Back to Journals » Journal of Inflammation Research » Volume 17
A Risk Score Based on Immune- and Oxidative Stress-Related LncRNAs Predicts Prognosis in Lung Adenocarcinoma: Insights from in vitro Experiments and Large-Scale Transcriptome Analysis
Authors Liu X, Zhao F, Wang X, Ma Z, Yan H, Lu X, Li S , Zhu H, Gao S
Received 30 June 2023
Accepted for publication 20 February 2024
Published 5 March 2024 Volume 2024:17 Pages 1453—1465
DOI https://doi.org/10.2147/JIR.S428287
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Xin Liu,1,* Fangchao Zhao,2,* Xiaodan Wang,2,* Zheng Ma,2 Hongjiang Yan,2 Xuchao Lu,2 Shujun Li,2 Haiyong Zhu,2,* Shaolin Gao2
1Second Department of Pulmonary and Critical Care Medicine, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China; 2Department of Thoracic Surgery, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shaolin Gao, Department of Thoracic Surgery, The Second Hospital of Hebei Medical University, No. 215 Heping West Road, Shijiazhuang, Hebei, People’s Republic of China, Tel/Fax +86-311-66003588, Email [email protected]
Background: Long non-coding RNAs (lncRNAs) were demonstrated to be key to cancer progression and highly associated with the tumor immune microenvironment. Oxidative stress and immune may modulate the biological behaviors of tumors. Therefore, biomarkers that combined oxidative stress, immune, and lncRNA can be a promising candidate bioindicator in clinical therapy of cancers.
Methods: Immune-related genes (IRGs) and oxidative stress-related genes (ORGs) were identified based on a detailed review of published literatures. The transcriptome data and clinical information of lung adenocarcinoma (LUAD) patients were obtained from TCGA database. Lasso and Cox regression analyses were conducted to develop a prognostic model. Additionally, the link between immune checkpoints, immune cells, and the prognostic model was investigated, and predict the sensitivity of immunotherapy.
Results: 2498 IRGs and 809 ORGs were extracted from previous studies, and 190 immune- and oxidative stress-related genes (IOGs) were acquired by overlapping the above genes. 658 immune- and oxidative stress-related lncRNAs (IOLs) were screened by Pearson correlation analysis. A total of 25 prognosis-related IOLs were screened by univariate regression analysis. Finally, LASSO Cox regression analysis was adopted for determining a 12-IOLs prognostic risk signature. The signature performance was confirmed in the training cohort and the testing cohort, and cases were classified into low- and high-risk groups by the risk score calculated from the signature. Patients in the high-risk group had poor prognoses and immunosuppression, while the risk score was significantly associated with tumor-infiltrating immune cells, immune checkpoint expression, and immunotherapy responses. In vitro experiments further confirmed the expression of key signature gene.
Conclusion: Our new IOLs-related prognostic signature can be reliable prognostic tools and therapeutic targets for LUAD patients.
Keywords: immune, oxidative stress, lung adenocarcinoma, prognostic model
Introduction
Based on data obtained from the National Cancer Center, it is evident that lung cancer holds the distinction of being the most prevalent malignant tumor and leading cause of cancer-related fatalities in China.1 In terms of tissue type, lung cancer can primarily be categorized into two main types: non-small cell lung cancer (NSCLC) and small cell lung cancer. Of these, NSCLC is the most common, with lung adenocarcinoma (LUAD) emerging as the primary histological subtype.2,3 Unfortunately, early-stage LUAD cases typically lack typical clinical symptoms and signs, resulting in most cases being diagnosed at middle and late stages and leading to a poor overall prognosis.4 Despite the increasing maturity and perfection of LUAD treatment technology and strategy in China, the overall 5-year survival rate remains low.5 Therefore, predictive biomarkers with high reliability and corresponding predictive models are essential for individualized treatment and early, accurate diagnosis of LUAD cases.
Oxidative stress occurs due to the accumulation of reactive nitrogen and reactive oxygen species (ROS) in cells and organs under certain special conditions, resulting in oxidative damage when the body’s oxidation-reduction balance is imbalanced and free radicals exceed the range of antioxidant scavenging capacity.6 Oxidative stress plays a crucial role in tumor metastasis/invasion, tumor progression, carcinogenesis, and acute/chronic inflammation in the tumor microenvironment (TME).7,8 Immune cells are highly sensitive to oxidative stress due to their high content of unsaturated fatty acids in their cell membranes, making them susceptible to peroxidation reactions. As a result, excess free radicals can decrease immune function, leading to immune system dysfunction.9 In summary, oxidative stress is closely related to tumor immunity.
Long non-coding RNAs (lncRNAs) account for 80% of the human transcriptome and are over 200 nucleotides in length.10 Initially considered “noise” of genomic transcription, lncRNAs were thought to have no biological function due to their lack of a valid open reading frame and encoding little or no protein.11 However, accumulating evidence has shown that lncRNAs, once thought to be “non-functional”, are extensively involved in essential processes such as cancer, immune cell differentiation, and immune system regulation12,13 and are associated with oxidative stress.14 Recent reports suggest that lncRNAs negatively or positively affect response to oxidative stress,14,15 implying that lncRNAs may be critical molecules involved in the oxidative stress field and can serve as biomarkers for oxidative stress-associated disorders.16,17 While previous studies have investigated the role of lncRNAs in LUAD, our research contributes to the field by identifying a novel prognostic risk signature based on immune- and oxidative stress-related lncRNAs (IOLs). The signature we developed has not been reported before and provides a reliable prognostic tool for LUAD patients. Additionally, our study provides new insights into the underlying mechanisms of IOLs in LUAD pathogenesis and demonstrates that LINC01352 knockdown inhibits LUAD cell proliferation, suggesting that IOLs may serve as potential therapeutic targets for LUAD treatment.
This study aimed to construct a novel lncRNA prognostic risk signature combining oxidative stress and immunity to better predict the prognosis of LUAD patients. We comprehensively analyzed lncRNA profiles, combined them with oxidative stress and immune profiles, and identified their clinical importance in constructing a prognostic signature. Furthermore, we discussed the differences in immune status between different risk subgroups and clustering subtypes.
Materials and Methods
Dataset and Preprocessing
The design of this study was shown in the flow chart. Normal tissue from healthy lung tissue away from the tumor area is sufficient, and there are no specific exclusion criteria. RNA-Seq data of 490 tumor specimens and 59 normal specimens were obtained from the TCGA database in TPM format. Specifically, we utilized the log2 transformation in R software of the RNA-Seq data to stabilize variance and meet the assumptions of downstream statistical analyses. The clinical information for this cohort was shown in Table S1.
 |
Flow Chart. |
From previous investigations, 2498 immune-related genes (IRGs) and 809 oxidative stress-related genes (ORGs) were extracted. An overlapping set of 190 immune- and oxidative stress-associated genes (IOGs) was obtained by overlapping the two sets of genes. Differential expression analysis of the IOGs between normal and LUAD specimens was performed using the “limma” package (v3.50.3). The criteria for significance were set at |log2 fold alteration (FC)| ≥ 1 and p < 0.05. Pearson correlation analyses (p < 0.001, correlation coefficient > 0.4) were performed for the confirmed differentially expressed IOGs and each lncRNA. Finally, 658 immune- and oxidative stress-related lncRNAs (IOLs) were selected for subsequent bioinformatics analysis.
Unsupervised Clustering Analysis
To identify consistent clustering patterns based on the expression of prognosis-related IOLs, unsupervised analyses were conducted. The determination of the number of clusters was accomplished using the “ConsensusClusterPlus” (v1.58.0) package. In order to assess the stability of the clusters, 100 replicates with a plum value of 0.8 were performed, as described in previous works.18,19 The difference in OS among different clusters was evaluated using the Kaplan-Meier (K-M) curve. As for potential confounding factors, adjustments were made for relevant clinical variables, such as age, gender, tumor stage.
Establishment and Verification of Prognostic Signature
The TCGA-LUAD cohort underwent a random split in a 1:1 ratio. To establish a prognostic signature in the training cohort, the least absolute shrinkage and choice operator (LASSO) Cox regression analyses were employed for the elimination of redundant genes. The risk score was then determined by summing the expression values of all differentially expressed IOLs, along with their corresponding coefficients. Based on the median risk score, cases were categorized into either a high-risk or low-risk group. The prognostic performance of the risk signature was assessed using K-M analyses and time-dependent receiver operation feature (ROC) analyses in both the testing and training cohorts. For verification of the risk scores in the immunotherapy cohort, the IMvigor-210 cohort, previously described in the literature, was utilized and risk scores were computed using the identical formula.
Immune Analysis
Various algorithms, including EPIC, XCELL, MCP-counter, QUANTISEQ, CIBERSORT, and TIMER,20 were used to estimate immune cell abundance and relativity between different risk subgroups. To estimate the abundance of immune cells and evaluate immune-related functions, we employed the ssGSEA algorithm.21 This algorithm allowed us to assess various immune cell populations, including but not limited to activated B cells, activated CD4+ T cells, activated CD8+ T cells, central memory CD4+ T cells, central memory CD8+ T cells, and others.21 Furthermore, to determine the immune microenvironment status, we utilized the ESTIMATE algorithm. This algorithm enabled the calculation of essential parameters such as tumor purity, stromal score, and immune score. These scores provided insights into the composition of the tumor microenvironment and its immune-related characteristics.
Vitro Assays
Human LUAD cell lines A549 and PC9, as well as the normal human bronchial epithelial cell line BEAS-2B, were procured from the American Type Culture Collection (ATCC) located in Manassas, VA, USA. These cell lines were cultured in RPMI 1640 media supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Each assay was performed in triplicate to ensure statistical reliability. Transfection with siRNAs designed by GenePharma (Shanghai, China) was performed using jetPRIME® transfection reagent (Polyplus Transfection, China). Real-time qPCR (RT-qPCR) was conducted using an ABI 7500 instrument and a kit from TAKARA (Japan) to assess LINC01352 expression in cells and tissue. The cell proliferation assay was performed on transfected A549 and PC9 cells using cell counting kit-8 (MedChemExpress, China) and measuring OD values at a wavelength of 450 nm. We seeded LUAD cells in 96-well plates at a density of 2000 cells per well and incubated them. After 24, 48, 72 and 96 hours, we removed the medium from the wells and added fresh medium containing CCK-8 solution (10 uL/well). We then incubated the cells at 37°C for 2 hours to allow the CCK-8 solution to be absorbed by living cells. Subsequently, we measured the absorbance of each well at 450 nm using a microplate reader. The absorbance at 450nm is proportional to the number of living cells in each well. To ensure the objectivity of the results, the experiments were conducted in a blinded manner in our lab. The researchers performing the measurements were unaware of the sample identities, thus eliminating bias. The siRNAs sequences of LINC01352 were as follows: si-1, 5’-UUGGAUAGAUGUAAAGAGA-3’ and si-2, 5’-AAGGCAUAAGAUCAAGGAGAA-3’. The qRT-PCR results were analysed using the 2−ΔΔCT method. The primers sequences were as follows: LINC01352, F: 5’-GTTGTGTCTTCTCAGCTGCC-3’, and R: 5’-CAACCCTGAACGACACACAG-3’. GAPDH, F: 5’-GAAGGTGAAGGTCGGAGTCAACG-3’, R: 5’-TGCCATGGGTGGAATCATATTGG-3’.
Statistical Analysis
All statistical analyses were performed using R software (v.4.0.1). Pairwise comparisons were conducted using the Wilcoxon test, and we applied the Benjamini-Hochberg procedure to control the false discovery rate (FDR) at 5% in order to account for multiple comparisons in the context of pairwise comparisons. More details on the statistical methods used for transcriptome data processing can be found in the above section. P < 0.05 was considered statistically significant.
Results
Identification of IOLs
A total of 2498 IRGs and 809 ORGs were extracted from previous studies. A Venn diagram analysis was performed, which generated 190 IOGs (Figure 1a). Differential gene expression analysis of the 190 IOGs in the TCGA-LUAD cohort showed that 15 genes were up-regulated while 20 genes were down-regulated (Figure 1b). A total of 658 IOLs were confirmed using 35 differentially expressed IOLs and all annotated lncRNAs for correlation analysis (Figure 1c).
Construction of LncRNAs-Based Cluster
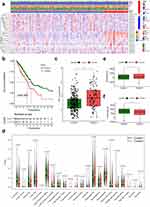
To identify prognosis-associated IOLs, univariate Cox regression analyses were performed after batch correction in the TCGA-LUAD cohort. Subsequently, 25 IOLs were screened for subsequent analysis (Figure 2a). The Wilcox detection showed that the 25 prognosis-associated IOLs were differentially expressed between LUAD and normal samples (Figure 2b). We used the R package “ConsensusClusterPlus” for consensus clustering analysis, when the number of clusters was 2, the clustering stability was the best (Figure 2d); therefore, we choose k = 2 to obtain two different clusters (C1: 390 individuals and C2: 100 individuals) (Figure 2c).
A composite heatmap of the different clusters and clinical information showed that the different clusters correlated with staging and grading (Figure 3a). K-M analysis (Figure 3b) demonstrated that the overall survival of cases in C1 was significantly better than that of cases in C2. Immune checkpoint inhibitors (ICIs) are very important treatments for LUAD patients in clinical practice; therefore, we evaluated whether different clusters were associated with ICI-related biomarkers. The outcomes exhibited cases in C1 featured higher PD-L1 expression and may have better efficacy (Figure 3c). We input the TCGA-LUAD expression matrix into the CIBERSORT algorithm and filtered the poor quality samples using p < 0.05. Finally, we found that in two cluster subtypes, NK cells resting, T cells CD4 memory activated, T cells CD4 memory resting, plasma cells, B cells naive, monocytes, mast cells activated and eosinophils infiltration were significantly different (Figure 3d). Moreover, the ESTIMATE algorithm showed that cases in C2 featured higher stromal scores (Figure 3e), while those in C1 featured higher immune scores (Figure 3f).
Construction and Validation of Risk Signature
To further minimize gene quantity in the signature, we randomly divided the TCGA-LUAD cohort (1:1) and conducted LASSO regression analysis on the 25 prognostic-associated IOLs in the training cohort (Figure 4a), as previously mentioned. Subsequently, 12 IOLs, along with their expression and regression coefficients, were identified and utilized to calculate the risk score for LUAD cases: 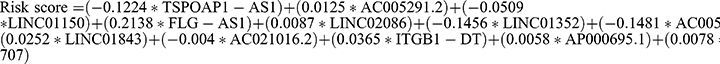 . Based on the median risk score of patients in the training cohort, cases were categorized into low and high-risk subgroups in each cohort. The AUC of the training cohort at one, three, and five years was 0.717, 0.742, and 0.677, respectively (Figure 4b), and the survival time of high-risk cases was significantly lower than that of low-risk cases (Figure 4c). In addition, the AUC of the testing cohort at one, three, and five years was 0.668, 0.635, and 0.633, respectively (Figure 4d), with the high-risk population exhibiting shorter survival times (Figure 4e).
. Based on the median risk score of patients in the training cohort, cases were categorized into low and high-risk subgroups in each cohort. The AUC of the training cohort at one, three, and five years was 0.717, 0.742, and 0.677, respectively (Figure 4b), and the survival time of high-risk cases was significantly lower than that of low-risk cases (Figure 4c). In addition, the AUC of the testing cohort at one, three, and five years was 0.668, 0.635, and 0.633, respectively (Figure 4d), with the high-risk population exhibiting shorter survival times (Figure 4e).
Analysis of Clinical Correlation
We plotted a composite heatmap between two risk subgroups and clinicopathological features in the TCGA-LUAD cohort, in which we found significant correlations between patients in different risk subgroups and staging, grading, T-stage, M-stage, gender, immune score, and cluster subtypes (Figure 5a). Stratification analyses were further performed by employing the clinicopathological features below: age (≥ 60 and < 60), gender, and tumor phase (I, III, and IV). The results indicated that the signature has prognostic significance between high- and low-risk patients. Cases in the high-risk group exhibited poorer OS than those in the low-risk group (Figure 5b–d). In sum, the outcomes testify the 12-IOLs risk signature is key to determining LUAD patient prognoses.
Immunity Analysis
To comprehensively explore the relationship between different subgroups and immune cell infiltration, we generated an immune cell infiltration heatmap using six algorithms (EPIC, XCELL, MCP-counter, QUANTISEQ, CIBERSORT, and TIMER) (Figure 6a). Interestingly, across the various algorithms, most of the tumor-infiltrating immune cells showed a negative correlation with the risk score. These findings suggest that the high-risk population is more likely to have a cold tumor state and a poor response to immunotherapy. To further investigate the link between the risk score and immune cells, we conducted a differential analysis and observed significant differences in the immune cell composition between the high and low-risk groups. Specifically, killer cells such as CD4+ T cells and CD8+ T cells were highly expressed in the low-risk group (Figure 6b). Meanwhile, we analyzed immune checkpoints in different risk groups and found that the low-risk group had higher mRNA expression (Figure 6c), which may suggested a better response to immunotherapy. In addition, the negative correlation between some IOLs involved in the risk signature with immune checkpoints explained the source of the above differences (Figure 6d). Finally, we validated our risk signature in the IMvigor-210 cohort with the same survival stratification ability (Figure S1a) and better immunotherapy corresponding to low-risk patients (Figure S1b and c).
Vitro Assays
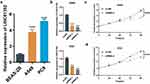
To identify the candidate molecule for cell function assays, we selected LINC01352. RT-qPCR analysis confirmed significant upregulation of LINC01352 in two LUAD cell lines (Figure 7a). Tow siRNAs were tested for transfection efficiency, with siRNA-2 demonstrating the highest efficiency (Figure 7b). The viability of A549 and PC9 cells was significantly suppressed after transfection with the corresponding siRNAs, as shown in the CCK-8 assay (Figure 7c and d).
Discussion
The high mortality rate of LUAD continues to pose a significant challenge for clinicians, necessitating the development of an effective and reliable prognostic model.22,23 Recent research has highlighted the role of oxidative stress in regulating tumor cell proliferation, invasion, and immune cell function, particularly immune cells involved in tumor metastasis.24,25 This suggests a potential link between oxidative stress patterns and antitumor immunity.26 However, previous studies mainly focused on constructing prognostic models using single oxidative stress or immune features, which often suffer from limitations in robustness and generalizability.27–30 In order to address these limitations, our study takes a comprehensive approach by incorporating both immune-related and oxidative stress-related lncRNAs. By encompassing multiscale clinical features, we aim to improve the accuracy and robustness of prognostic signatures.
Based on the expression of 25 prognosis-related IOLs in LUAD, patients were categorized into two molecular subtypes, with C1 exhibiting a better prognosis than C2. Furthermore, immune infiltration levels were examined between the two clusters, and C1 was found to be enriched in activated immune cells. These findings suggest that the higher infiltration levels of activated immune cells in C1 may partially explain the better prognoses of LUAD patients. Moreover, LUAD with distinct prognoses and immune microenvironments were better classified by the two IOLs subtypes, indicating that these IOLs may be associated with the formation of a complex immune microenvironment and an influence on LUAD progression. This evidence facilitates our understanding of the crosstalk between IOLs, immune cell infiltration, and LUAD progression. ICIs are very important treatments for LUAD patients in clinical practice; therefore, we evaluated whether different clusters were associated with ICI-related biomarkers. The outcomes exhibited cases in C1 featured higher PD-L1 expression and may have better efficacy. Moreover, the ESTIMATE algorithm exhibited that cases in C2 possessed higher stromal scores, while patients in C1 had higher immune scores.
Subsequently, to establish a clinically relevant prognostic signature, we employed a LASSO-Cox regression analysis to screen the 25 identified prognosis-related IOLs mentioned above. Through this analysis, we selected the optimal putative genes for signature creation. Following rigorous validation, we successfully developed a novel prognostic signature consisting of 12 genes: TSPOAP1-AS1, AC005291.2, LINC01150, FLG-AS1, LINC02086, LINC01352, AC005884.1, LINC01843, AC021016.2, ITGB1-DT, AP000695.1, and LINC00707. Utilizing this signature, we were able to classify LUAD patients into high-risk and low-risk categories, which corresponded to unfavorable and favorable prognoses, respectively, in both the training and testing cohorts. Notably, the AUC values of ROC curves demonstrated the outstanding predictive performance of the developed signature. To our knowledge, this is the first lncRNA prognostic risk signature that combines oxidative stress and immunity.
To identify the candidate molecule for cell function assays, we selected LINC01352. The upregulation of LINC01352 was confirmed through real-time qPCR analysis. Of the two siRNAs tested, siRNA-2 showed higher transfection efficiency. The viability of A549 and PC9 cells, LUAD cell lines, was significantly reduced following transfection with these siRNAs, as demonstrated in the CCK-8 assay. It was found that the long non-coding RNA LINC01352 is significantly downregulated in HCC cells and clinical samples by the HBV X protein (HBx). This downregulation of LINC01352, which acts as an endogenous sponge, increases the expression of miR-135b (32157216). This leads to activate Wnt/β-catenin signaling and facilitating tumor progression. Moreover, in AML, high expression of LINC01352 was observed in leukemia stem cells (LSCs) and correlated with poor patient prognosis (34295821).
Tumor progression and response to immunotherapy have been increasingly recognized to be closely tied to immune infiltration.31 In the context of our IOLs signature, we observed a significant association between the signature and immune functions, immune cell infiltration, and immune checkpoint expression. Notably, oxidative stress within the tumor microenvironment has been reported to recruit immune cells.24 Analyses revealed a negative correlation between the risk score derived from our signature and the abundance of tumor-infiltrating immune cells, indicating that higher risk scores were associated with immunosuppression. These findings suggest that the high-risk population is more likely to have a cold tumor state and a poor response to immunotherapy. Similar results were obtained for differential analysis, where killer cells such as CD4+ T cells and CD8+ T cells were lowly expressed in the high-risk group. These findings suggest that immune suppression is a key factor contributing to poorer prognoses observed in high-risk patients.
Immunotherapy strategies targeting immune checkpoints, such as the PD-1/PD-L1 pathway, have emerged as promising approaches for the treatment of LUAD.32 However, only a small proportion of LUAD patients, approximately less than 10%, exhibit long-term responses to anti-PD-1 immunotherapy, and these responders generally have improved survival outcomes compared to nonresponders.33 Unfortunately, there remains a scarcity of reliable markers for predicting immunotherapeutic outcomes.34 In this study, we compared the levels of 41 immune checkpoints between the low- and high-risk groups identified by our prognostic signature. Interestingly, we found that the low-risk group showed higher expression of most immune checkpoints. This finding suggests a greater potential for patients at low risk to benefit from immunotherapy. It is noteworthy that the expression levels of the majority of immune checkpoint genes analyzed were inversely correlated with the risk score derived from our prognostic signature. These findings underline the potential utility of our signature in improving the identification of LUAD patients who may have a favorable response to immunotherapy. Encouragingly, we validated our risk signature in the IMvigor-210 cohort with the same survival stratification ability and better immunotherapy corresponding to low-risk patients. These results suggest that patients with a low risk score, as determined by our IOLs signature, may exhibit increased sensitivity to immunotherapy compared to those with a high risk score. Therefore, implementing risk stratification based on our signature could potentially provide significant benefits for metastatic LUAD patients by accurately identifying individuals who are more likely to respond positively to immunotherapy.
However, it is important to acknowledge the limitations of our study. Firstly, our study design was retrospective in nature, and therefore, prospective experiments and clinical validation using larger sample sizes are necessary to further establish the validity of our prognostic signature. Secondly, although our bioinformatic analyses yielded robust results, further experimental evidence is needed to elucidate the regulatory networks and underlying mechanisms of the IOLs in LUAD. Thirdly, while our findings indicate a therapeutic potential for the low-risk group through ICI-based immunotherapy, further exploration is warranted to determine the optimal treatment protocols for these patients. Moreover, while bioinformatics algorithms involving immunology can provide valuable insights into the overall immune cell landscape, the estimation results should be interpreted with caution and validated using complementary experimental techniques such as immunohistochemistry or flow cytometry.
Conclusion
In conclusion, our study has provided novel insights into the relationship between IOLs and LUAD. We have successfully developed a prognostic signature based on 12 carefully selected IOLs, demonstrating its excellent predictive performance in LUAD patients. This signature holds promise for identifying cold and hot tumors, optimizing immune surveillance status, and potentially guiding individualized therapeutic interventions aimed at enhancing the survival outcomes of LUAD patients. Overall, our findings have important implications for advancing personalized treatment strategies in the management of LUAD.
Study Approval
All datasets in the present study were downloaded from public databases. These public databases allowed researchers to download and analyze public datasets for scientific purposes. The current research follows the TCGA data access policies and publication guidelines. Users can download relevant data for free, our study is based on open-source data, there are no ethical issues and other conflicts of interest. Inclusion of the Human cell lines in the study were approved by The Second Hospital of Hebei Medical University review board.
Acknowledgments
We would like to acknowledge reviewers and editors for their helpful comments on this paper.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Hebei Provincial Government sponsored the project of training excellent talents in clinical medicine (No. 303-2022-27-37 to Shujun Li) and Hebei Medical Research Project Plan (No. 20210373 to Xin Liu).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398(10299):535–554. doi:10.1016/S0140-6736(21)00312-3
3. Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol. 2021;33(1):40–46. doi:10.1097/CCO.0000000000000703
4. Brody H. Lung cancer. Nature. 2020;587(7834):S7. doi:10.1038/d41586-020-03152-0
5. Singh T, Fatehi Hassanabad M, Fatehi Hassanabad A. Non-small cell lung cancer: emerging molecular targeted and immunotherapeutic agents. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188636. doi:10.1016/j.bbcan.2021.188636
6. Gupta RK, Patel AK, Shah N, et al. Oxidative stress and antioxidants in disease and cancer: a review. Asian Pac J Cancer Prev. 2014;15(11):4405–4409. doi:10.7314/apjcp.2014.15.11.4405
7. Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. doi:10.1038/nrd4002
8. Sosa V, Moliné T, Somoza R, Paciucci R, Kondoh H, LLeonart ME. Oxidative stress and cancer: an overview. Ageing Res Rev. 2013;12(1):376–390. doi:10.1016/j.arr.2012.10.004
9. Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi:10.1016/j.freeradbiomed.2010.09.006
10. Zhao F, Li Y, Dong Z, et al. Identification of a risk signature based on lactic acid metabolism-related LncRNAs in patients with esophageal squamous cell carcinoma. Front Cell Dev Biol. 2022;10:845293. doi:10.3389/fcell.2022.845293
11. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi:10.1038/s41580-020-00315-9
12. Zhang L, Peng D, Sood AK, Dang CV, Zhong X. Shedding light on the dark cancer genomes: long noncoding RNAs as novel biomarkers and potential therapeutic targets for cancer. Mol Cancer Ther. 2018;17(9):1816–1823. doi:10.1158/1535-7163.MCT-18-0124
13. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177–198. doi:10.1146/annurev-immunol-041015-055459
14. Kim C, Kang D, Lee EK, Lee JS. Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxid Med Cell Longev. 2017;2017:2062384. doi:10.1155/2017/2062384
15. Fuschi P, Carrara M, Voellenkle C, et al. Central role of the p53 pathway in the noncoding-RNA response to oxidative stress. Aging. 2017;9(12):2559–2586. doi:10.18632/aging.101341
16. Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi:10.1126/science.1112014
17. Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–19. doi:10.1101/gr.4200206
18. Ma S, Wang Y, Li W, et al. Integrated analysis identities Rho GTPases related molecular map in patients with gastric carcinoma. Sci Rep. 2023;13(1):21443. doi:10.1038/s41598-023-48294-z
19. Ye F, Hu Y, Gao J, et al. Radiogenomics map reveals the landscape of m6A methylation modification pattern in bladder cancer. Front Immunol. 2021;12:722642. doi:10.3389/fimmu.2021.722642
20. Sturm G, Finotello F, Petitprez F, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35(14):i436–i445. doi:10.1093/bioinformatics/btz363
21. Jia Q, Wu W, Wang Y, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9(1):5361. doi:10.1038/s41467-018-07767-w
22. Deng X, Xiong W, Jiang X, et al. LncRNA LINC00472 regulates cell stiffness and inhibits the migration and invasion of lung adenocarcinoma by binding to YBX1. Cell Death Dis. 2020;11(11):945. doi:10.1038/s41419-020-03147-9
23. Pan J, Fang S, Tian H, et al. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer. 2020;19(1):9. doi:10.1186/s12943-020-1133-9
24. Kuo CL, Ponneri Babuharisankar A, Lin YC, et al. Mitochondrial oxidative stress in the tumor microenvironment and cancer immunoescape: foe or friend? J Biomed Sci. 2022;29(1):74. doi:10.1186/s12929-022-00859-2
25. Wang X, Xu D, Chen B, et al. Delicaflavone represses lung cancer growth by activating antitumor immune response through N6-methyladenosine transferases and oxidative stress. Oxid Med Cell Longev. 2022;2022:8619275. doi:10.1155/2022/8619275
26. Bhattacharyya S, Saha J. Tumour, oxidative stress and host T cell response: cementing the Dominance. Scand J Immunol. 2015;82(6):477–488. doi:10.1111/sji.12350
27. Wang Y, Xu J, Fang Y, et al. Comprehensive analysis of a novel signature incorporating lipid metabolism and immune-related genes for assessing prognosis and immune landscape in lung adenocarcinoma. Front Immunol. 2022;13:950001. doi:10.3389/fimmu.2022.950001
28. Wang H, Tian RF, Liang X, et al. A four oxidative stress gene prognostic model and integrated immunity-analysis in pancreatic adenocarcinoma. Front Oncol. 2023;12:1015042. PMID: 36713541; PMCID: PMC9880292. doi:10.3389/fonc.2022.1015042
29. Liu X, Zhang X, Liu C, Mu W, Peng J, Song K. Immune and inflammation: related factor alterations as biomarkers for predicting prognosis and responsiveness to PD-1 monoclonal antibodies in cervical cancer. Discov Oncol. 2022;13(1):96. doi:10.1007/s12672-022-00560-8
30. Zhang D, Lu W, Zhuo Z, Mei H, Wu X, Cui Y. Construction of a breast cancer prognosis model based on alternative splicing and immune infiltration. Discov Oncol. 2022;13(1):78. doi:10.1007/s12672-022-00506-0
31. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 2020;17(8):807–821. doi:10.1038/s41423-020-0488-6
32. Zhou F, Qiao M, Zhou C. The cutting-edge progress of immune-checkpoint blockade in lung cancer. Cell Mol Immunol. 2021;18(2):279–293. doi:10.1038/s41423-020-00577-5
33. Rebuzzi SE, Leonetti A, Tiseo M, Facchinetti F. Advances in the prediction of long-term effectiveness of immune checkpoint blockers for non-small-cell lung cancer. Immunotherapy. 2019;11(12):993–1003. doi:10.2217/imt-2019-0107
34. Lahiri A, Maji A, Potdar PD, et al. Lung cancer immunotherapy: progress, pitfalls, and promises. Mol Cancer. 2023;22(1):40. doi:10.1186/s12943-023-01740-y
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.