRNA and lncRNA molecules have important significance in the diagnosis and treatment of tumors and can be used as new molecular markers for tumor prognosis. However, EGFR 19 downstream molecules, especially miRNAs, lncRNAs, circRNAs and mRNA-related regulatory networks, have not been thoroughly investigated in NSCLC.
In this study, we conduct exploratory research on lncRNA/miRNA/circRNA and mRNA expression with two groups of lung adenocarcinoma tissues, including EGFR exon-19 E746-A750 deletion group and EGFR exon-19 wild-type group, using the transcript microarray technology. Meanwhile, we screen out the hub genes related to the EGFR-19-D patient. Accordingly we discuss the association of hub genes function with EGFR exon-19 deletion, tumorigenesis and metastasis.
Materials and Methods
Clinical Specimens
All patients were collected from the Affiliated Hospital of Jiangnan University (Table 1). The medical ethics committee of the affiliated hospital of Jiangnan University reviewed this study. This study complies with the Declaration of Helsinki. They sanctioned the collection of the specimens, and the patients gave written informed consent. The clinical specimens included three cases of EGFR19 deletion and three cases of EGFR19 non-deletion lung cancer. None of lung cancer patient received any treatment before surgery, including radiotherapy and chemotherapy. All specimens of lung cancer were obtained by the chief pathologist after operation. They were immediately frozen in liquid nitrogen and stored at −80℃. All the tissues were confirmed histologically.
 |
Table 1 Infortation of Clinical Specimens for Microarray |
RNA Isolation, cDNA Construction and Illumina Deep Sequencing
Total RNAs of all specimens was extracted using the Ribo-Zero Reagent (Invitrogen, USA) according to the instruction manual. The TruseqTM RNA sample prep kit (Illumina, San Diego, CA, USA) was used to constructed cDNA library following the manufacture’s instruction. Briefly, oligo (dT) magnetic beads were used to the purification of poly (A)+RNA from total RNA through shearing into short fragments, and Truseq RNA sample preparation kit was used to cDNA library synthesis according to the manufacturer’s instructions (Illumina). RNA was quantified using the NanoDrop (Thermo Scientific NanoDrop 2000), and RNA integrity number (RIN) >7 were used for cDNA library construction. After quantitation, the samples were clustered (TruSeq paired-end cluster kit v3-cBot-HS; Illumina) and sequenced on the HiSeq 2000 platform (100 bp, TruSeq SBS kit v3-HS 200 cycles; Illumina).
GO, KEGG Pathway Analysis
KOBAS 2.0 is an open web server for annotation and identification of enriched pathways.14 We performed possible GO and KEGG enrichment pathways predictions through KOBAS annotation. The significant items were screened out under the conditions of p<0.05 and FDR <0.05.
PPI Network Construction and Hub Gene Analysis
To screen out the interaction networks between the differentially expressed mRNAs, we used the StringApp plugin in Cytoscape 3.7.1 to import the protein–protein interaction (PPI) network from the STRING database. Then. the Cytohubba plugin was used to screen out the hub genes. The top four hub genes were screened out by degree score=3 in the Cytohubba plugin. Then the top four hub genes related to mRNA–miRNA pair, mRNA–circRNA pairs, and mRNA–ncRNA pairs were selected. Network visualization was achieved by Cytoscape 3.7.1.
Statistical Analysis
The screened differentially expression genes were analyzed by independent-sample t-test, and the results were expressed as the mean±standard deviation (SD). P value <0.05 were consider statistically significant, and SPSS 17.0 was used to statistical analysis. The heat map and other graphs in the article are made by R-project.
Results
The lncRNAs Expression Profiling Regulated by EGFR-19-D Exon Mutation
The lncRNA expression profiling was performed in EGFR-19-D exon mutation biopsies and non-EGFR mutation lung cancer biopsies (Supplementary 1). A total of 44 lncRNAs were significantly induced, and 36 lncRNAs were reduced with the EGFR-19 exon mutation (Figure 1A). Among these 80 lncRNAs, 17 lncRNAs were non-protein coding genes and 52 lncRNAs were no-name genes. These genes were mainly involved in signaling pathways, such as the“complement and coagulation cascades”,“Glutathione metabolism”,“Taurine and hypotaurine metabolism” (Figure 1B). Recently, Qing Tang’s15 study indicated that compared to normal people, complement and coagulation cascades pathway played an important role in the NSCLC patient.
The miRNAs Expression Profiling Regulated by EGFR-19-D Exon Mutation
The miRNA expression profiling was performed in lung cancer between EGFR-19-D group and EGFR wild-type group (Supplementary 2). Among these differentially expressed miRNAs, 10 miRNAs were down-regulated significantly in EGFR-19-D exon mutation lung cancer, while 23 miRNAs were up-regulated compared to EGFR wild-type group (Figure 1C). These genes were mainly involved in signaling pathways, such as the “MAPK signaling pathway”, “ErbB signaling pathway”, “EGFR tyrosine kinase inhibitor resistance”, “Endocrine resistance”, “Ras signaling pathway”, “Spliceosome” (Figure 1D). Among these, MAPK signaling pathway and Ras signaling pathway were top2 mainly gene enrichment pathway. KEGG (Kyoto Encyclopedia of Genes and Genomes) database analysis shows that EGFR mutations mainly activate signaling pathway including RAS-ERK signaling pathway, PI3K-Akt signaling pathway and PLCG-ERK signaling pathway.
The circRNAs Expression Profile Regulated by EGFR-19-D Exon Mutation
Seven circRNAs expression were up-regulated and 10 circRNAs were down-regulated significantly in the EGFR-19-D exon procession (Figure 2A). These differentially expressed circRNAs were enriched in the pathway such as “Cell adhesion molecules (CAMs)”, “Synaptic vesicle cycle”, “Viral myocarditis”, “Autoimmune thyroid disease”, “Type I diabetes mellitus”, “Graft−versus−host disease”, “Allograft rejection”, “Propanoate metabolism”, and “Citrate cycle (TCA cycle)” (Figure 2B). Besides, the total number of circRNAs was shown in the Supplementary 3. The location of circRNAs on the chromosome was listed in Supplementary Figure 1. In the end, the top 10 deregulated RNAs are shown in Table 2.
 |
Table 2 Top 10 Significantly Deregulated Genes in Two Groups of Patients |
The mRNAs Expression Profile Regulated by EGFR-19-D Exon Mutation
The mRNA expression profiling was performed in EGFR-19-D exon mutant biopsies and non-EGFR mutant lung cancer biopsies (Supplementary 4). A total of 38 mRNAs were significantly induced, and 35 mRNAs were reduced by EGFR-19-D exon mutation (Figure 2C). These genes were mainly involved in signaling pathways, such as the “Complement and coagulation cascades”, “Glutathione metabolism”, and “Taurine and hypotaurine metabolism” (Figure 2D). Besides, we constructed the PPI network to explore the major protein in the EGFR-19-D exon mutation (Figure 3). We found that CASR, FGG, HOXB9, OPRK1 were the hub genes in the network (Figure 4A). Among these genes, the gene cluster of CASR and OPRK1 interacting with crosstalk is more obvious. It indicated that there were three HOXB9-circRNA pairs, two HOXB9-lincRNA pairs and four HOXB9-miRNA pairs (Figure 4B). Six miRNAs, four circRNAs and nine lincRNAs were related to gene CASR (Figure 4C). Besides, there were 8 FGG-lincRNA pairs, 1 FGG-circRNA pair and 4 OPRK1-circRNAs, 18 OPRK1-lincRNA pairs, and 5 OPRK1-miRNA pairs (Figure 4D).
The ceRNA Network Constructed by the Screened RNAs
Total RNAs were transferred from ENSEMAL ID to SYMBOL. The RNAs which could match symbol name were screened out. In total, 1224 RNA–RNA pairs were screened out (Figure 5), including 15 circRNA, 31 lincRNAs, 34 miRNAs, and 71 mRNAs. The total number of RNA–RNA pairs was shown in Supplementary 5.
Corroboration of the 18 RNA Expressions with RT-qPCR
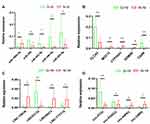
RT-qPCR was performed on five pairs of lung cancer tissues with EGFR-19 exon mutation and EGFR-19 exon wild-type, including five miRNAs (Figure 6A), five mRNAs (Figure 6B), four lncRNAs (Figure 6C) and four circRNAs (Figure 6D). Eighteen RNA expressions were statistically different (p<0.05), of which miR-300, CLCA2, CYP24A1, CASR, LINCO1133, LINC00473, and circITCH were significantly different (p<0.01). And the results were consistent with the sequencing results.
Discussion
In the past study, GSH metabolism was thought to be one major mechanism of chemo-resistance in the NSCLC.16 In addition, Pool et al17 deemed that the GSK metabolism pathway might be essential for tumor response to EGFR-TKI afatinib alone or combined with Rapamycin. Presently, EGFR-TKIs are effective in the lung cancer patients who have a deletion mutation of exon 19 and L858R mutation of exon 21.18 Canonical ligand-dependent EGFR signaling pathway contains the Ras/MAPK pathway, the PI3K/AKT pathway, and the phospholipase C (PLC)/protein kinase C (PKC) signaling cascade.19
We found four new hub genes, which were not directly contained in the canonical pathway were related to the EGFR-19 exon E746-A750 fragment deletion. In our study, CaSR was up-regulated in the EGFR-19 exon deletion group compared to the EGFR-19 exon wild-type. When we analyzed whether there was any relationship between these CaSR and EGFR-19 exon E746-A750 mutation, we found that CaSR interacted with G proteins that could regulate the EGFR downstream signaling molecular pathway.20 Moreover, CaSR was activated by signals from glutathione and stimulated the CaSR trans-activated expression of EGFR, causing phosphorylation of ERK and secretion of PTHrP.21,22 Besides, CaSR could induce calcium overload and subsequent mitogen-activated protein kinase (MAPK) pathways.23 In our study, 6 miRNAs, 4 circRNAs and 17 lncRNAs were predicted to bind to CaSR. The function of has-miR-499b-5p is unknown, but its family member miR-499a has been well studied. MiR-449a inhibits the expression of MAP2K1 by directly targeting its 3ʹUTR and modulates the activity of the MEK1/ERK1/2/c-Jun pathway by auto-regulating the feedback loop.24 Has-miR-551a is a medium for 5-FU to acquire drug resistance.25 Zhang26 data indicated that LINC-CECR7 was negatively associated with OS Hepatocellular Carcinoma. CircRNA-CEP70 acted as a novel circRNA targeting the miR-561/E2F8 that regulated SH-SY5Y cell proliferation and differentiation.27 Wang et al28 identified that Axl-altered miR-548b played a crucial role in the gefitinib-induced apoptosis and EMT of gefitinib-resistant lung cancer cells by targeting CCNB1.
FGG belongs to the fibrinogen family, which is the critical gene in the complement and coagulation cascades pathway.29 High expression of fibrinogen causes tumor metastasis and recurrence in the patients with esophageal squamous cell carcinoma, especially combined with miRNA.30 Thus, decreased expressed FGG might attenuate the processes of pathologic thrombosis and angiogenesis associated with cancer cells. Moreover, we found that FGG might bind to circRNA-TM9SF3, LOC105376382. Yet, the function of LINC02678 is unknown.
HOXB9 is a homeobox transcription factor, which plays an important role in carcinoma development. Nguyen et al31 unveiled that HOXB9 acted as promoters of cancer cell metastasis and mediators of chemotactic invasion and colony outgrowth in lung adenocarcinoma. HOXB9 was down-regulated in our study. As a consequence, we speculated that EGFR-19 exon deletion lung cancer cell metastasis is related to HOXB9. Besides, Siu32 study demonstrated that down-regulated miR-203 could induce the bone metastasis and TKI resistance, which EGFR pathway activated via altered expression of EGFR ligand (EREG and TGFA) and anti-apoptotic proteins (API5 BIRC2 and TRIAP1). MiR-873 could act as a negative regulator of tumor proliferation and metastasis.33
OPRK1 is the κ opioid receptor, which encodes G protein-coupled opioid receptors. The activation of EGFR trans-regulates the function of G protein-coupled opioid receptors through GRK2 (G protein-coupled receptor kinases 2) phosphorylation and activation.34 GRK2 acts as a mediator of cross-talk from RTK to GPCR (G protein-coupled receptor) signaling pathway. In the past study, OPRK1 was concentrated on the alcohol and drug dependence.35 Zhang et al36 reported that was down-regulated in HCC (hepatocellular carcinoma) tissues. Multiple Scancer’s study37,38 indicated that miR-136-5p was deregulated. MiRNA-136-5p was down-regulated in hepatocellular carcinoma, while it was up-regulated in lung adenocarcinoma. So far, no study has mentioned the relationship between miR-136-5p and OPRK1. Kumar et al39 indicated that the putative targets of miR-300 were enriched in the TGF-beta signaling pathway, moreover up-regulation of miR-300 may induce cisplatin resistance.
In our analysis, we found that LINC01750, LINC00885, LINC01331, LINC01335, LINC01332, AC011752.1, LOC442497, AL3354766.2, AC354766.2, RIMS1, METP1, PTPN13, CEP70 and CENE-1 were the cross-linked targets between OPRK1 and CaSR. The interaction between OPRK1 and CaSR needs further study. However, we believe that the two genes play an important role in the expression and metastasis of EGFR-19 exon mutation in lung carcinoma.
Conclusion
EGFR exon-19 E746-A750 fragment deletion is prone to have a longer overall survival and/or progression-free survival with treatment of gefitinib or erlotinib compared to EGFR exon 19 wild-type. These hub genes may potentially give a new strategy for EGFR-19-D treatment. The ceRNA network may provide a new exploration about EGFR-19-D, which one point may be the newly response to EGFR-19-D patients.
Additional Information
Supplementary information accompanies this paper in Supplementary 1–5, Supplementary Figure 1, Tables 1 and 2. Supplementary 6 listed the primers involved in this study.
Acknowledgments
This study was supported by the grants of National Natural Science Fund (81701511) and grants of Wuxi Young Medical Talents Program (QNRC016, QNRC017).
Disclosure
The authors declare no conflicts of interest.
References
1. Sharma SV, Bell DW, Jeffrey S, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi:10.1038/nrc2088
2. Wang T, Nelson RA, Bogardus A, Grannis FW. Five-year lung cancer survival: which advanced stage nonsmall cell lung cancer patients attain long-term survival? Cancer. 2010;116(6):1518–1525. doi:10.1002/cncr.24871
3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi:10.1016/S0025-6196(11)60735-0
4. Sequist LV, Joshi VA, Jänne PA, et al. Response to treatment and survival of patients with non-small cell lung cancer undergoing somatic EGFR mutation testing. Oncologist. 2007;12(1):90. doi:10.1634/theoncologist.12-1-90
5. Tetsuya M, Yasushi Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98(1):1817–1824. doi:10.1111/j.1349-7006.2007.00607.x
6. Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38(3):323–332. doi:10.1016/j.molcel.2010.03.013
7. Ma X, Conklin DJ, Li F, et al. The oncogenic microRNA miR-21 promotes regulated necrosis in mice. Nat Commun. 2015;6:7151. doi:10.1038/ncomms8151
8. Li JW, Gao C, Wang YC, et al. A bioinformatics method for predicting long noncoding RNAs associated with vascular disease. Sci China Life Sci. 2014;57(8):852–857. doi:10.1007/s11427-014-4692-4
9. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
10. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
11. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
12. Wang P, Zhi H, Zhang Y, et al. miRSponge: a manually curated database for experimentally supported miRNA sponges and ceRNAs. Database. 2015;2015:bav098.
13. Liu T, Chi H, Chen J, et al. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene. 2017;631:29–38. doi:10.1016/j.gene.2017.08.008
14. Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–W322. doi:10.1093/nar/gkr483
15. Tang Q, Zhang H, Kong M, Mao X, Cao X. Hub genes and key pathways of non-small lung cancer identified using bioinformatics. Oncol Lett. 2018;16(2):2344–2354. doi:10.3892/ol.2018.8882
16. Ankita B, Celeste SM. Glutathione metabolism in cancer progression and treatment resistance. J Cell Biol. 2018;217(7):2291–2308.
17. Pool M, de Boer HR, Hooge MNL, van Vugt M, de Vries EGE. Harnessing integrative omics to facilitate molecular imaging of the human epidermal growth factor receptor family for precision medicine. Theranostics. 2017;7(7):2111–2133. doi:10.7150/thno.17934
18. Toyooka S, Mitsudomi T, Soh J, et al. Molecular oncology of lung cancer. Gen Thorac Cardiovasc Surg. 2011;59(8):527–537. doi:10.1007/s11748-010-0743-3
19. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi:10.1016/j.cell.2010.06.011
20. Yamamura A, Guo Q, Yamamura H, et al. Enhanced Ca(2+)-sensing receptor function in idiopathic pulmonary arterial hypertension. Circ Res. 2012;111(4):469–481. doi:10.1161/CIRCRESAHA.112.266361
21. Boudot HL, Thiem U, Geraci S, et al. Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulin-mediated osteoprotegerin downregulation. Oncotarget. 2017;8(34):56460–56472. doi:10.18632/oncotarget.16999
22. Bandyopadhyay S, Tfelt-Hansen J, Chattopadhyay N. Diverse roles of extracellular calcium-sensing receptor in the central nervous system. J Neurosci Res. 2010;88(10):2073–2082. doi:10.1002/jnr.22391
23. Kong WY, Tong LQ, Zhang HJ, et al. The calcium-sensing receptor participates in testicular damage in streptozotocin-induced diabetic rats. Asian J Androl. 2016;18(5):803–808. doi:10.4103/1008-682X.160885
24. You J, Zhang Y, Li Y, et al. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res. 2015;5(9):2730–2744.
25. Kang H, Kim C, Ji E, et al. The MicroRNA-551a/MEF2C axis regulates the survival and sphere formation of cancer cells in response to 5-fluorouracil. Mol Cells. 2019;42(2):175–182. doi:10.14348/molcells.2018.0288
26. Zhang J, Fan D, Jian Z, Chen GG, Lai PB, Guan X-Y. Cancer specific long noncoding RNAs show differential expression patterns and competing endogenous RNA potential in hepatocellular carcinoma. PLoS One. 2015;10(10):e0141042. doi:10.1371/journal.pone.0141042
27. Yang M, Xiang G, Yu D, et al. Hsa_circ_0002468 regulates the neuronal differentiation of SH-SY5Y cells by modulating the MiR-561/E2F8 axis. Med Sci Monit. 2019;25:2511–2519. doi:10.12659/MSM.915518
28. Wang Y, Xia H, Zhuang Z, Miao L, Chen X, Cai H. Axl-altered microRNAs regulate tumorigenicity and gefitinib resistance in lung cancer. Cell Death Dis. 2014;5(5):e1227. doi:10.1038/cddis.2014.186
29. Yin SJ, Luo YQ, Zhao CP, et al. Antithrombotic effect and action mechanism of Salvia miltiorrhiza and Panax notoginseng herbal pair on the zebrafish. Chin Med. 2020;15:35. doi:10.1186/s13020-020-00316-y
30. Du J, Zhang L. Analysis of salivary microRNA expression profiles and identification of novel biomarkers in esophageal cancer. Oncol Lett. 2017;14(2):1387–1394. doi:10.3892/ol.2017.6328
31. Nguyen DX, Chiang AC, Zhang XH, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138(1):51–62. doi:10.1016/j.cell.2009.04.030
32. Siu MK, Abou-Kheir W, Yin JJ, et al. Loss of EGFR signaling regulated miR-203 promotes prostate cancer bone metastasis and tyrosine kinase inhibitors resistance. Oncotarget. 2014;5(11):3770–3784. doi:10.18632/oncotarget.1994
33. Tang L, Chen Y, Tang X, Wei D, Xu X, Yan F. Long noncoding RNA DCST1-AS1 promotes cell proliferation and metastasis in triple-negative breast cancer by forming a positive regulatory loop with miR-873-5p and MYC. J Cancer. 2020;11(2):311–323. doi:10.7150/jca.33982
34. Chen Y, Long H, Wu Z, Jiang X, Ma L, Assoian R. EGF transregulates opioid receptors through EGFR-mediated GRK2 phosphorylation and activation. Mol Biol Cell. 2008;19(7):2973–2983. doi:10.1091/mbc.e07-10-1058
35. Edenberg HJ, Wang J, Tian H, et al. A regulatory variation in OPRK1, the gene encoding the kappa-opioid receptor, is associated with alcohol dependence. Hum Mol Genet. 2008;17(12):1783–1789. doi:10.1093/hmg/ddn068
36. Zhang Q, Matsuura K, Kleiner DE, Zamboni F, Alter HJ, Farci P. Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J Transl Med. 2016;14(1):328. doi:10.1186/s12967-016-1085-4
37. Boo L, Ho WY, Mohd Ali N, et al. Phenotypic and microRNA transcriptomic profiling of the MDA-MB-231 spheroid-enriched CSCs with comparison of MCF-7 microRNA profiling dataset. PeerJ. 2017;5:e3551. doi:10.7717/peerj.3551
38. Hua D, Zhi-Hua Y, Dong-Yue W, et al. Downregulation of miR-136-5p in hepatocellular carcinoma and its clinicopathological significance. Mol Med Rep. 2017;16(4):5393–5405. doi:10.3892/mmr.2017.7275
39. Kumar S, Kumar A, Shah PP, Rai SN, Panguluri SK, Kakar SS. MicroRNA signature of cis-platin resistant vs. cis-platin sensitive ovarian cancer cell lines. J Ovarian Res. 2011;4(1):17. doi:10.1186/1757-2215-4-17
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.