Back to Journals » International Journal of Nanomedicine » Volume 18
A Progressively Disassembled DNA Repair Inhibitors Nanosystem for the Treatment of BRCA Wild-Type Triple-Negative Breast Cancer
Authors Fang W , Wang J, Ma X, Shao N, Ye K, Zhang D, Shi C, Luo L
Received 10 July 2023
Accepted for publication 5 October 2023
Published 24 October 2023 Volume 2023:18 Pages 6001—6019
DOI https://doi.org/10.2147/IJN.S426639
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Weimin Fang,1,2,* Jinghao Wang,3,* Xiaocong Ma,1,2 Ni Shao,1,2 Kunlin Ye,1,2 Dong Zhang,1,2 Changzheng Shi,1,2 Liangping Luo1,2
1Medical Imaging Center, the First Affiliated Hospital of Jinan University, Guangzhou, Guangdong, People’s Republic of China; 2The Guangzhou Key Laboratory of Molecular and Functional Imaging for Clinical Translation, Jinan University, Guangzhou, Guangdong, People’s Republic of China; 3Department of Pharmacy, the First Affiliated Hospital of Jinan University, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Changzheng Shi, Medical Imaging Center, The Guangzhou Key Laboratory of Molecular and Functional Imaging for Clinical Translation, The First Affiliated Hospital of Jinan University, Guangzhou, Guangdong, People’s Republic of China, Email [email protected] Liangping Luo, Medical Imaging Center, The Guangzhou Key Laboratory of Molecular and Functional Imaging for Clinical Translation, The First Affiliated Hospital of Jinan University, Guangzhou, Guangdong, People’s Republic of China. Department of Radiology, The Fifth Affiliated Hospital of Jinan University, Guangzhou, Guangdong, People’s Republic of China, Email [email protected]
Background: Olaparib, a poly (adenosine diphosphate-ribose) polymerase (PARP) inhibitor has demonstrated promising efficacy in patients with triple-negative breast cancer (TNBC) carrying breast cancer gene (BRCA) mutations. However, its impact on BRCA wild-type (BRCAwt) TNBC is limited. Hence, it is crucial to sensitize BRCAwt TNBC cells to olaparib for effective clinical practice. Novobiocin, a DNA polymerase theta (POLθ) inhibitor, exhibits sensitivity towards BRCA-mutated cancer cells that have acquired resistance to PARP inhibitors. Although both of these DNA repair inhibitors demonstrate therapeutic efficacy in BRCA-mutated cancers, their nanomedicine formulations’ antitumor effects on wild-type cancer remain unclear. Furthermore, ensuring effective drug accumulation and release at the cancer site is essential for the clinical application of olaparib.
Materials and Methods: Herein, we designed a progressively disassembled nanosystem of DNA repair inhibitors as a novel strategy to enhance the effectiveness of olaparib in BRCAwt TNBC. The nanosystem enabled synergistic delivery of two DNA repair inhibitors olaparib and novobiocin, within an ultrathin silica framework interconnected by disulfide bonds.
Results: The designed nanosystem demonstrated remarkable capabilities, including long-term molecular storage and specific drug release triggered by the tumor microenvironment. Furthermore, the nanosystem exhibited potent inhibitory effects on cell viability, enhanced accumulation of DNA damage, and promotion of apoptosis in BRCAwt TNBC cells. Additionally, the nanosystem effectively accumulated within BRCAwt TNBC, leading to significant growth inhibition and displaying vascular regulatory abilities as assessed by magnetic resonance imaging (MRI).
Conclusion: Our results provided the inaugural evidence showcasing the potential of a progressively disassembled nanosystem of DNA repair inhibitors, as a promising strategy for the treatment of BRCA wild-type triple-negative breast cancer.
Keywords: olaparib, novobiocin, tumor microenvironment, disulfide bonds, drug release, magnetic resonance
Introduction
Breast cancer is the leading cause of morbidity and mortality in women worldwide, accounting for 30% of new cancer cases and 15% of deaths.1,2 Triple-negative breast cancer (TNBC) is a highly heterogeneous breast cancer type characterized by a lack of hormone receptors and a poor clinical prognosis.3,4 Although chemotherapy is still the first-line treatment for most patients with TNBC, adverse side effects limit its application.5,6 In addition, about 10–15% of TNBC patients have mutations in BRCA genes, which also seem to be one of the driving factors of TNBC.7,8 Meanwhile, BRCA-regulated DNA homologous recombination (HR) repair is often absent or lost in cancer types with BRCA mutations, which are sensitive to some DNA repair inhibitors.9–11
In the realm of combatting TNBC, a cohort of highly selective DNA repair inhibitors has emerged as efficacious agents for the precise modulation of DNA repair signaling pathways.12 Cancer cells with BRCA mutations is the detriment to the HR DNA repair pathway, resulting in insufficient repair of double-strand breaks (DSB). Within this context, it is noteworthy that members of the poly (ADP-ribose) polymerase (PARP) family have been identified as pivotal participants in the DNA damage response (DDR).13,14 Of paramount significance, PARP inhibitors have risen to prominence as one of the most promising and innovative strategies in the realm of targeted cancer therapy. These inhibitors can induce synthetic lethality in HR repair deficient cells, offering an efficacious means to impede the proliferation and survival of cancer cells.15 Olaparib, the first PARP inhibitor approved for BRCA-mutated breast cancer, has shown promising efficacy in patients with metastatic breast cancer or germline BRCA-mutated breast cancer, including some TNBC patients.16,17 However, the overall mutation rate of BRCA1/2 genes in TNBC patients is low and the monotherapy application of PARP inhibitors can develop drug resistance.18,19 Besides, PARP inhibitors can promote the repair failure of DSB to induce cancer cell death in breast cancers with BRCA mutations, which also show modest activity in BRCAwt breast cancer.20,21 Although olaparib showed great promise for the treatment of BRCA-mutant cancers, more than 40% of BRCA-mutant patients do not respond to PARP inhibitors or develop resistance after treatment.22,23 Furthermore, the skilled HR repair present in BRCAwt TNBC is naturally insensitive to PARP inhibitors.24,25 Currently, several research strategized that sensitize BRCAwt TNBC to PARP inhibitors by modulating the DDR, such as inactivation of the prolyl-isomerase, inhibition of DDR proteins or enhancer of zeste homologue 2 (EZH2).26–28 Furthermore, recent studies indicated that depletion of the DNA polymerase theta (POLθ) can kill BRCA1/2 gene-dominated HR deficient tumors and has a synergistic effect on PARP inhibitors.29–31 Therefore, there is still a great need to develop strategies to increase the sensitivity of PARP inhibitors which has important therapeutic implications for the treatment of BRCAwt TNBC.
Nanomedicine has made substantial strides in advancing cancer treatment through its ability to enable precise drug delivery to tumor sites, concurrently reducing side effects. Within the domain of cancer therapeutic diagnostics, nanocarriers designed for drug delivery and targeted therapies have garnered considerable interest.32,33 Furthermore, rapid nanoscale drug delivery systems (NDDS) development can exhibit high stability under physiological conditions, multifunctionality and suitable biocompatibility trends.28,34,35 At the same time, drug delivery into cells must be efficiently loaded by NDDS, not leaked before being transported into the cell, and a reasonable drug release rate.36,37 Silica is an NDDS with better storage performance, which can maintain the internal drug for a long time without releasing it.38–40 Some suitable modifications to the linkage between the surfaces of the NDDS enable the acidification and redox degradation strategies of cells, such as triggering redox-responsive degradation behavior of disulfide bonds and glutathione (GSH).41–44 Compared with traditional NDDS, the degradable disulfide bridged NDDS exhibited a faster drug release rate and could be degraded under reducing conditions. Considering the notable disparity in GSH concentrations between tumor extracellular and intracellular fluids, coupled with the coexistence of low pH and high GSH concentrations within the cytoplasm of tumor cells, the utilization of disulfide bond oxidation-responsive stimulation holds distinct advantages for TNBC.45–47 The unique advantages of redox gating-based NDDS are expected to be promising vehicles for the delivery of PARP inhibitors to BRCAwt TNBC. Despite the resistance of BRCAwt TNBC to olaparib, we can take advantage of NDDS as a new strategy to affect the sensitivity of olaparib to BRCAwt TNBC.
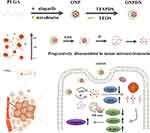
Herein, we designed a tumor microenvironment (TME) responsive progressively disassembled nanosystem (ONPDN) that can synergistically deliver two DNA repair inhibitors via a two-step approach (Scheme 1). Olaparib and novobiocin were co-loaded into polylactic-co-glycolic acid (PLGA) vesicles as the core of the nanosystem (ONP). Subsequently, an ultrathin silica framework with disulfide bond bridges was constructed on the surface of PLGA. This chemically gated ultrathin silica framework stored encapsulated molecules well until the disulfide bonds were reduced. The ultrathin silica framework was selectively disassembled with GSH and acidic pH responses. ONPDN exerted the synergistic anticancer effects of two DNA repair inhibitors and exhibited considerable anticancer effects in the BRCAwt TNBC xenograft nude mice model. Additionally, the accumulation capacity and vasomodulatory effect of the nanosystem at the tumor site were monitored by Magnetic Resonance Imaging (MRI). We analyzed the therapeutic effect of ONPDN and explored the synergistic effect between olaparib and novobiocin. In summary, we revealed the great potential of the progressively disassembled nanosystem to enhance the efficacy of olaparib in the treatment of BRCA wild-type TNBC.
Materials and Methods
Preparation of ONP
70 mg of poly (lactic-co-glycolic acid) (PLGA) (Aladdin, China), along with 20 mg of olaparib (Aladdin, China), and 20 mg of novobiocin (Aladdin, China) were completely dissolved in 10 mL of acetone. The above solution was slowly added to 40 mL of ultrapure water containing 0.2 g of cetyltrimethylammonium bromide (CTAB) (Macklin, China) while being gently shaken during the dropwise addition. A white emulsion was gradually formed as these solutions were mixed. The resulting white emulsion was subjected to ultrasonication using an ultrasonic disruptor operating at 600 W (ultrasonic for 2 seconds, stopped for 1 second) for a total duration of 10 minutes, all while placed in an ice bath. Subsequently, the mixture was left in a fume hood at room temperature for 24 hours to allow complete acetone evaporation. Following this, the mixture was centrifuged at 8000 rpm for 15 minutes to collect the precipitate, which was then washed 3 times with ultrapure water to collect ONP.
Preparation of ONPDN
The prepared ONP was dispersed into 60 mL ultrapure water under an ultrasonic bath. The above solution was added with 4 mL of isopropanol and 1.5 mL of 25% ammonia water after stirring at 200 rpm for 2 hours. Then 0.2 mL tetraethyl orthosilicate (TEOS) (Aladdin, China), 0.05 mL 3-Aminopropyltriethoxysilane (APTES) (Aladdin, China), and 0.05 mL bis(triethoxysilylpropyl) disulfide (TESPDS) (Aladdin, China) were added to the solution and stirred for 12 hours. The solution was subsequently centrifuged at 12,000 rpm for 10 minutes to gather the precipitate, and this precipitate underwent three washes with ultrapure water to yield the final ONPDN. Furthermore, TESPDS was excluded from the procedure for ONNP preparation. The protocols for generating OPDN and ONPDN share similarities, differing primarily in the absence of novobiocin during the vesicle preparation stage. Notably, the preparation of OPDN does not involve the inclusion of olaparib in the vesicle generation process. PDN, on the other hand, is derived from vesicles devoid of DNA repair inhibitors.
Characterization
Morphological analysis of ONPDN conducted using a transmission electron microscope (TEM) (JEM 2100F, JEOL, Japan). The zeta potential of the nanoparticles was determined with a Zetasizer Nano-ZS (Malvern, UK). The TEM was equipped with energy dispersive X-ray spectroscopy (EDS) for elemental analysis. The encapsulation efficiency and loading capacity of olaparib and novobiocin were measured by ultraviolet-visible spectrophotometer (UV-vis) (UV-2700, HIMADZU, Japan). The loading amount was calculated by drug content of nanosystem and the loading rate was calculated by comparing it to the initial addition of free drug.
Drug Storage and TME Responsive Release of ONPDN
To assess the drug storage capacity of ONPDN, 500 mg of ONPDN was added to phosphate buffered saline (PBS) at pH=7.4. The solution containing the nanoparticles was placed in a shake with 80 rpm and maintained at 37°C. At various time points (0, 1, 2, 3, 5, and 7 days), 300 μL of supernatant was collected and replenished with the corresponding volume of PBS. The supernatant was subjected to UV-vis absorbance at 256nm and 312nm. To evaluate the TME responsive release of ONPDN, 500 mg of ONPDN was added to 0, 5, and 10 nM GSH solutions at pH=5.8. Then solutions were placed in a shaker at 37 °C, 80 rpm. Each 300 μL of supernatant was collected after 0, 0.5, 1, 2, and 6 hours and the corresponding volume of GSH solution was added to the original solution. The supernatant was detected by UV-vis absorbance at 256 nm and 312 nm.
Cells and Animals
These cells were cultivated in either Dulbecco’s Modified Eagle Medium (DMEM) or Roswell Park Memorial Institute (RPMI) 1640 medium, both sourced from Gibco, USA. The culture media, DMEM and RPMI 1640, were supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 50 units/mL of streptomycin and 100 units/mL of penicillin. Cell cultures were maintained in an incubator set at 37°C, with a controlled atmosphere of 5% CO2 and a relative humidity of 95%. Female BALB/c nude mice, aged 3–4 weeks, were obtained from Vital River Laboratories, China. Ethical approval for all animal experiments was granted by the Ethics Committee of Jinan University, and the study adhered to the “Guidelines for Ethical Review of Laboratory Animal Welfare” (GB/T 35892–2018) to ensure the well-being of all animals involved in the research.
Cell Viability Assay
Cells were seeded in 96-well plates at a density of 2×103 cells per well and cultured for 24 hours. Subsequently, the culture medium was replaced with a drug-containing medium in each well (100 μL per well). For the free drug group, the ratio of olaparib and novobiocin (ON) was adjusted to match the ODPDN loading ratio, which was 2:1. After a 48-hour incubation, 30 μL of MTT solution (5 mg/mL) was introduced to each well and allowed to incubate for 3 hours. Following this, the upper-layer solution was aspirated from the 96-well plate, and 150 μL of dimethyl sulfoxide (DMSO) was added to each well. The absorbance at 570 nm was determined using a multifunctional microplate reader.
Cellular Uptake Assay
MDA-MB-231 cells (2×105 cells/well) were seeded in a 2 cm confocal dish and cultured for 24 hours. 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (Sigma–Aldrich, USA) was used to stain cell nuclei. Then free coumarin 6 and ONPDN (labeled by coumarin 6) were added. The coumarin 6 concentration of the two groups was 5 μg/mL. Pictures were taken by fluorescence microscopy (LSM 880, Zeiss, Germany) after 60 minutes.
Reactive Oxygen Species (ROS) Detection
MDA-MB-231 cells (2×104 cells/well) were cultured in 96-well plates for 24 hours. Afterward, the supernatant was replaced with 100 μL of dichloro-dihydro-fluorescein diacetate (DCFH-DA) probe (10 μM) (Sigma-Aldrich, USA). The cells were then incubated for 30 minutes at 37°C. Subsequently, different drug groups, including PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), or ONPDN (G6), were added to the plate. The concentration of olaparib in all groups was 20 μg/mL, while novobiocin was at 10 μg/mL. The change in fluorescence over a 2-hour period was recorded using a microplate reader (Excitation (Ex) = 488nm, emission (Em) = 525nm). Additionally, fluorescence images of the various groups were captured using confocal laser microscopy after 2 hours.
The Gamma-H2A Histone Family Member X (γ-H2AX) Detection
MDA-MB-231 cells (1×105 cells/well) were cultured in confocal dishes for 24 hours. The supernatant was replaced with a complete medium containing PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), or ONPDN (G6), with the concentration of olaparib set at 20 μg/mL and novobiocin at 10 μg/mL. Following a 2-hour incubation, the cells were washed with PBS, and then fixed and washed three times. A blocking reagent was added for 20 minutes. The cells were subsequently incubated with a γ-H2AX antibody (Abcam, UK) for 1 hour, followed by the addition of anti-rabbit 488 (Abcam, UK). Finally, images were observed and captured using a laser confocal fluorescence microscope.
Cell Cloning Test
MDA-MB-231 cells (500 cells/well) were cultured in 6-well plates for 24 hours. Subsequently, the supernatant was replaced with a complete medium containing PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), or ONPDN (G6), with the concentration of olaparib set at 20 μg/mL and novobiocin at 10 μg/mL. After 24 hours, the supernatant was replaced with fresh medium every 3 days. Cells were stained with crystal violet after 14 days, and pictures of the cell colonies were captured and the number of cell colonies was analyzed using ImageJ.
Dead-Live Cell Staining Assay
MDA-MB-231 cells (2×105 cells/well) were cultured in 6-well plates for 24 hours. Subsequently, the supernatant was replaced with a complete medium containing PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), or ONPDN (G6), with the concentration of olaparib set at 20 μg/mL and novobiocin at 10 μg/mL. After 24 hours, the medium was removed, and the cells were washed three times with PBS. A staining solution containing 2 μM of Calcein-AM and 4.5 μM of pyridinium iodide (PI) (Sigma-Aldrich, USA) was added to the plate, followed by incubation in the dark for 15 minutes. Finally, a confocal laser microscope was used to observe the cells (Ex=490/535nm, Em=515/617nm).
Magnetic Resonance Imaging (MRI)
A clinical 3T MRI scanner (Discovery 750, GE Medical, USA) equipped with a wrist coil was used for routine and functional serial scans, including R2* imaging, T2 weighted imaging (T2WI), and intravoxel incoherent motion (IVIM) - diffusion weighted imaging (DWI). Eight nude mice with a tumor volume of 150 mm3 were used for R2* imaging. Prior to scanning, the tail veins of the nude mice were injected with either free Fe3O4 or ONPDN (labeled with Fe3O4) at a dosage of 10 mg Fe/kg. Abdominal anesthesia using 2% pentobarbital sodium solution (40 mg/kg) was administered to the mice. R2* imaging of the tumor area was conducted at 2, 6, 12, 24, and 48 hours post-administration, with the following scanning sequences: Time to echo (TE) = 3.4, 9.3, 15.2, 21.2, 27.1, 33, 38.9, 44.8, 50.7, 56.6, 62.5, 68.5, 72.4, 80.3, 86 ms. Repetition time (TR) = 3.4, 9.3, 86.2 ms, scanning field of view (FOV) = 7×6 cm2, layer thickness/distance = 2.0/0.2, matrix size= 192×128, and number of excitations (NEX) = 2. R2* raw data were processed using the GE adw4.5 post-processing workstation and the Functool software package for region of interest (ROI) analysis.
T2WI and IVIM-DWI scans were conducted at the conclusion of the nude mice treatment. The scanning sequences and parameters were as follows: Fast spin echo (FSE) sequences were employed for T2WI, with TR = 4500 ms, TE = 60 ms, matrix = 96×128, slice thickness = 2.0 mm, slice distance = 0.2 mm, FOV = 7.0 cm × 5.0 cm, and NEX = 4. For IVIM-DWI, a multiple b-value model was utilized, encompassing 13 b values: 0, 25, 50, 75, 100, 150, 200, 400, 600, 800, 1000, 1200, and 1500 s/mm2. The NEX corresponding to each b value was set at 1, 2, 2, 3, 3, 3, 4, 4, 4, 6, 6, 8, and 10, with diffusion gradients applied along the X, Y, and Z axes axially.
Single-shot, echo-planar imaging pulse sequences were employed in conjunction with chemical shift-selective saturated fat suppression techniques. TR/TE = 3000/101.7 ms, matrix = 128×96, layer thickness = 2.0 mm, layer spacing = 0.2 mm and FOV=10 cm × 8.0 cm.
The relevant parameters of IVIM-DWI, including the slow apparent diffusion coefficient (ADC), fast ADC, and the fraction of fast ADC, were analyzed using the GE adw4.5 post-processing workstation. Pseudo-color images were generated using the Functool package, with T2WI images and b-value = 0 images serving as reference images for ROI measurements in IVIM-DWI. Measurements from all tumor section levels were averaged for analysis.
Evaluation of ONPDN Antitumor Efficacy in vivo
MDA-MB-231 cells in the logarithmic growth phase were harvested and prepared as a cell suspension at a concentration of 3×107 cells/mL using PBS. Subsequently, nude mice were injected with 100 μL of the cell suspension beneath the right fourth pair of mammary glands and were regularly monitored in a specific pathogen-free environment. When the tumor volume of nude mice reached 50–100 mm3, a total of 48 nude mice were randomly divided into six groups. Out of these, 24 nude mice were utilized for treatment observation, MRI evaluation, and were ultimately sacrificed at the end of the treatment regimen. The remaining 24 nude mice were observed for survival analysis.
The different groups of nude mice received injections of 200 μL of various substances, including PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), or ONPDN (G6). These injections were administered every 3 days (on days 0, 3, 6, 9, and 12). The concentration of olaparib was maintained at 20 mg/kg, and novobiocin was administered at 10 mg/kg. Throughout the treatment period, the body weight and tumor volume of the nude mice were measured and recorded every 3 days. Tumor volume (V) was calculated using the formula V = (L × W2)/2 (L = tumor long diameter, W = tumor wide diameter). The cut-off point for survival curves was plotted when the tumor grew to 2000 mm3 or the vital signs of nude mice were abnormal. On day 15 of the treatment cycle, the nude mice were sacrificed after inhaling excessive CO2. Tissue samples from the tumors, heart, lung, liver, kidney, and spleen of nude mice were extracted, fixed in 4% paraformaldehyde, and subsequently used for either H&E staining or immunofluorescence staining.
Hematoxylin and Eosin Stain (H&E) Staining and Immunofluorescence Staining Analysis
The immunofluorescent staining protocol for tumor tissue involved several steps: Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) staining was utilized to detect tumor cell apoptosis. Ki-67 immunofluorescent staining was employed to assess tumor proliferation. PARP immunofluorescence staining was conducted to evaluate PARP expression. Dihydroethidium (DHE) immunofluorescence staining was utilized to evaluate ROS expression. CD31 immunofluorescence staining was performed to assess microvessel density. γ-H2AX immunofluorescent staining was used to determine the extent of DNA damage accumulation.
Cleaved caspase-3 (clCasp3) immunofluorescence staining was employed to evaluate the expression of clCasp3 apoptotic protein. All reagents required for H&E staining and immunofluorescence staining of selected sections were procured from Servicebio, China. Fluorescence microscopy was used to visualize histopathology, and images were captured in areas with positive staining. Subsequently, the measurement and calculation of the positive expression area in the immunofluorescence images were performed using ImageJ.
Hematology Analysis
Sera were obtained from the orbital venous blood of the nude mice and subsequently utilized for blood analysis. The parameters that were measured include albumin (ALB), urea nitrogen (BUN), creatine kinase (CK), lactate dehydrogenase (LDH), creatinine (CREA), and aspartate aminotransferase (AST).
Statistical Analysis
Statistical analysis and graph generation were conducted using the statistical software packages SPSS 22.0 (USA) and Prism 8.0 (GraphPad, USA). To determine the statistical significance between groups, Student’s t-test and one-way analysis of variance (ANOVA) were employed. Kaplan-Meier survival analysis was utilized to assess the survival rate of the nude mice. All quantitative measurements were conducted in triplicate at minimum. Numerical results are presented as mean ± standard deviation (SD). Statistical significance was established at a P-value <0.05 (* indicating P<0.05, ** indicating P<0.01, and *** indicating P<0.001).
Results and Discussion
Synthesis and Characterization of ONPDN
In this study, we employed the nanoprecipitation method to prepare PLGA nanoparticles co-loaded with olaparib and novobiocin, denoted as ONP (Figure 1A). Subsequently, we synthesized ONP encapsulated within an ultrathin silica framework, forming ONPDN, where disulfide bond bridges were introduced onto the surface of ONP. The morphology of ONPDN, as illustrated in Figure 1B, exhibited a characteristic core-shell structure when examined by TEM. This observation confirmed the presence of the anticipated ultrathin outer silica framework. ImageJ results revealed that the average thickness of the silicon dioxide coating on ONNP (without disulfide bonds) measured approximately 8nm. This dimension was related to the initial particle size of ONP (182nm). Importantly, the introduction of disulfide bonds did not significantly alter the particle size of ONPDN (191 nm) (Figure 1C). Further characterization via Zetasizer indicated a change in the zeta potential of ONPDN, increasing from −24.92 mV (for ONP) to approximately −11.85 mV. This finding served as evidence of the successful modification of the silica shell, now containing disulfide bonds (Figure 1D). Analysis of the TEM-EDS elemental spectrum confirmed that silicon (Si), oxygen (O), and sulfur (S) elements within ONPDN were predominantly distributed on the ultrathin silica framework. Additionally, the dispersed presence of carbon (C), nitrogen (N), and sodium (Na) elements further confirmed the successful loading of olaparib and novobiocin into ONPDN (Figure 1E). The loading amounts and rates of olaparib and novobiocin in ONPDN were quantitatively determined using UV-vis spectroscopy. Olaparib was found to have a loading amount of 68 μg/mg, with a loading rate of 12.96%. Similarly, novobiocin exhibited a loading amount of 34 μg/mg, with a loading rate of 6.48%. Moreover, it is worth noting that both ONP, ONNP, and ONPDN exhibited colloidal stability in PBS and PBS supplemented with 10% FBS. This observation suggests that the colloidal stability of silica was well-maintained even after the introduction of disulfide bridges (Figure S1A and B).
Evaluation of Drug Storage and Release in ONPDN
Subsequently, we assessed the drug storage performance of ONPDN. As depicted in Figure 1F, the remaining drug content within ONPDN exhibited minimal variation during the initial 2 days of testing. Even at the end of the 7-day evaluation period, the residual drug content remained above 77.8%, underscoring the outstanding drug storage capabilities conferred by the ultrathin SiO2-coated framework with PLGA as the core encapsulation material. Considering the more acidic and reducing environment of tumor tissues than normal tissues, we proceeded to investigate release behavior of ONPDN in solutions with different glutathione concentrations under acidic conditions. As illustrated in Figures 1G and S2, ONPDN exhibited similar release rates for olaparib (86.7%) and novobiocin (86.4%) within 1 hour when exposed to a 10 nM glutathione solution. Notably, at the same pH, even in a 5 nM glutathione solution, both olaparib (65.3%) and novobiocin (65%) displayed rapid release efficiencies. To validate these release dynamics, TEM imaging was conducted, revealing observable morphological alterations in ONPDN when subjected to acidic glutathione solutions. These changes included the structural collapse of the silica shell, progressive degradation of the shell, and eventual formation of flocs over time, as evidenced in Figure S3. Overall, our findings suggest that ONPDN maintains stability within non-specific environments but swiftly self-disassembles in response to acidic and glutathione-rich conditions, facilitating rapid drug release.
In vitro Antitumor Activity of ONPDN
Two BRCA-mutant TNBC cell lines (MDA-MB-436 and HCC1937), a BRCAwt ovarian cancer cell line (SKOV3), and a BRCAwt TNBC cell line (MDA-MB-231) were tested for in vitro anticancer activity of ONPDN.28,48–50 Both OPDN (with no novobiocin added) and ONPDN demonstrated higher inhibitory effects on the proliferation of BRCAwt cancer cells (SKOV3 and MDA-MB-231), as depicted in Figure S4A. Notably, when compared to free olaparib and the free combined drug group (ON), ONPDN exhibited a remarkable enhancement in antitumor activity against BRCAwt cancer cells, with MDA-MB-231 showing the highest sensitivity among them.
To delve deeper into the toxic responses of different drugs on MDA-MB-231 cells, we conducted a 48-hour MTT experiment (Figure S4B and C). The results revealed that each drug group displayed a dose-dependent tumor cell killing ability. Significantly, ONPDN exhibited a pronounced anti-cancer effect on MDA-MB-231 cells when compared to the free drug group (ON) (P < 0.001). Subsequently, we employed MDA-MB-231 cells, representative of BRCAwt TNBC, to further investigate the anticancer mechanism of ONPDN.
We employed coumarin 6 to label ONPDN, resulting in green fluorescence, which demonstrated the uptake of ONPDN by MDA-MB-231 cells (Figure S5). In earlier studies, it was reported that olaparib could elevate intracellular ROS levels, and novobiocin derivatives exhibited similar effects.51–53 As illustrated in Figures 2A and S6A, ONPDN (G6) demonstrated a significant capacity to upregulate intracellular ROS content in MDA-MB-231 cells when compared to the free drug group (G3). Furthermore, continuous monitoring of ROS levels for each drug group over a 2-hour period revealed that the ROS levels in the ONPDN group began to increase sharply within 20 minutes and reached their peak at approximately 60 minutes (Figure S6B). One of the mechanisms of action for PARP inhibitors is their ability to hinder the repair of DNA damage sites. To assess this effect, γ-H2AX fluorescence staining experiments were conducted to detect DNA damage.54,55 As depicted in Figure 2B, ONPDN exhibited a notable accumulation of DSB within MDA-MB-231 cells. The results stemming from ROS and γ-H2AX assessments collectively suggest that ONPDN not only inhibits DNA repair but also induces additional damage, leading to the accumulation of DNA lesions.
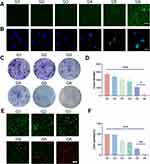 |
Figure 2 Analysis of intracellular ROS, γ-H2AX, cell clones, and calcein-AM/propidium iodide (PI) double staining. (A) ROS analysis of MDA-MB-231 cells treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5) or ONPDN (G6). Scale bar = 50 μm. (B) γ-H2AX analysis of MDA-MB-231 cells treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5) or ONPDN (G6). Scale bar = 20 μm. (C) Cell clone analysis of MDA-MB-231 cells treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5) or ONPDN (G6). (D) Statistical analysis of the clone number in different groups from Figure 2C. (E) Calcein-AM/propidium iodide (PI) double staining of MDA-MB-231 cells treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5) or ONPDN (G6). Scale bar = 50 μm. (F) Statistical analysis of the cell viability of MDA-MB-231 cells in different groups from Figure 2E. Data are expressed as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. |
Furthermore, we conducted cell cloning experiments to evaluate the impact of these drugs on the proliferation of MDA-MB-231 cells. As presented in Figure 2C and D, ONPDN proved to be more effective in suppressing the proliferative capacity of MDA-MB-231 cells when compared to the control group (G1) or the free drug group (G3). Additionally, we performed dual fluorescent staining with calcein-AM and PI on MDA-MB-231 cells from various treatment groups after a 24-hour exposure period. It became evident that ONPDN exhibited a more potent cytotoxic effect than the other groups (see Figure 2E and F). In summary, these data suggested that this enhanced effect can be attributed to the delivery of DNA repair inhibitors through progressively disassembled nanosystem. The nanosystem significantly elevate ROS levels within cancer cells, facilitating the accumulation of DNA damage, which ultimately leads to the inhibition of cancer cell growth and the induction of apoptosis.
In vivo Antitumor Efficacy of ONPDN Evaluation
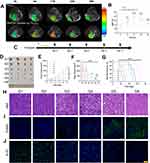
The utilization of Fe3O4 as a MR agent for tracking nanosystems has been established in previous studies, given its capacity to modulate the magnetic signal during MRI.56–59 In this study, we incorporated Fe3O4 into ONPDN to enable the monitoring of intratumoral drug accumulation. Subsequently, we administered ONPDN (labeled with Fe3O4) or free Fe3O4 through intravenous injection into nude mice bearing orthotopic MDA-MB-231 xenografts. We monitored the nude mice at various time intervals using MRI. As depicted in Figure 3A and B, the R2* signal of MRI in the tumor region exhibited a rapid increase within 2 hours post-injection of ONPDN, reaching its peak at approximately 12 hours. This observation strongly suggested that ONPDN effectively facilitated the accumulation of drugs within the tumor area, potentially resulting in higher drug concentrations.
The in vivo anti-tumor efficacy of various drug groups was subsequently assessed in female BALB/c orthotopic MDA-MB-231 tumor-bearing nude mice, as outlined in the experimental scheme presented in Figure 3C. The nude mice were randomly divided into six groups: PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), and ONPDN (G6). The interval between drug treatments was set at 3 days. Photographs (Figure 3D) and tumor volumes (Figure 3F) vividly illustrated that the G6 exhibited the smallest tumors compared to the other groups at the conclusion of the treatment period. Furthermore, both G5 and G6 effectively restrained tumor growth during the treatment period, surpassing the G1, G2 and G3 (Figure 3E). These findings underscored the remarkable anti-tumor efficacy of ONPDN. Additionally, survival analysis of nude mice within the different drug groups demonstrated that ONPDN yielded a superior survival rate compared to the other groups (Figure 3G).
To further evaluate the effects of ONPDN on BRCAwt TNBC, we conducted H&E, TUNEL, and Ki-67 staining analyses on tumor samples from various groups. H&E staining, as shown in Figure 3H, revealed that tumor progression was most effectively inhibited in the G6. TUNEL staining demonstrated that the apoptosis rate of tumor cells in G6 was the highest (Figure 3I). Furthermore, Ki-67 staining analysis revealed that G6 displayed the lowest cell density within the tumor area (Figure 3J). These results collectively indicated that ONPDN possessed substantial tumor growth inhibitory properties and promotes apoptosis in BRCAwt TNBC.
Immunofluorescence Staining
We subsequently conducted an analysis of immunofluorescence staining images following drug treatment in the different groups. PARP fluorescence staining, as depicted in Figure 4A and B, revealed that both the G5 and G6 significantly inhibited PARP, suggesting that the progressively disassembled of the system may lead to the accumulation of more PARP inhibitors. DHE immunofluorescence was employed to assess ROS levels. Figure 4A and C indicated that ROS levels in G5 and G6 were markedly higher compared to the other groups, implying that the synergistic delivery of olaparib and novobiocin beneficially elevates ROS levels. Notably, in addition to its role in DNA repair inhibition, novobiocin also acts as a heat shock protein (HSP) inhibitor with potential angiostatic function.60–64 CD31 immunofluorescence images, shown in Figure 4A and D, demonstrated that the microvessel density in the G4 and G6 was significantly lower than in the other groups. Furthermore, analysis of the γ-H2AX immunofluorescence images from different groups revealed that G6 exhibited superior DNA damage accumulation compared to the other groups (Figure 4A and E). Additionally, we assessed the immunofluorescence staining images of cleaved caspase-3 (clCasp3) in different groups to evaluate the antitumor effect of ONPDN, and clCasp3 levels in G6 were significantly higher than in the other groups (Figure 4A and F). In summary, these results indicate that the synergistic delivery of olaparib and novobiocin through the progressively disassembled system effectively inhibits DNA repair, reduces microvessel density, upregulates ROS levels, and ultimately promotes DNA damage and apoptosis in BRCAwt TNBC.
 |
Figure 4 Immunofluorescence analyses of tumor tissues from nude mice treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5) or ONPDN (G6). (A) Immunofluorescence staining images of tumor tissues from nude mice treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), or ONPDN (G6) for PARP, DHE, CD31, γ-H2AX, and cleaved caspase-3 (clCasp3). Scale bar = 100 μm. (B) PARP (C) ROS (D) CD31 (E) γ-H2AX (F) clCasp3 signals in the tumor were quantified according to the data in Figure 4A. Data are expressed as mean ± SD (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001. |
MRI Evaluation of ONPDN
MRI serves as a valuable non-invasive tool for the initial assessment of the effectiveness of anti-tumor drug treatments.65,66 Furthermore, IVIM-DWI enables the evaluation of cellular density and vascular perfusion within tissues.67,68 We conducted MRI assessments on different groups of nude mice (n=4) treated with PBS (G1), PDN (G2), olaparib and novobiocin (G3), NPDN (G4), OPDN (G5), and ONPDN (G6) at the conclusion of the treatment regimen (Figure 5A). The efficacy of ONPDN in inhibiting tumor growth was strikingly demonstrated by T2WI, surpassing the effectiveness of the other treatment groups. We further conducted a detailed analysis of the slowADC, fastADC, and the fraction of fastADC in the different groups. Remarkably, as shown in Figure 5A and B, the slowADC in the G6 exhibited an increase compared to the other groups. Additionally, the fast ADC and the fraction of fast ADC suggested that ONPDN had the potential to modulate vascular perfusion (Figure 5A, C, and D). Therefore, the MRI evaluation results at the end of the treatment period underscored the excellent therapeutic efficacy of ONPDN in BRCAwt TNBC as well.
Biosafety Evaluation of ONPDN
We conducted a preliminary biosafety assessment of ONPDN, which included monitoring the body weight changes of different treatment groups throughout the treatment period. Furthermore, we performed H&E staining and conducted a blood biochemical analysis at the conclusion of the treatment regimen. During the treatment period, the body weight of nude mice in the various treatment groups exhibited a slight increase compared to their initial weight, with no significant differences observed among the groups (Figure S7). As illustrated in Figure 6A, major organs including the heart, liver, spleen, lungs, and kidneys from different treatment groups did not exhibit any noticeable signs of lethal damage. Additionally, blood biochemical indicators remained relatively stable across all treatment groups, as depicted in Figure 6B. In summary, these initial findings suggest that ONPDN does not display acute toxicity within the assessed parameters.
Discussion
PARP inhibitors have undergone extensive investigation as a therapeutic avenue for BRCA-mutated TNBC. However, their efficacy remains constrained when applied to BRCAwt TNBC. Furthermore, the therapeutic potential of PARP inhibitors is hindered by phenomena such as increased drug efflux, PARP1 mutations, and the restoration of HR proficiency or replication fork stability.69 Hence, it is imperative to explore and develop efficacious combination therapy strategies that can surmount the barriers posed by DNA repair pathways, ultimately leading to a more efficient induction of cell death in BRCAwt TNBC.
In this study, we have developed a TME-responsive nanosystem, ONPDN, as a delivery vehicle for DNA repair inhibitors under MRI on breast cancer. Our initial observations reveal that ONPDN boasts a remarkably appropriate particle size, measuring at a mere 191 nm. The presence of an ultra-thin silica framework on its surface prevents premature leakage of the drug before it reaches the intended tumor site. Furthermore, ONPDN exhibits rapid drug release within the glutathione rich TME, facilitated by the presence of disulfide bonds within the silica structure. Figure 1F and G illustrated the ONPDN’s drug release kinetics. Under neutral, glutathione-free conditions, the drug release remains minimal, with less than 30% of the drug being liberated over 7 days. In stark contrast, within the tumor-specific microenvironment, ONPDN demonstrates an expedited drug release profile, surpassing 80% in 1 hour. Notably, a study conducted by Chowdhury et al demonstrated that tannic acid nano-formulations released 95.52% of the drug within 6 hours.70 The rapid release of DNA repair enzyme inhibitors in specific TME is of paramount importance and may facilitate the accumulation of DNA damage that ultimately leads to cancer cell death.
In our investigation, ONPDN exhibited the remarkable ability to augment the cytotoxicity of DNA repair enzyme inhibitors in both BRCAwt cancer cells. Intriguingly, a study by Lee et al conducted a comprehensive assessment of DNA repair protein expression and repair capacity within four BRCA1/2 wild-type TNBC cell lines, thereby unveiling notable variations in HR proficiency among these cellular models.69 Furthermore, the synergistic combination of therapeutic agents proved highly effective in mitigating potential crosstalk between DNA repair pathways, thus substantially enhancing the capacity to induce cell death. Figures 2A and B vividly illustrated ONPDN’s capacity to induce DNA damage and impede repair mechanisms by elevating ROS levels. This effect culminated in the accumulation of a greater number of unrepaired DNA damage sites when compared to the free drug group.
A previous study showed that the combined application of therapeutic agents leads to a significant reduction in breast cancer cell viability.71 It’s worth noting that this combined treatment did not exhibit a significant increase in phosphorylated H2AX levels.
Presently, PARP inhibitors are no longer limited solely to the treatment of BRCA-deficient tumors; their scope has expanded to encompass tumors bearing deletion mutations within genes associated with HR, such as phosphatase and tensin homolog (PTEN).72 A study showed that AZD2461 exerted a more pronounced inhibitory effect on tumor cells characterized by significantly diminished PTEN expression, as opposed to those with elevated PTEN levels.12 These findings underscore the notion that HR deficiencies stemming from mutations in various genes render tumor cells susceptible to DNA repair inhibitors. Furthermore, given the diverse spectrum of mutations found in various types of solid tumors, the development of tailored therapeutic strategies targeting distinct gene-driven DNA repair pathways may hold promise for the treatment of a wide array of solid tumor malignancies.
The TME assumes a pivotal role in the pathogenesis and advancement of various cancer types, rendering it a promising target for cancer molecular imaging. Neovascularization within the TME represents a hallmark of tumor progression and metastasis, underscoring its significance in cancer biology.73 One of the primary challenges in cancer therapy pertains to the constrained perfusion of the tumor vascular network, resulting in diminished intra-tumoral pH levels and escalated tissue fluid pressure within the TME. This phenomenon creates a significant barrier to effective drug penetration. As delineated in Figure 3A and B, ONPDN exhibited the capability to track the accumulation of nanosystems within the tumor site at distinct time intervals, facilitated by the magnetic attributes of Fe3O4. This phenomenon reached its zenith at approximately 12 hours, surpassing the signal intensity observed in the free drug group and maintaining superiority for up to 48 hours. Remarkably, magnetic nanoparticles hold potential as therapeutic agents for cancer treatment, as evidenced by a study utilizing ternary metal alloy nanoparticles that demonstrated cytotoxicity against breast cancer cells while sparing normal human cells.74
Conclusion
In this study, we successfully synthesized ONPDN, which possesses the capability to progressively disassemble under acidic and glutathione-rich conditions. This progressive disassembly of the nanosystem allows for enhanced drug accumulation and rapid release. ONPDN effectively synergizes the delivery of olaparib and novobiocin into BRCAwt TNBC, leading to significant growth inhibition in orthotopic tumor-bearing nude mice. Furthermore, our results reveal that olaparib, when incorporated into the progressively disassembling system, effectively restrains the growth of BRCAwt TNBC, suggesting a potential sensitizing effect of novobiocin on olaparib. Given that PARP inhibitors are no longer restricted solely to BRCA-deficient tumors, the concept of combining different DNA repair inhibitors may also have relevance for other HR-deficient tumors. In conclusion, ONPDN represents a promising therapeutic approach utilizing DNA repair enzyme inhibitors for the enhanced treatment of BRCAwt TNBC with intact HR repair mechanisms.
Abbreviations
ADC, apparent diffusion coefficient; ALB, albumin; ANOVA, one-way analysis of variance; APTES, 3-Aminopropyltriethoxysilane; AST, aspartate aminotransferase; BRCA, breast cancer gene; BUN, urea nitrogen; CK, creatine kinase; clCasp3, Cleaved caspase-3; CREA, creatinine; CTAB, cetyltrimethylammonium bromide; DAPI, 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride; DCFH-DA, dichloro-dihydro-fluorescein diacetate; DDR, DNA damage repair response; DHE, dihydroethidium; DMEM, Dulbecco’s Modified Eagle Medium; DMSO, dimethyl sulfoxide; DSB, double-strand breaks; DWI, diffusion weighted imaging; EDS, energy dispersive spectroscopy; Em, emission; Ex, excitation; EZH2, enhancer of zeste homologue 2; FOV, field of view; FSE, fast spin echo; GSH, glutathione; LDH, lactate dehydrogenase; γH2AX, gamma-H2A histone family member X; H&E, Hematoxylin and eosin stain; HR, homologous recombination; HSP, heat shock protein; MRI, magnetic resonance imaging. NDDS, nanoscale drug delivery systems; NEX, number of excitations; PARP, poly (adenosine diphosphate-ribose) polymerase; PBS, phosphate buffered saline; PI, pyridinium iodide; PLGA, polylactic-co-glycolic acid; POLθ, polymerase theta; PTEN, phosphatase and tensin homolog; ROI, region of interest; ROS, reactive oxygen species; RPMI, Roswell Park Memorial Institute; SD, standard deviation; TE, time to echo; TEM, transmission electron microscope; TEOS, tetraethyl orthosilicate; TESPDS, bis(triethoxysilylpropyl) disulfide; TME, tumor microenvironment; TNBC, triple-negative breast cancer; TR, repetition time; TUNEL, deoxynucleotidyl transferase dUTP nick-end labeling.
Acknowledgments
We acknowledge funding by the National Natural Science Foundation of China (82271943) and Science and Technology Planning Project of Guangzhou (2023A03J0566).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi:10.3322/caac.21763
2. Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397:1750–1769. doi:10.1016/S0140-6736(20)32381-3
3. Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Rev Clin Oncol. 2022;19:91–113. doi:10.1038/s41571-021-00565-2
4. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi:10.1038/nrclinonc.2016.66
5. Waks AG, Winer EP. Breast Cancer Treatment: a Review. JAMA. 2019;321:288–300. doi:10.1001/jama.2018.19323
6. Nedeljkovic M, Damjanovic A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells. 2019;8. doi:10.3390/cells8090957
7. Badve S, Dabbs DJ, Schnitt SJ, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi:10.1038/modpathol.2010.200
8. Caramelo O, Silva C, Caramelo F, et al. Efficacy of different neoadjuvant treatment regimens in BRCA-mutated triple negative breast cancer: a systematic review and meta-analysis. Hered Cancer Clin Pract. 2022;20:34. doi:10.1186/s13053-022-00242-0
9. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi:10.1038/nature03445
10. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi:10.1038/nature03443
11. Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi:10.1126/science.aam7344
12. Sargazi S, Saravani R, Zavar Reza J, et al. Novel Poly(Adenosine Diphosphate-Ribose) Polymerase (PARP) Inhibitor, AZD2461, Down-Regulates VEGF and Induces Apoptosis in Prostate Cancer Cells. Iran Biomed J. 2019;23:312–323. doi:10.29252/.23.5.312
13. Slade D. PARP and PARG inhibitors in cancer treatment. Genes Dev. 2020;34:360–394. doi:10.1101/gad.334516.119
14. Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi:10.1158/0008-5472.CAN-12-2753
15. Patel M, Nowsheen S, Maraboyina S, Xia F. The role of poly(ADP-ribose) polymerase inhibitors in the treatment of cancer and methods to overcome resistance: a review. Cell Biosci. 2020;10:35. doi:10.1186/s13578-020-00390-7
16. Robson M, Im SA, Senkus E, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med. 2017;377:523–533. doi:10.1056/NEJMoa1706450
17. Eikesdal HP, Yndestad S, Elzawahry A, et al. Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann Oncol. 2021;32:240–249. doi:10.1016/j.annonc.2020.11.009
18. Sharma P, Klemp JR, Kimler BF, et al. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat. 2014;145:707–714. doi:10.1007/s10549-014-2980-0
19. Hartman AR, Kaldate RR, Sailer LM, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer. 2012;118:2787–2795. doi:10.1002/cncr.26576
20. Evans KW, Yuca E, Akcakanat A, et al. A Population of Heterogeneous Breast Cancer Patient-Derived Xenografts Demonstrate Broad Activity of PARP Inhibitor in BRCA1/2 Wild-Type Tumors. Clin Cancer Res. 2017;23:6468–6477. doi:10.1158/1078-0432.CCR-17-0615
21. Gelmon KA, Tischkowitz M, Mackay H, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a Phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi:10.1016/S1470-2045(11)70214-5
22. Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi:10.1016/S0140-6736(10)60893-8
23. Luo L, Keyomarsi K. PARP inhibitors as single agents and in combination therapy: the most promising treatment strategies in clinical trials for BRCA-mutant ovarian and triple-negative breast cancers. Expert Opin Investig Drugs. 2022;31:607–631. doi:10.1080/13543784.2022.2067527
24. Dong Y, Liao H, Fu H, et al. pH-Sensitive Shell-Core Platform Block DNA Repair Pathway To Amplify Irreversible DNA Damage of Triple Negative Breast Cancer. ACS Appl Mater Interfaces. 2019;11:38417–38428. doi:10.1021/acsami.9b12140
25. Yusoh NA, Tiley PR, James SD, et al. Discovery of Ruthenium(II) Metallocompound and Olaparib Synergy for Cancer Combination Therapy. J Med Chem. 2023;66:6922–6937. doi:10.1021/acs.jmedchem.3c00322
26. Wang C, Qu L, Li S, et al. Discovery of First-in-Class Dual PARP and EZH2 Inhibitors for Triple-Negative Breast Cancer with Wild-Type BRCA. J Med Chem. 2021;64:12630–12650. doi:10.1021/acs.jmedchem.1c00567
27. Yusoh NA, Leong SW, Chia SL, et al. Metallointercalator [Ru(dppz)(2)(PIP)](2+) Renders BRCA Wild-Type Triple-Negative Breast Cancer Cells Hypersensitive to PARP Inhibition. ACS Chem Biol. 2020;15:378–387. doi:10.1021/acschembio.9b00843
28. Luo ML, Zheng F, Chen W, et al. Inactivation of the Prolyl Isomerase Pin1 Sensitizes BRCA1-Proficient Breast Cancer to PARP Inhibition. Cancer Res. 2020;80:3033–3045. doi:10.1158/0008-5472.CAN-19-2739
29. Zhou J, Gelot C, Pantelidou C, et al. A first-in-class Polymerase Theta Inhibitor selectively targets Homologous-Recombination-Deficient Tumors. Nat Cancer. 2021;2:598–610. doi:10.1038/s43018-021-00203-x
30. Ceccaldi R, Liu JC, Amunugama R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi:10.1038/nature14184
31. Mateos-Gomez PA, Gong F, Nair N, et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi:10.1038/nature14157
32. Davodabadi F, Mirinejad S, Fathi-Karkan S, et al. Aptamer-functionalized quantum dots as theranostic nanotools against cancer and bacterial infections: a comprehensive overview of recent trends. Biotechnol Prog. 2023:e3366. doi:10.1002/btpr.3366
33. Razlansari M, Jafarinejad S, Rahdar A, et al. Development and classification of RNA aptamers for therapeutic purposes: an updated review with emphasis on cancer. Mol Cell Biochem. 2023;478:1573–1598. doi:10.1007/s11010-022-04614-x
34. Vallet-Regi M, Schuth F, Lozano D, Colilla M, Manzano M. Engineering mesoporous silica nanoparticles for drug delivery: where are we after two decades? Chem Soc Rev. 2022;51:5365–5451. doi:10.1039/d1cs00659b
35. Wang D, Nie T, Huang C, et al. Metal-Cyclic Dinucleotide Nanomodulator-Stimulated STING Signaling for Strengthened Radioimmunotherapy of Large Tumor. Small. 2022;18:e2203227. doi:10.1002/smll.202203227
36. Zeng L, Wang H, Shi W, et al. Aloe derived nanovesicle as a functional carrier for indocyanine green encapsulation and phototherapy. J Nanobiotechnology. 2021;19:439. doi:10.1186/s12951-021-01195-7
37. Shi L, Jin Y, Lai S, et al. Redox-responsive carrier based on fluorinated gemini amphiphilic polymer for combinational cancer therapy. Colloids Surf B Biointerfaces. 2022;216:112551. doi:10.1016/j.colsurfb.2022.112551
38. Guo J, De May H, Franco S, et al. Cancer vaccines from cryogenically silicified tumour cells functionalized with pathogen-associated molecular patterns. Nat Biomed Eng. 2022;6:19–31. doi:10.1038/s41551-021-00795-w
39. Wang D, Zhang M, Zhang Y, et al. Intraparticle Double-Scattering-Decoded Sonogenetics for Augmenting Immune Checkpoint Blockade and CAR-T Therapy. Adv Sci. 2022;9:e2203106. doi:10.1002/advs.202203106
40. You Y, Zhao Z, He L, et al. Long‐Term Oxygen Storage Nanosystem for Near‐Infrared Light‐Triggered Oxygen Supplies to Antagonize Hypoxia‐Induced Therapeutic Resistance in Nasopharyngeal Carcinoma. Adv Funct Mater. 2020;30. doi:10.1002/adfm.202002369
41. Yang Y, Sun B, Zuo S, et al. Trisulfide bond-mediated doxorubicin dimeric prodrug nanoassemblies with high drug loading, high self-assembly stability, and high tumor selectivity. Sci Adv. 2020;6. doi:10.1126/sciadv.abc1725
42. Li S, Saw PE, Lin C, et al. Redox-responsive polyprodrug nanoparticles for targeted siRNA delivery and synergistic liver cancer therapy. Biomaterials. 2020;234:119760. doi:10.1016/j.biomaterials.2020.119760
43. Wu J, Williams GR, Niu S, et al. A Multifunctional Biodegradable Nanocomposite for Cancer Theranostics. Adv Sci. 2019;6:1802001. doi:10.1002/advs.201802001
44. Li J, Xu R, Lu X, He J, Jin S. A simple reduction-sensitive micelles co-delivery of paclitaxel and dasatinib to overcome tumor multidrug resistance. Int J Nanomedicine. 2017;12:8043–8056. doi:10.2147/IJN.S148273
45. Chen W, Glackin CA, Horwitz MA, Zink JI. Nanomachines and Other Caps on Mesoporous Silica Nanoparticles for Drug Delivery. Acc Chem Res. 2019;52:1531–1542. doi:10.1021/acs.accounts.9b00116
46. Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi:10.1093/jn/134.3.489
47. Zhao Q, Geng H, Wang Y, et al. Hyaluronic acid oligosaccharide modified redox-responsive mesoporous silica nanoparticles for targeted drug delivery. ACS Appl Mater Interfaces. 2014;6:20290–20299. doi:10.1021/am505824d
48. Dhoonmoon A, Nicolae CM, Moldovan GL. The KU-PARP14 axis differentially regulates DNA resection at stalled replication forks by MRE11 and EXO1. Nat Commun. 2022;13:5063. doi:10.1038/s41467-022-32756-5
49. Hijaz M, Chhina J, Mert I, et al. Preclinical evaluation of olaparib and metformin combination in BRCA1 wildtype ovarian cancer. Gynecol Oncol. 2016;142:323–331. doi:10.1016/j.ygyno.2016.06.005
50. Wu Z, Bai Y, Jin J, et al. Discovery of novel and potent PARP/PI3K dual inhibitors for the treatment of cancer. Eur J Med Chem. 2021;217:113357. doi:10.1016/j.ejmech.2021.113357
51. Liu Q, Gheorghiu L, Drumm M, et al. PARP-1 inhibition with or without ionizing radiation confers reactive oxygen species-mediated cytotoxicity preferentially to cancer cells with mutant TP53. Oncogene. 2018;37:2793–2805. doi:10.1038/s41388-018-0130-6
52. Zhang N, Huang L, Tian J, et al. A novel synthetic novobiocin analog, FM-Nov17, induces DNA damage in CML cells through generation of reactive oxygen species. Pharmacol Rep. 2016;68:423–428. doi:10.1016/j.pharep.2015.11.002
53. Wu L, Chen X, Huang L, et al. A Novobiocin Derivative, XN4, Inhibits the Proliferation of Chronic Myeloid Leukemia Cells by Inducing Oxidative DNA Damage. PLoS One. 2015;10:e0123314. doi:10.1371/journal.pone.0123314
54. Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi:10.1038/leu.2010.6
55. Lee Y, Wang Q, Shuryak I, Brenner DJ, Turner HC. Development of a high-throughput gamma-H2AX assay based on imaging flow cytometry. Radiat Oncol. 2019;14:150. doi:10.1186/s13014-019-1344-7
56. Liu D, He H, Kong F, et al. A versatile metal–organic nanoplatform in combination with CXCR4 antagonist and PD-L1 inhibitor for multimodal synergistic cancer therapy and MRI-guided tumor imaging. Nano Today. 2022;47. doi:10.1016/j.nantod.2022.101689
57. Ren J, Tang X, Wang T, et al. A Dual-Modal Magnetic Resonance/Photoacoustic Imaging Tracer for Long-Term High-Precision Tracking and Facilitating Repair of Peripheral Nerve Injuries. Adv Healthc Mater. 2022;11:e2200183. doi:10.1002/adhm.202200183
58. Wang D, Zhou J, Fang W, et al. A multifunctional nanotheranostic agent potentiates erlotinib to EGFR wild-type non-small cell lung cancer. Bioact Mater. 2022;13:312–323. doi:10.1016/j.bioactmat.2021.10.046
59. Xiao Z, You Y, Liu Y, et al. NIR-Triggered Blasting Nanovesicles for Targeted Multimodal Image-Guided Synergistic Cancer Photothermal and Chemotherapy. ACS Appl Mater Interfaces. 2021;13:35376–35388. doi:10.1021/acsami.1c08339
60. Marcu MG, Schulte TW, Neckers L. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J Natl Cancer Inst. 2000;92:242–248. doi:10.1093/jnci/92.3.242
61. Ghosh S, Liu Y, Garg G, et al. Diverging Novobiocin Anti-Cancer Activity from Neuroprotective Activity through Modification of the Amide Tail. ACS Med Chem Lett. 2016;7:813–818. doi:10.1021/acsmedchemlett.6b00224
62. Calderwood SK, Gong J. Heat Shock Proteins Promote Cancer: it’s a Protection Racket. Trends Biochem Sci. 2016;41:311–323. doi:10.1016/j.tibs.2016.01.003
63. Richardson PG, Mitsiades CS, Laubach JP, et al. Inhibition of heat shock protein 90 (HSP90) as a therapeutic strategy for the treatment of myeloma and other cancers. Br J Haematol. 2011;152:367–379. doi:10.1111/j.1365-2141.2010.08360.x
64. Zhang PC, Liu X, Li MM, et al. AT-533, a novel Hsp90 inhibitor, inhibits breast cancer growth and HIF-1alpha/VEGF/VEGFR-2-mediated angiogenesis in vitro and in vivo. Biochem Pharmacol. 2020;172:113771. doi:10.1016/j.bcp.2019.113771
65. Liu Y, Wu J, Jin Y, et al. Copper(I) Phosphide Nanocrystals for In Situ Self‐Generation Magnetic Resonance Imaging‐Guided Photothermal‐Enhanced Chemodynamic Synergetic Therapy Resisting Deep‐Seated Tumor. Adv Funct Mater. 2019;29. doi:10.1002/adfm.201904678
66. Khan M, Boumati S, Arib C, et al. Doxorubicin (DOX) Gadolinium-Gold-Complex: a New Way to Tune Hybrid Nanorods as Theranostic Agent. Int J Nanomedicine. 2021;16:2219–2236. doi:10.2147/IJN.S295809
67. Shi C, Liu D, Xiao Z, et al. Monitoring Tumor Response to Antivascular Therapy Using Non-Contrast Intravoxel Incoherent Motion Diffusion-Weighted MRI. Cancer Res. 2017;77:3491–3501. doi:10.1158/0008-5472.CAN-16-2499
68. Sheng Y, Dang X, Zhang H, et al. Correlations between intravoxel incoherent motion-derived fast diffusion and perfusion fraction parameters and VEGF- and MIB-1-positive rates in brain gliomas: an intraoperative MR-navigated, biopsy-based histopathologic study. Eur Radiol. 2023. doi:10.1007/s00330-023-09506-2
69. Lee KJ, Mann E, Wright G, et al. Exploiting DNA repair defects in triple negative breast cancer to improve cell killing. Ther Adv Med Oncol. 2020;12:1758835920958354. doi:10.1177/1758835920958354
70. Chowdhury P, Nagesh PKB, Hatami E, et al. Tannic acid-inspired paclitaxel nanoparticles for enhanced anticancer effects in breast cancer cells. J Colloid Interface Sci. 2019;535:133–148. doi:10.1016/j.jcis.2018.09.072
71. Sargazi S, Kooshkaki O, Zavar Reza J, et al. Mild antagonistic effect of Valproic acid in combination with AZD2461 in MCF-7 breast cancer cells. Med J Islam Repub Iran. 2019;33:29. doi:10.34171/mjiri.33.29
72. Mendes-Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi:10.1002/emmm.200900041
73. Siminzar P, Tohidkia MR, Eppard E, et al. Recent Trends in Diagnostic Biomarkers of Tumor Microenvironment. Mol Imaging Biol. 2023;25:464–482. doi:10.1007/s11307-022-01795-1
74. Alikhanzadeh-Arani S, Almasi-Kashi M, Sargazi S, et al. CoNiZn and CoNiFe Nanoparticles: synthesis, Physical Characterization, and In Vitro Cytotoxicity Evaluations. Appl Sci. 2021;11. doi:10.3390/app11125339
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.