Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 16
A Novel Circ_Arf3/miR-452-5p/Mbnl1 Axis Regulates Proliferation and Expression of Fibrosis-Related Proteins of Mouse Mesangial Cells Under High Glucose
Authors Wang Q, Zhu Y, Dong Q, Zhang L, Zhang W
Received 6 December 2022
Accepted for publication 31 March 2023
Published 11 July 2023 Volume 2023:16 Pages 2105—2116
DOI https://doi.org/10.2147/DMSO.S400530
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Konstantinos Tziomalos
Qiong Wang, Yanting Zhu, Qianlan Dong, Linping Zhang, Wei Zhang
Kidney Disease and Dialysis Center, Shaanxi Provincial People’s Hospital, Xi’an City, Shaanxi, People’s Republic of China
Correspondence: Wei Zhang, Kidney Disease and Dialysis Center, Shaanxi Provincial People’s Hospital, No. 256 West Youyi Road, Beilin District, Xi’an, Shaanxi, 710068, People’s Republic of China, Tel +86 2985251331, Email [email protected]
Background: Diabetic nephropathy (DN) is a serious microvascular complication of diabetes that may lead to chronic renal failure and end-stage renal disease. Circular RNAs (circRNAs) play important roles in DN progression. However, the action of circRNA ADP ribosylation factor 3 (circ_Arf3) in high glucose (HG)-induced change is still unclear.
Methods: Mouse mesangial cells (MCs) were treated with 30 mM HG as a DN cell model in vitro. Quantitative real-time polymerase chain reaction (qRT-PCR) was employed to examine the expression levels of circ_Arf3, microRNA (miR)-452-5p and muscleblind like splicing regulator 1 (Mbnl1). The proliferation of HG-treated MCs was assessed using 5 Ethynyl 2′ deoxyuridine (EdU) and cell counting kit-8 (CCK-8) assays, and the levels of proliferation and fibrosis-related proteins and Mbnl1 were detected by Western blot. Dual-luciferase reporter and RNA pull-down assays were utilized to determine the relationship between miR-452-5p and circ_Arf3 or Mbnl1.
Results: Our results discovered that circ_Arf3 and Mbnl1 were lowly expressed in HG-treated MCs, while miR-452-5p expression was up-regulated. Moreover, circ_Arf3 was mainly located in the cytoplasm and had a ring-like stable structure. Functional assays demonstrated that overexpression of circ_Arf3 prevented cell proliferation and fibrous formation in HG-treated MCs. Circ_Arf3 could sponge miR-452-5p, and the effect of circ_Arf3 overexpression was reversed by enhanced expression of miR-452-5p. Mbnl1 was a direct target of miR-452-5p. Knockdown of Mbnl1 abolished the suppressive effects of miR-452-5p inhibitor on proliferation and fibrosis-related protein expression in HG-treated MCs. Moreover, circ_Arf3 regulated Mbnl1 through miR-452-5p.
Conclusion: Overexpression of circ_Arf3 prevents cell proliferation and fibrous formation in HG-treated MCs by regulating the expression of Mbnl1 via miR-452-5p.
Keywords: circ_Arf3, miR-452-5p, Mbnl1, diabetes nephropathy
Introduction
Diabetic nephropathy (DN) is a serious glomerular complication caused by diabetes.1 DN is the dominant causative agent of end-stage renal disease globally, and approximately 40% of people with diabetes may develop end-stage kidney disease.2,3 In the clinical setting, the main pathological features of DN are proteinuria, mesangial cells (MCs) hyperplasia, extracellular matrix (ECM) accumulation and renal fibrosis and so on.4 It is well known that tethered cells and foot cell dysfunction are closely involved in DN development.5 A previous study showed that MCs exposed to a high glucose (HG) environment could cause abnormal expression of fibroin and cytokines, which in turn affected the initiation and progression of renal fibrosis.6 Therefore, it is highly important to investigate the potential targets and mechanisms of MCs under HG treatment for the therapy and prevention of DN.
Circular RNAs (circRNAs) are new endogenous RNAs with covalently closed loops formed by reverse splicing of pre-mRNAs.7 MicroRNAs (miRNAs) are a set of small non-coding RNAs of roughly 20 nucleotides negatively, which control the expression of downstream genes.8,9 Some circRNAs contain miRNA response elements (MREs) and can reduce miRNA activity by functioning as miRNA sponges.10 Importantly, circRNAs repress the activity of certain miRNAs to exert critical functions in the development and progression of human diseases, including DN.11,12 As reported by Wang et al, circ_0037128 expression is up-regulated in HG-treated MCs, and circ_0037128 is involved in regulating DN progression by sponging miR-17-3p to adjust the expression level of AKT serine/threonine kinase 3 (AKT3).13 Circ_0003928 regulates HG-induced HK-2 cell viability loss and apoptosis by sponging miR-151-3p.14 Circ_0080425 is aberrantly expressed in DN, and circ_0080425 modulates cell proliferation and fibrosis by targeting miR-24-3p.15 Circ_0000064 has also been shown to promote MC proliferation and exacerbate fibrosis by targeting miR-143 in DN.16 Conversely, circ_010383 is underexpressed in DN, and increased expression of circ_010383 exerts an inhibitory effect on proteinuria and renal fibrosis through operating as a miR-135a sponge.17
MiRNAs have been identified to serve crucial regulatory functions in the pathogenesis of various human diseases, including DN.18 As an example, miR-337 can contribute to DN progression by causing podocyte injury.19 Silencing of miR-133b and miR-199b reduces renal fibrosis induced by TGF-β1 through the modulation of sirtuin 1, thereby participating in DN progress.20 Of interest, miR-452-5p, a frequently dysregulated miRNA in various disorders,21,22 can participate in the modulation of HG-induced levels of immune and antioxidant factors in HK-2 cells.23 In our preliminary survey for the circRNAs targeted miR-452-5p using online program starbase, we found four circRNAs: circRNA ADP ribosylation factor 3 (circ_Arf3), circRNA nuclear FMR1 interacting protein 2 (circ_Nufip2), circRNA ataxin 2 (circ_Atxn2), and circRNA myosin heavy chain 9 (circ_Myh9) that are closely associated with human disease,24,25 and our data showed that only circ_Arf3 is aberrantly expressed in HG-treated MCs (Supplementary Figure 1). Circ_Arf3 (also called circ_0000650 based on the circRNA ID) is formed by the exonic circularization of Arf3 pre-mRNA. Intriguingly, Gao et al reported that circ_Arf3 was implicated in the pathogenesis of osteosarcoma (OS), where it is expressed at high levels in OS tissues and cells.25 Zhou et al uncovered the implication of circ_Arf3 in the pathogenesis of chronic hepatitis B through the miR-6873-3p/TGFβ2 cascade.26 However, the action of circ_Arf3 in HG-induced changes in MCs is still unclear. Furthermore, no studies proved the relationship between circ_Arf3 and miR-452-5p in DN pathogenesis.
Based on these reasons, we herein investigated the effect and underlying mechanism of circ_Arf3 in HG-induced mouse MC proliferation and expression of fibrosis-related proteins, with the hope that such research might provide novel insights for the implications of circRNAs in the causal mechanisms of DN development and progress.
Methods
Cell Lines and Cell Transfection
Mouse mesangial cells (MCs) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and propagated in Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Genetimes, Shanghai, China) in an incubator at 37°C, 5% CO2 volume fraction and 97% humidity. For HG stimulation, cells were treated with 30 mM glucose (Genetimes) for 24 h. The cells grown in normal glucose (5.5 mM) were defined as the control group.
Mouse MCs were seeded in 6-well plates and then were transfected using Lipofectamine 3000 (Promega, Madison, WI, USA). pcDNA3.1(+)-based circ_Arf3 overexpression vector (circ_Arf3) and its pcDNA control (vector), miR-452-5p mimic (miR-452-5p) and its control (miR-NC), miR-452-5p inhibitor (anti-miR-452-5p) and its control (anti-miR-NC), and the small interfering RNA (siRNA) of Mbnl1 (si-Mbnl1) and matched control (si-NC) were synthesized by RiboBio Ltd. (Guangzhou). After 6 h of transfection, the medium was replaced by fresh growth media, and the transfection efficiency was verified by qRT-PCR after 24 h transfection.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) and circRNA Localization
Extraction of total RNA from cultured mouse MCs was performed using Trizol Reagent (Invitrogen, Carlsbad, CA, USA), and high quality of RNA (OD260/280=1.8–2.1) was used for complementary DNA (cDNA) synthesis. For circ_Arf3, Arf3, and Mbnl1 analyses, reverse transcription of RNA to cDNA was done using the PrimeScript Reverse Transcription Reagent Kit (TaKaRa, Beijing, China), and diluted cDNA was amplified by qRT-PCR using SYBR Green Kit (TaKaRa) with specific primers. For miR-452-5p analysis, the miScript Reverse Transcription Kit and miScript SYBR Green PCR Kit were used as recommended by the manufacturer (Qiagen, Hilden, Germany). The 2−ΔΔCt method was taken to calculate the relative expression of the target RNAs. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 snRNA was served as an internal reference, and the forward and reverse primers are displayed in Table 1.
 |
Table 1 Primer Sequences Used for PCR |
For circRNA localization, nuclear and cytoplasmic RNA of mouse MCs were firstly isolated and extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). Afterward, the expression of circ_Arf3 in nuclear and cytoplasmic RNA was measured using qRT-PCR.
Ribonuclease R (RNase R) Degradation and Actinomycin D Treatment Assays
To authenticate the circular structure of circ_Arf3, total RNA from MCs (10 μg) was incubated with or without 30 U of RNase R (Tiangen Biochemical Technology, Beijing, China) for 15 min at 37°C, and the levels of circ_Arf3 and linear Arf3 mRNA were measured by qRT-PCR. For actinomycin D treatment, cells were cultured with or without 2 μg/mL actinomycin D (Sigma-Aldrich, St. Louis, MO, USA) for 0, 8, 16 and 24 h. Then, cells were harvested, and qRT-PCR was performed to detect the levels of circ_Arf3 and linear Arf3 mRNA.
5-Ethynyl-2′-Deoxyuridine (EDU) Assay
After transfection, MCs were seeded into 96-well plates at a density of 2×103 cells per well and exposed to HG for 24 h. The percentage of positive cells was detected by the EdU incorporation assay kit (RiboBio). Briefly, treated MCs were incubated with EdU solution and stained with 1× Apollo567. Afterward, the cells were then stained using DAPI. Photographs were taken and analyzed under a fluorescence microscope (Nikon, Japan).
Cell Counting Kit-8 Assay (CCK-8)
Untransfected or transfected MCs (2×103 cells per well) were plated into 96-well plates. The cells were then treated with or without HG for 24 h, followed by the addition of 10 μL CCK-8 reagent (Thermo Fisher Scientific) per well. After 2 h incubation, the absorbance of the cells in each well was measured using a microplate reader (Thermo Fisher Scientific) at 450 nm. The number of the viable cells was proportional to the absorbance.
Western Blot Assay
Total protein was extracted by Radioimmunoprecipitation lysate protein extraction reagent (RIPA, Thermo Fisher Scientific) containing phenylmethylsulfonyl fluoride (PMSF). Determination of protein concentration was carried out using bicinchoninic acid (BCA) Protein Assay Kit (Beyotime, Shanghai, China). The 10% SDS polyacrylamide gel electrophoresis (SDS-PAGE) was employed to separate the proteins, and then the proteins were transferred onto polyvinylidene difluoride (PVDF, Millipore, Billerica, MA, USA) membranes. The membranes were probed with primary antibodies overnight at 4℃ and then incubated with the secondary antibody at 37℃. Signals were developed using the enhanced electrochemiluminescence (ECL) method and the grey values of the bands were analyzed using Quantity One 4.6.2 software. All the antibodies were gained from Abcam (Cambridge, MA, USA), including anti-proliferating cell nuclear antigen (anti-PCNA, 1:1000, ab92552, Abcam), anti-alpha smooth muscle actin (anti-α-SMA, 1:1000, ab108424, Abcam), anti-collagen I (anti-Col I, 1:1000, ab34710, Abcam), anti-fibronectin (anti-FN, 1:1000, ab174833, Abcam), anti-Col IV (1:400, ab6586, Abcam), anti-GAPDH (1:2500, ab9485, Abcam), anti-Mbnl1 (1:1000, ab108519, Abcam), and goat Anti-Rabbit IgG coupled horseradish peroxidase (HRP) (1:2000, ab6721 Abcam).
Dual-Luciferase Reporter Assay
The fragments of circ_Arf3 and Mbnl1 3’untranslated region (3’UTR) harboring the wild-type or mutant-type miR-452-5p binding sequence were ligated into the psiCHECK-2 vector (Promega) to produce wild-type luciferase vectors for circ_Arf3 and Mbnl1 (circ_Arf3-WT and Mbnl1-WT) and mutant luciferase vectors (circ_Arf3-MUT and Mbnl1-MUT), respectively. MCs were cultured in 6-well plates and transfected with the luciferase reporter (200 ng) and either miR-452-5p mimic (30 nM) or miR-NC mimic using Lipofectamine 3000. Forty-eight hours later, the luciferase activity was detected by Dual-Luciferase Reporter Assay Kit (GeneCopoeia, Rockville, MD, USA). Renilla luciferase activity was normalized to firefly activity.
RNA Pull-Down Assay
Lysates of mouse MCs extracted in RIPA buffer were incubated with biotinylated RNA probes including Bio-circ_Arf3-WT, Bio-circ_Arf3-MUT, Bio-Mbnl1-WT, Bio-Mbnl1-MUT or control Bio-NC (all from Abiocenter, Beijing, China) at 4°C for 4 h before adding M-280 streptavidin magnetic beads (Invitrogen) for 4 h. Beads were harvested and washed five times. Bead-bound RNA was purified by TRIzol, and the expression of miR-452-5p was detected by qRT-PCR.
Statistical Analysis
SPSS.22.0 software (IBM, SPSS, Chicago, IL, USA) was used for statistical analysis. The data were expressed as mean ± standard deviation, and the comparison between 2 groups was done by independent samples t-test, while the comparison in multiple groups was done by one-way or two-way analysis of variance (ANOVA). P < 0.05 was deemed statistically significant.
Results
Circ_Arf3 Was Down-Regulated in MCs Treated with HG
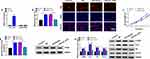
MCs were treated with 30mM HG for 24 h followed by qRT-PCR to examine the expression of circ_Arf3. The results presented that the expression of circ_Arf3 in HG-treated MCs was distinctly lower than that in the control group (Figure 1A). To further explore circ_Arf3, its subcellular localization was examined. The findings of the nucleus/cytoplasm separation assay indicated that most circ_Arf3 was expressed in the nucleus (Figure 1B). RNase R degradation and actinomycin D treatment assays were performed to monitor the structural stability of circ_Arf3. From the outcomes, we concluded that the half-life of circ_Arf3 after actinomycin D treatment was up to 24 h, whereas the half-life of its linear control Arf3 mRNA was only 8 h (Figure 1C). Furthermore, Figure 1D illustrated that Arf3 mRNA expression was reduced after RNase R treatment, while circ_Arf3 expression remained almost unchanged after RNase R treatment. The results for RNase R and actinomycin D assays showed that circ_Arf3 had a stable ring structure compared to linear mRNA.
Circ_Arf3 Overexpression Reduced HG-Induced Proliferation and Increase of Fibrosis-Related Proteins in Mouse MCs
The transfection efficiency of circ_Arf3 overexpression plasmid was investigated by qRT-PCR, which demonstrated that circ_Arf3 expression was significantly up-regulated by circ_Arf3 overexpression plasmid transfection, and the Afr3 mRNA level was not affected by circ_Arf3 overexpression plasmid (Figure 2A). Subsequently, EdU assays for cell proliferation displayed a substantially higher number of EdU-positive cells in the HG-treated group as compared to the control group, while transfection of circ_Arf3 in HG-treated MCs reversed this promotion (Figure 2B). Similarly, HG enhanced cell viability of MCs, while the transfection of circ_Arf3 resumed the HG-induced cell viability (Figure 2C). The Western blot results suggested that HG increased the protein expression of PCNA versus the control group and the addition of circ_Arf3 in HG-treated MCs decreased the protein level of PCNA in comparison with the HG + vector group (Figure 2D). To investigate the effect of circ_Arf3 on fibrosis, we examined the expression of DN fibrosis-related proteins α-SMA, Col I, FN and Col IV using Western blot. The results showed that the protein levels of α-SMA, Col I, FN and Col IV were elevated in MCs with HG treatment, and this enhancement was eliminated by transfection with circ_Arf3 overexpression plasmid (Figure 2E). Overall, overexpression of circ_Arf3 reduced HG-induced proliferation and fibrosis-related protein expression in mouse MCs.
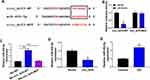
Circ_Arf3 Targeted miR-452-5p
Online software starbase predicted that miR-452-5p was a potential target of circ_Arf3. The binding sites between miR-452-5p and circ_Arf3 are shown in Figure 3A. The correlation between miR-452-5p and circ_Arf3 was confirmed by dual-luciferase reporter assay, which showed that the luciferase activity of the wild-type circ_Arf3 reporter plasmid was attenuated by transfection of miR-452-5p mimic; when the predicted target sequence was mutated, little reduction was found in the presence of miR-452-5p mimic (Figure 3B). The RNA pull-down assay further validated that miR-452-5p was enriched by Bio-circ_Arf3, suggesting that miR-452-5p might direct bind to circ_Arf3 (Figure 3C). Moreover, miR-452-5p was down-regulated by circ_Arf3 overexpression (Figure 3D). Also, miR-452-5p expression was up-regulated by HG treatment in MCs (Figure 3E). Overall, these data suggested that the miR-452-5p was targeted by circ_Arf3.
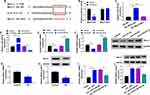
MiR-452-5p Overexpression Reversed the Effect of circ_Arf3 Up-Regulation on HG-Induced MC Proliferation and Fibrosis-Related Protein Expression
The transfection efficiency of miR-452-5p mimic was examined using qRT-PCR assay, which showed that transfection with circ_Arf3 overexpression plasmid reduced miR-452-5p level, while co-transfection with miR-452-5p mimic abated circ_Arf3 overexpression-driven miR-452-5p inhibition (Figure 4A). Cell proliferation and viability were tested by the EdU and CCK-8 assays, respectively. The results showed that the inhibitory effects of circ_Arf3 up-regulation on HG-induced MC proliferation and viability were eliminated by miR-452-5p restoration (Figure 4B and C). Moreover, miR-452-5p restoration abolished circ_Arf3 up-regulation-imposed PCNA expression inhibition in HG-treated MCs (Figure 4D). Additionally, the protein levels of α-SMA, Col I, FN and Col IV were increased in HG-treated MCs, and transfection with circ_Arf3 suppressed the expression of the above proteins, while co-transfection of miR-452-5p and circ_Arf3 reversed the inhibitory effect induced by circ_Arf3 in HG-treated MCs (Figure 4E). The above data indicated that increased expression of miR-452-5p reversed the effects of Arf3 overexpression on cell proliferation and fibrosis-related protein expression in HG-treated MCs.
MiR-452-5p Directly Targeted Mbnl1
StarBase 3.0 predicted the target binding of Mbnl1 to miR-452-5p, and Figure 5A showed the complementary sequence. Subsequently, dual-luciferase reporter assay displayed that co-transfection of miR-452-5p and Mbnl1-WT significantly reduced luciferase activity, whereas the luciferase activity in cells co-transfected with miR-452-5p and Mbnl1-MUT was unaltered (Figure 5B). Similarly, the relationship between Mbnl1 and miR-452-5p was further authenticated in the RNA pull-down assay (Figure 5C). Afterward, qRT-PCR showed that transfection of miR-452-5p increased the expression of miR-452-5p, while transfection of anti-miR-452-5p decreased the expression of miR-452-5p (Figure 5D). In addition, miR-452-5p restrained Mbnl1 mRNA expression, while anti-miR-452-5p facilitated the expression of Mbnl1 (Figure 5E and F). We also found the low expression of Mbnl1 in HG-treated cells by qRT-PCR and Western blot (Figure 5G and H). Moreover, overexpression of circ_Arf3 elevated the mRNA and protein expression of Mbnl1, and this enhancement was abrogated by increased expression of miR-452-5p (Figure 5I and J). In short, miR-452-5p could target and regulate Mbnl1.
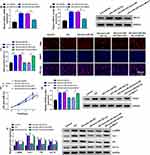
The miR-452-5p/Mbnl1 Axis Regulated HG-Induced Proliferation and Fibrosis-Related Protein Expression in Mouse MCs
qRT-PCR and Western blot were conducted to investigate the expression of Mbnl1. The results showed that anti-miR-452-5p promoted the mRNA and protein expression of Mbnl1, while co-transfection with si-Mbnl1 abated this promotion (Figure 6A and B). EdU and CCK-8 results exhibited that the addition of anti-miR-452-5p in HG-treated MCs repressed cell proliferation and viability, which was reversed by si-Mbnl1 (Figure 6C and D). Furthermore, Western blot revealed that co-transfection of si-Mbnl1 and anti-miR-452-5p in HG-treated MCs reinstated the inhibitory effect of anti-miR-452-5p on the expression of PCNA, α-SMA, Col I, FN and Col IV proteins (Figure 6E and F). Additionally, increased expression of Mbnl1 by an expression plasmid suppressed cell proliferation and viability, reduced PCNA expression, as well as downregulated the levels of α-SMA, Col I, FN and Col IV proteins (Supplementary Figure 2). Hence, we concluded that miR-452-5p down-regulation attenuated HG-induced MC proliferation and fibrosis-related protein expression by up-regulating Mbnl1.
Discussion
The competing endogenous RNA (ceRNA) networks may link the function of protein-coding mRNA to non-coding RNAs, and studies have highlighted that ceRNA crosstalk through shared miRNAs has been linked to a variety of human diseases, including DN.27,28 Here, we showed, for the first time, that circ_Arf3 acted as a ceRNA to regulate Mbnl1 expression by sponging miR-452-5p, thereby regulating HG-induced proliferation and fibrosis-related protein expression in mouse MCs.
Many circRNAs have established important roles in the pathogenesis of DN.29 For instance, circ_LARP4 was down-regulated in DN cell models, and overexpression of circ_LARP4 restrained cell proliferation rates and contributed to apoptosis.30 Peng et al reported that circ_010383 was expressed at low levels in DN, which supported renal fibrosis in DN.31 Circ_Arf3, a relatively unexplored circRNA, has been reported to contribute to OS progression through the miR-1299/cyclin-dependent kinase 6 (CDK6) axis.25 However, no reports showed whether circ_Arf3 is involved in the pathogenesis of DN. Using HG-induced in vitro DN cell model, we first identified that circ_Arf3 expression was down-regulated in HG-induced MCs. Through functional analyses, we uncovered that overexpression of circ_Arf3 reduced HG-induced proliferation and fibrosis-related protein expression in mouse MCs. Additionally, in line with reported other circRNAs,32,33 circ_Arf3 was validated to have a higher tolerance to exonuclease and actinomycin D. Subcellular fractionation assay showed the main cytoplasmic localization of circ_Arf3 in MCs, providing a possibility that circ_Arf3 might bind and target miRNAs, which are known to be present in the cytoplasm in the RNA-silencing complex (RISC).34
Further, we first identified that circ_Arf3 targeted miR-452-5p by binding to miR-452-5p. MiR-452-5p has been identified as a strong oncogenic driver in colorectal cancer and hepatocellular carcinoma, where it is present at high levels.35,36 Conversely, miR-452-5p is down-regulated in human prostate cancer.37 Moreover, miR-452-5p works as a modulator of pancreatic endocrine dysfunction in diabetic Zucker rats.38 Importantly, miR-452-5p reduction protects HK-2 renal tubular cells against HG-induced pyroptosis and oxidative stress, implying the potential role of miR-452-5p in DN pathogenesis.23 Our findings first uncovered that circ_Arf3 repressed HG-induced proliferation and fibrosis-related protein expression in mouse MCs by targeting miR-452-5p. Xie et al demonstrated that long non-coding RNA (lncRNA) GAS5 exerts an inhibitory influence in HG-induced oxidative stress, inflammation and pyroptosis in renal tubular cells by sponging miR-452-5p.23
Mbnl1 is a crucial RNA-binding protein, and the importance of Mbnl1 in human pathologic processes has become increasingly clear in recent years. For instance, it has been demonstrated that Mbnl1 exerts an important function in leukemia and non-small cell lung cancer.39,40 Moreover, Mbnl1 participates in myocardial remodeling induced by isoproterenol.41 Importantly, Mbnl1 is underexpressed in senescent renal tubular epithelial cells and is able to participate in the cell senescence in DN through the modulation of miR-130a-3p/STAT3 cascade, suggesting its role as a promising therapeutic agent for DN treatment.42 In this paper, we first ascertained that Mbnl1 was a direct miR-452-5p target. Our results also demonstrated that Mbnl1 expression was down-regulated in HG-treated MCs, and knockdown of Mbnl1 reversed the impact of miR-452-5p inhibitor on HG-induced cell proliferation and fibrosis-related protein expression. Similarly, Yao et al reported that miR-654-3p regulated the downstream gene mitogen-activated protein kinase 6 (MAPK6) to affect podocyte damage in DN.43 More importantly, we first pointed to the ceRNA activity of circ_Arf3 to regulate Mbnl1 expression through miR-452-5p.
Such experiments are limited at present by the lack of in vivo investigation using DN animal models. Future work will be performed to determine the novel circ_Arf3/miR-452-5p/Mbnl1 ceRNA crosstalk as a potential target for DN therapy using in vivo DN animal models. Because circ_Arf3 is a relatively unidentified circRNA, not much information is available in public databases about its expression and role in DN patients. A future challenge will be to determine circ_Arf3 expression in serum samples and kidney tissues of DN patients to elucidate whether abnormal expression of circ_Arf3 is associated with DN progression.
In conclusion, we presented that circ_Arf3 positively modulated Mbnl1 by functioning as a ceRNA for miR-452-5p, thereby affecting HG-induced cell proliferation and fibrosis-related protein expression in mouse MCs (Figure 7).
Abbreviations
DN, Diabetic nephropathy; circRNAs, Circular RNAs; circ_Arf3, circRNA ADP ribosylation factor 3; HG, high glucose; MCs, mesangial cells; qRT-PCR, quantitative real-time polymerase chain reaction; Mbnl1, muscleblind like splicing regulator 1; EdU, 5-Ethynyl-2′-deoxyuridine; CCK-8, cell counting kit-8; ECM, extracellular matrix; AKT3, AKT serine/threonine kinase 3; miRNAs, microRNAs; MREs, miRNA response elements; ATCC, American Type Culture Collection; DMEM; Dulbecco’s modified Eagle’s medium; siRNA, small interfering RNA; cDNA, complementary DNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RNase R, Ribonuclease R; SDS-PAGE, SDS polyacrylamide gel electrophoresis; PCNA, proliferating cell nuclear antigen; α-SMA, alpha smooth muscle actin; Col I, collagen I; FN, fibronectin; HRP, horseradish peroxidase; 3′UTR, 3′untranslated region; CDK6, cyclin-dependent kinase 6; MAPK6, mitogen-activated protein kinase 6.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Shimizu M, Wada T. [Nephrotic syndrome in diabetic nephropathy and diabetes]. Nihon Jinzo Gakkai Shi. 2014;56:500–509. Japanese.
2. Natesan V, Kim SJ. Diabetic nephropathy - a review of risk factors, progression, mechanism, and dietary management. Biomol Ther. 2021;29:365–372. doi:10.4062/biomolther.2020.204
3. Qi C, Mao X, Zhang Z, Wu H. Classification and differential diagnosis of diabetic nephropathy. J Diabetes Res. 2017;2017:8637138. doi:10.1155/2017/8637138
4. Chen C, Gong W, Li C, et al. Sphingosine kinase 1 mediates AGEs-induced fibronectin upregulation in diabetic nephropathy. Oncotarget. 2017;8(45):78660–78676. doi:10.18632/oncotarget.20205
5. Tung C-W, Hsu Y-C, Shih Y-H, Chang P-J, Lin C-L. Glomerular mesangial cell and podocyte injuries in diabetic nephropathy. Nephrology. 2018;23(Suppl 4):32–37. doi:10.1111/nep.13451
6. Hu W, Han Q, Zhao L, Wang L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-β1. J Cell Physiol. 2019;234(2):1469–1476. doi:10.1002/jcp.26959
7. Hsiao K-Y, Sun HS, Tsai S-J. Circular RNA – new member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141. doi:10.1177/1535370217708978
8. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(01):3–11. doi:10.1055/s-0034-1397344
9. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
10. Li D, Yang Y, Li ZQ, Li LC, Zhu XH. Circular RNAs: from biogenesis and function to diseases. Chin Med J. 2019;132(20):2457–2464. doi:10.1097/CM9.0000000000000465
11. Wang L, Long H, Zheng Q, et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1):119. doi:10.1186/s12943-019-1046-7
12. Ge X, Xi L, Wang Q, et al. Circular RNA Circ_0000064 promotes the proliferation and fibrosis of mesangial cells via miR-143 in diabetic nephropathy. Gene. 2020;758:144952. doi:10.1016/j.gene.2020.144952
13. Wang Q, Cang Z, Shen L, et al. circ_0037128/miR-17-3p/AKT3 axis promotes the development of diabetic nephropathy. Gene. 2021;765:145076. doi:10.1016/j.gene.2020.145076
14. An L, Ji D, Hu W, et al. Interference of Hsa_circ_0003928 alleviates high glucose-induced cell apoptosis and inflammation in HK-2 cells via miR-151-3p/Anxa2. Diabetes Metab Syndr Obes. 2020;13:3157–3168. doi:10.2147/DMSO.S265543
15. Liu H, Wang X, Wang ZY, Li L. Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol. 2020;235:4520–4529. doi:10.1002/jcp.29329
16. Glinoer D. [The medical treatment of endocrine exophthalmos]. Bull Soc Belge Ophtalmol. 1988;226:59–64. French.
17. Peng F, Gong W, Li S, et al. circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 2021;70(2):603–615. doi:10.2337/db20-0203
18. Kaur P, Kotru S, Singh S, Munshi A. miRNA signatures in diabetic retinopathy and nephropathy: delineating underlying mechanisms. J Physiol Biochem. 2022;78(1):19–37. doi:10.1007/s13105-021-00867-0
19. Zhao SM, Zhang T, Qiu Q, et al. MiRNA-337 leads to podocyte injury in mice with diabetic nephropathy. Eur Rev Med Pharmacol Sci. 2019;23(19):8485–8492. doi:10.26355/eurrev_201910_19161
20. Sun Z, Ma Y, Chen F, et al. miR-133b and miR-199b knockdown attenuate TGF-β1-induced epithelial to mesenchymal transition and renal fibrosis by targeting SIRT1 in diabetic nephropathy. Eur J Pharmacol. 2018;837:96–104. doi:10.1016/j.ejphar.2018.08.022
21. Zongqiang H, Jiapeng C, Yingpeng Z, et al. Exosomal miR-452-5p induce M2 macrophage polarization to accelerate hepatocellular carcinoma progression by targeting TIMP3. J Immunol Res. 2022;2022:1032106. doi:10.1155/2022/1032106
22. Deng M, Hu J, Tong R, et al. miR-452-5p regulates the responsiveness of intestinal epithelial cells in inflammatory bowel disease through Mcl-1. Exp Ther Med. 2021;22:813. doi:10.3892/etm.2021.10245
23. Xie C, Wu W, Tang A, Luo N, Tan Y. lncRNA GAS5/miR-452-5p reduces oxidative stress and pyroptosis of high-glucose-stimulated renal tubular cells. Diabetes Metab Syndr Obes. 2019;12:2609–2617. doi:10.2147/DMSO.S228654
24. Song XH, He N, Xing Y-T, et al. A novel age-related circular RNA Circ-ATXN2 inhibits proliferation, promotes cell death and adipogenesis in rat adipose tissue-derived stromal cells. Front Genet. 2021;12:761926. doi:10.3389/fgene.2021.761926
25. Gao AM, Yuan C, Hu AX, Liu XS. circ_ARF3 regulates the pathogenesis of osteosarcoma by sponging miR-1299 to maintain CDK6 expression. Cell Signal. 2020;72:109622. doi:10.1016/j.cellsig.2020.109622
26. Zhou TC, Li X, Chen L-J, et al. Differential expression profile of hepatic circular RNAs in chronic hepatitis B. J Viral Hepat. 2018;25:1341–1351. doi:10.1111/jvh.12944
27. Karreth FA, P P. Pandolfi, ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi:10.1158/2159-8290.CD-13-0202
28. Qi X, Zhang D-H, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52(10):710–718. doi:10.1136/jmedgenet-2015-103334
29. Mou X, Chenv JW, Zhou DY, et al. A novel identified circular RNA, circ_0000491, aggravates the extracellular matrix of diabetic nephropathy glomerular mesangial cells through suppressing miR101b by targeting TGFbetaRI. Mol Med Rep. 2020;22(5):3785–3794. doi:10.3892/mmr.2020.11486
30. Wang X, Bai M. CircTM7SF3 contributes to oxidized low-density lipoprotein-induced apoptosis, inflammation and oxidative stress through targeting miR-206/ASPH axis in atherosclerosis cell model in vitro. BMC Cardiovasc Disord. 2021;21(1):51. doi:10.1186/s12872-020-01800-x
31. Peng F, Gong W, Li S, et al. circRNA_010383 acts as a sponge for miR-135a and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 2020:db200203. doi:10.2337/db200203
32. Zheng L, Liang H, Zhang Q, et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol Cancer. 2022;21(1):41. doi:10.1186/s12943-022-01495-y
33. Chen W, Cen S, Zhou X, et al. Circular RNA CircNOLC1, upregulated by NF-KappaB, promotes the progression of prostate cancer via miR-647/PAQR4 axis. Front Cell Dev Biol. 2021;8:624764. doi:10.3389/fcell.2020.624764
34. Stavast CJ, Erkeland SJ. The non-canonical aspects of microRNAs: many roads to gene regulation. Cells. 2019;8(11):1465. doi:10.3390/cells8111465
35. Lin X, Han L, Gu C, et al. MiR-452-5p promotes colorectal cancer progression by regulating an ERK/MAPK positive feedback loop. Aging. 2021;13(5):7608–7626. doi:10.18632/aging.202657
36. Zheng J, Cheng D, Wu D, et al. MiR-452-5p mediates the proliferation, migration and invasion of hepatocellular carcinoma cells via targeting COLEC10. Per Med. 2021;18(2):97–106. doi:10.2217/pme-2020-0027
37. Gao L, Zhang L-J, Li S-H, et al. Role of miR-452-5p in the tumorigenesis of prostate cancer: a study based on the Cancer Genome Atl(TCGA), Gene Expression Omnibus (GEO), and bioinformatics analysis. Pathol Res Pract. 2018;214(5):732–749. doi:10.1016/j.prp.2018.03.002
38. Su T, Hou J, Liu T, et al. MiR-34a-5p and miR-452-5p: the novel regulators of pancreatic endocrine dysfunction in diabetic Zucker rats? Int J Med Sci. 2021;18(14):3171–3181. doi:10.7150/ijms.62843
39. Itskovich SS, Gurunathan A, Clark J, et al. MBNL1 regulates essential alternative RNA splicing patterns in MLL-rearranged leukemia. Nat Commun. 2020;11(1):2369. doi:10.1038/s41467-020-15733-8
40. Cao G, Tan B, Wei S, et al. Down-regulation of MBNL1-AS1 contributes to tumorigenesis of NSCLC via sponging miR-135a-5p. Biomed Pharmacother. 2020;125:109856. doi:10.1016/j.biopha.2020.109856
41. Xu Y, Liang C, Luo Y, Zhang T. MBNL1 regulates isoproterenol-induced myocardial remodelling in vitro and in vivo. J Cell Mol Med. 2021;25(2):1100–1115. doi:10.1111/jcmm.16177
42. Jiang X, Ruan X-L, Xue Y-X, et al. Metformin reduces the senescence of renal tubular epithelial cells in diabetic nephropathy via the MBNL1/miR-130a-3p/STAT3 pathway. Oxid Med Cell Longev. 2020;2020:8708236. doi:10.1155/2020/8708236
43. Yao T, Zha D, Hu C, Wu X. Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene. 2020;747:144661. doi:10.1016/j.gene.2020.144661
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.