Back to Journals » Infection and Drug Resistance » Volume 13
A Novel blaCTX-M-65-Harboring IncHI2 Plasmid pE648CTX-M-65 Isolated from a Clinical Extensively-Drug-Resistant Escherichia coli ST648
Received 8 July 2020
Accepted for publication 9 September 2020
Published 30 September 2020 Volume 2020:13 Pages 3383—3391
DOI https://doi.org/10.2147/IDR.S269766
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Yang Lü,1 Haiquan Kang,2 Jianming Fan3
1State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, People’s Republic of China; 2Department of Laboratory Medicine, The Affiliated Hospital of Xuzhou Medical University, Xuzhou 221002, People’s Republic of China; 3The Laboratory of Toxicology, College of Public Health, Zhengzhou University, Zhengzhou 450001, People’s Republic of China
Correspondence: Jianming Fan Tel +86-13783617236
Email [email protected]
Background: An ESBL, carbapenemase- and MCR-1-producing Escherichia coli ST648 strain was isolated from the urine sample of a patient in a Chinese tertiary hospital in 2016.
Methods: The strain was fully sequenced by GridION X5 platform of Oxford Nanopore Technology.
Results: The sequence analysis showed that the extended-spectrum β-lactamases CTX-M-65 and OXA-1, the carbapenemase NDM-5, the MCR-1 were encoded, respectively, by three different resistance plasmids. The pE648CTX-M-65-carrying blaCTX-M-65 was a novel conjugative plasmid belonging to IncHI2 type; except for the blaCTX-M-65, it also carried resistance genes ble, floR, sul1, aph(4)-Ia, aac(3)-VI, aac(6ʹ)-II, blaOXA-1, catB, arr3 and tetA. Besides, an IncX4 plasmid pE648MCR-1-carrying mcr-1 and an IncX3 plasmid pE648NDM-5-carrying blaNDM-5 were also identified.
Conclusion: The three transferable resistance plasmids coexisting in the E. coli ST648 isolate indicated the high risk to disseminate the extensively-drug-resistance among Enterobacteriaceae.
Keywords: CTX-M-65, MCR-1, NDM-5, extensively-drug-resistance, Escherichia coli
Backgrounds
The global increase in carbapenemase-producing Enterobacteriaceae has resulted in increased use of colistin with the inevitable risk of emerging pan-drug-resistant Gram-negative bacteria.1 MCR-1-producing carbapenem-resistant Enterobacteriaceae (CRE) isolates pose a significant threat to global health. In 2016, two Escherichia coli isolates ST648 and ST156 coproducing MCR-1 and NDM-5 were first reported from a duck sample in China.2 Subsequently, the coexistence of MCR-1 resistance and the NDM-5 has been reported in E. coli and Klebsiella pneumoniae isolates cultured from patients in the USA and China; E. coli isolates, such as ST206, ST167, ST156 and ST405, harboring mcr-1 and blaNDM-5 have been reported causing intra-abdominal, bloodstream and urinary tract infections of patients.3–6 So, Feng et al concluded that the dissemination of mcr-1 colistin resistance gene is ongoing by clonal expansion among different sequence types of E. coli. MCR-1-producing CRE isolates represent a great concern for public health.7
In the reported E. coli ST648 isolate coproducing MCR-1 and NDM-5 cultured from a duck, the blaNDM-5 was found on an IncX3 plasmid while the mcr-1 gene was located on an IncHI2 plasmid, besides, the isolate also harbored extended-spectrum β-lactamases (ESBL) genes blaTEM-1 and blaCTX-M-55, fosfomycin resistance gene fosA3 and quinolone resistance gene aac(6ʹ)-Ib.2 E. coli ST648 strain, a predominant multidrug-resistant clone observed worldwide in humans, companion animals, livestock, and wild birds, is frequently associated with various β-lactamases, including ESBLs, NDM and KPC.8–11 So the epidemic multiresistant E. coli ST648 clone might raise a potential threat to human health.2 The first clinical E. coli ST648 isolate with mcr-1 and blaCTX-M-15 located on different plasmids was cultured from the microbiota of diarrhea patients in China in 2016, which NDM-1 variants NDM-5 or NDM-7 were not found in it.7,12 Here we firstly reported the clinical E. coli ST648 isolate coharboring ESBL genes blaCTX-M-65 and blaOXA-1; the carbapenemase gene blaNDM-5; the colistin-resistant gene mcr-1 located, respectively, in three different resistance plasmids.
The blaCTX-M-9-group gene blaCTX-M-65 has become one of the dominant CTX-M types in animal isolates in China since 2007;13,14 then it has rapidly spread and become the most common ESBL type instead of blaTEM and blaSHV in the acquired infection of the hospital and the community in China.15,16 The IncFII plasmids were the main vectors to transmit the blaCTX-M-65 among Enterobacteriaceae in China.17 The blaCTX-M-65 genes often coexist with genes encoding 16S rRNA methylases such as armA and rmtB, or with fosA3 that confers resistance to fosfomycin.17–19 The ISEcp1-blaCTX-M-65-IS903-iroN structure is a typical transposition unit in which blaCTX-M-65 locates.17 But a novel conjugative blaCTX-M-65-carrying plasmid designated pE648CTX-M-65 (241,735bp) belonging to IncHI2 type was firstly reported here, which the blaCTX-M-65 located in a different mosaic structure. Besides, a mcr-1-carrying plasmid designated pE648MCR-1 (33,277bp) and a blaNDM-5-carrying plasmid designated pE648NDM-5 (47,827bp) were also identified. This represents the clinical clone of E. coli ST648 can spread extensively-drug-resistance among Enterobacteriaceae.
Materials and Methods
Bacterial Isolates and Identification
The isolate was cultured from the urine specimen of a 55-year-old male patient suffered from recurrent urinary tract infection, along with bladder cancer, hypertension and coronary disease in a tertiary hospital of Xuzhou, China, in 2016. The patient had not been travelling and not engaged in animal breeding. The urinary tract infection was treated by ceftazidime and levofloxacin at the beginning. The bacterial species identification was performed using BioMérieux Vitek 2, Bruker MALDI Biotyper and 16S rRNA gene sequencing. The sequence type was identified by using the genome sequence to query the PubMLST database using the software written by Dr Torsten Seemann.20 The seven house-keeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) were assigned an allele number and a sequence type (ST) was determined for the isolate according to the allele profile. The phylogenetic tree of the five E. coli ST648 strains available as full genome sequences in Genbank was inferred using maximum likelihood as part of the software kSNP3 on all complete genomes.21
Antimicrobial Resistance
The minimum inhibitory concentrations (MICs) of antimicrobial agents were determined using the microdilution broth method according to the 2020 CLSI guidelines. The antimicrobial susceptibility profiles were interpreted by the 2020 CLSI guidelines and EUCAST available version (http://www.eucast.org/). The ESBL genes, carbapenemase genes and the mcr-1 gene were screened for by PCR with specific primers,1,22 as shown in Supplementary materials Table S1.
Plasmid Transfer
Conjugal transfer experiment was performed with Escherichia coli J53 Azir as the recipient strain. Overnight cultures of the bacteria were diluted to 1.5×108 cells/mL. Donor and recipient cells were mixed at 1:10 donor-to-recipient ratio, after 18 h of incubation of donor-recipient mixtures on blood plates at 35°C, cells were washed by normal saline solution and diluted to be cultured selectively on MacConkey agar plates supplemented with sodium azide (100 μg/mL) and ceftriaxone (10 μg/mL), or imipenem (1 μg/mL), or colistin (2 μg/mL) for 24 h. Transconjugants were confirmed by PCR amplifying blaCTX-M-9-group, blaNDM and mcr-1 with corresponding primers as shown in Table S1.
Whole Genome Sequencing and Data Analysis
Genomic DNA of the E. coli isolate was extracted using a QIAGEN Genomic-tip 500/G (Cat No.10262, QIAGEN, Duesseldorf, Germany). We applied an Oxford Nanopore Technology’s (ONT) GridION X5 sequencing technology for the sequencing.23,24 The strategy yielded a total of 4724.41 Mb raw data and a total of 4723.08 Mb filtered data were finally obtained after removing reads with mean_qscore <7 and length <1000 bp. The filtered reads were de novo assembled using the Canu package (version 1.3) with default parameters and the assembled data were fixed by Pilon (version 1.22) with default parameters, using the data obtained from the Illumina sequencing performed on an Illumina HiseqX10 platform (IlluminaInc., SanDiego, CA, USA) as the reference. The finally resulting genomes including the single circular chromosome and the circular plasmids were annotated using the best-placed reference protein set (GeneMarkS+) in the NCBI Prokaryotic Genome Annotation Pipeline (version. 3.3) and the RAST tool (version 2).25–27 The complete genome and plasmids sequences of the E. coli isolate 201,609 were submitted to GenBank under accession number CP048107 (Chromosomal genome sequence), MN200941 (Plasmid sequence of pE648CTX-M-65), MN200942 (Plasmid sequence of pE648NDM-5) and MN200943 (Plasmid sequence of pE648MCR-1).
Results and Discussion
Antibiotic Resistance of the E. coli Isolate
The isolated E. coli 201,609 was resistant to piperacillin, amoxicillin/clavulanic acid, ampicillin/sulbactam, piperacillin/tazobactam, cefoxitin, ceftazidime, cefotaxime, cefepime, aztreonam, imipenem, meropenem, ertapenem, amikacin, gentamycin, ciprofloxacin, levofloxacin, sulfamethoxazole, tetracycline and colistin, but susceptible to fosfomycin and tigecycline; see Table 1. We screened for the resistance genes by PCR as mentioned in Materials and Methods, the blaCTX-M-9-group, blaOXA-1, blaNDM and mcr-1 genes were successfully amplified. In the conjugation experiment, we also detected the blaCTX-M-9, blaNDM and mcr-1 genes, respectively, by PCR in the recipient E. coli J53 Azir, indicating resistance plasmids were transferable.
 |
Table 1 Antimicrobial Drug Susceptibility Profiles |
Genome Sequence of the E. coli Isolate
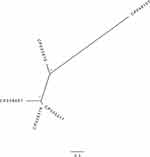
The complete genome of the E. coli isolate 201,609 was 5.53Mb which contained a 5.20Mb circular chromosome and three resistance plasmids named pE648CTX-M-65, pE648NDM-5 and pE648MCR-1, respectively. The GC percentage of the genome is 50.2%. The average depth of the chromosome (the raw data/the assembled data; per reference start position) was 175Х for Nanopore. The coverage of the circular chromosome (length alignment to plasmid_ databank/the whole length) was 19.58%, while the coverage of the three resistance plasmids was over 95%. The sequence type was identified as ST648 according to the allele profile of the seven house-keeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) which were assigned an allele number (92, 4, 87, 96, 70, 58, 2). The relatedness of the isolate in this study (Genbank ID: CP048107) to another four multiple antibiotic-resistant E. coli ST648 strains whose genomes were available in Genbank was shown in Figure 1. The four resistance strains were, respectively, isolated from a hospital of Beijing in 2009 (Genbank ID: CP008697), a hospital of Greek in 2018 (Genbank ID: CP035318 and CP035317) and a wild bird of Germany in 2012 (Genbank ID: CP023815). The phylogenetic tree generated using kSNP3.0 indicated the isolate in this study had a more close evolutionary distance to the one isolated from a wild bird compared with the another three isolates from hospitals.28
The sequencing results also showed that the E. coli ST648 isolate 201,609 harbored three resistance plasmids, the pE648CTX-M-65 carrying the ESBL genes blaCTX-M-65 and blaOXA-1 (Figure S1), the pE648NDM-5 carrying the carbapenemase gene blaNDM-5 (Figure S2) and the pE648MCR-1 carrying the colistin resistance gene mcr-1 (Figure S3). The annotated genes shown in the Figures S1–S3 were provided as the supplementary file: Annotated genes of Figures S1–S3.
Overview and Comparative Analysis of pE648CTX-M-65
The pE648CTX-M-65 is a 241,735 bp circular plasmid containing 373 putative ORFs (215 hypothetical proteins), the average GC content is 46.5%. The pE648CTX-M-65 is classified to IncHI2 type according to PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder-1.0/).29 The sequence alignment revealed the backbone of pE648CTX-M-65 might derive from the mcr-1-carrying plasmid pGD80-2 (GenBank ID KY075659) isolated from E. coli GD80 strain in China (Figure 2). Compared with the pGD80-2 which has two drug-resistance regions, the pE648CTX-M-65 has only one multiple-drug resistance region (MRR); and the genome was inserted into two IS903B, two IS629, one IS679 and one IS2 as shown in Figure 3. The MRR of pE648CTX-M-65, carried resistance genes ble, blaCTX-M-65, floR, sul1, aph(4)-Ia, aac(3)-VI, aac(6ʹ)-II, blaOXA-1, catB, arr3 and tetA. Here, the blaCTX-M-65 located in the IS26-blaCTX-M-65-ΔIS903 structure and was adjacent to the floR which was flanked by IS1006-ΔISVsa3 at its 5ʹ end and ISVsa3 at its 3ʹ end. An integrase-deficient class 1 integron with the conserved qacEΔ1 and sul1, carrying the antibiotic resistance gene cassettes aac(6ʹ)-II, blaOXA-1, catB and arr3 was flanked by two inversely oriented IS26. Here, the IS26 plays an important role in the accumulation of diverse antimicrobial resistance genes and the rearrangement of multidrug-resistance regions. The aph(4)-Ia and aac(3)-VI were flanked by IS26 and ISEc59, while the sul1 was flanked by ISAba1 and ISVsa3. The transposon flanked by three copies of IS26, also combined phenol degradation genes dmpK and dmpL flanked by ΔTn5393 and ISAba1, suggesting active occurrence of recombination and horizontal transfer of resistance genes under the environmental pressures.
 |
Figure 2 Sequence alignment of the pGD80-2 with the pE648CTX-M-65 generated by the Brig software. |
Overview and Comparative Analysis of pE648NDM-5 and pE648MCR-1
The pE648NDM-5-carrying blaNDM-5 is a 47,827 bp circular plasmid containing 73 putative ORFs (35 hypothetical proteins), the average GC content is 47%. It is classified into IncX3 type according to the PlasmidFinder. The direct spread of NDM-1 or NDM-5 by IncX3-type plasmids among Enterobacteriaceae was very common in China.30,31 Here, the pE648NDM-5 has 96% identity with the other two plasmids, the pNDM5_IncX3 (GenBank ID KU761328) isolated from a clinical K. pneumoniae and the pP855-NDM5 (GenBank ID MF547508) in an E. coli ST7511 strain isolated from a pig,3,31 as shown in Figure 4; indicating the epidemicity of blaNDM-5-carrying IncX3 plasmids among Enterobacteriaceae.
 |
Figure 4 Sequence alignment of the pE648NDM-5 with the pNDM5_IncX3 and the pP855-NDM5 generated by the Brig software. |
The mcr-1-carrying pE648MCR-1 is a 33,277 bp circular plasmid containing 54 putative ORFs (20 hypothetical proteins), the average GC content is 41.8%. It is classified into IncX4 type, which was also found very epidemic among mcr-1-carrying Enterobacteriaceae.32–34 Genome alignments indicated the pE648MCR-1 reported here was nearly identical to the pMCR1_IncX4 (GenBank ID KU761327) isolated from a clinical K. pneumoniae and the pGD65-4 (GenBank ID KY075660) in an E. coli isolated from swine,32,35 as shown in Figure 5. Besides, very similar plasmids (>95% identity) had been extensively reported in Salmonella enterica, K. pneumoniae and E. coli.3,31,33,35–37
 |
Figure 5 Sequence alignment of the pE648MCR-1 with the pMCR1_IncX4 and the pGD65-4 generated by the Brig software. |
Here, the mcr-1 and blaNDM-5 were located on different transferable plasmids, while a IncX3-X4 hybrid pCQ02-121 (Genbank ID KU647721) coharboring the mcr-1 and blaNDM-5 in an E. coli ST156 strain was isolated from a Chinese pet cat in 2015.38 The author proposed that the stable hybrid pCQ02-121 origin from the pNDM5_IncX3 (GenBank ID KU761328) and pMCR1_IncX4 (GenBank ID KU761327) isolated from a clinical K. pneumoniae ST25 strain in China by two IS26-mediated recombinations. Here, the sequence analysis of the pE648NDM-5 and the pE648MCR-1 which were highly homologous to the pNDM5_IncX3 and the pMCR1_IncX4, respectively, shown in Figures 4 and 5, was a more direct proof to support the recombination conclusions on the origin of a IncX3-X4 hybrid pCQ02-121, as shown in Figure 6. The majority (nt 8203–42968) of pCQ02-121 was almost identical to the pE648NDM-5 (nt 6389–41126) except for 4 mismatches and 22 gaps; while the region spanning nt 42958–9026 matched to the pE648MCR-1 (nt 27885–8994), only with 4 mismatches and 2 gaps.
Conclusions
The coexistence of ESBL, carbapenemase and MCR-1 leads to the dissemination of the extensively-drug-resistance among Enterobacteriaceae, which will inevitably become global. The finding of the extensively-drug-resistant E. coli ST648 201,609 strain stresses the need to monitor the use of colistin in treatment of both human beings and animals, as well as the need to surveil and restrict its further dissemination.
Ethical Approval
The urine specimen was part of the routine hospital laboratory procedure. The use of human specimens and all related experimental protocols was approved by the Committee on Human Research of the indicated institutions, and was carried out in accordance with the approved guidelines.
Funding
This work was supported by National Natural Science Foundation of China (No. 31372453) in the design of the research, sequencing and interpretation of data; and also supported by Six talent peaks project of Jiangsu Province (WSN-091) by sample collection and analysis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
2. Yang RS, Feng Y, Lv XY, et al. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (cairina moschata). Antimicrob Agents Chemother. 2016;60(11):6899–6902. doi:10.1128/AAC.01365-16
3. Du H, Chen L, Tang Y, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis. 2016;16(3):287–288. doi:10.1016/S1473-3099(16)00056-6
4. Yu H, Qu F, Shan B, et al. Detection of the mcr-1 colistin resistance gene in carbapenem-resistant enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother. 2016;60(8):5033–5035. doi:10.1128/AAC.00440-16
5. Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio. 2016;7(4). doi:10.1128/mBio.01191-16.
6. Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int J Antimicrob Agents. 2018;51(2):273–275. doi:10.1016/j.ijantimicag.2017.09.005
7. Zhang H, Seward CH, Wu Z, et al. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Sci Bull (Beijing). 2016;61(11):875–878. doi:10.1007/s11434-016-1086-y
8. Ewers C, Bethe A, Stamm I, et al. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother. 2014;69(5):1224–1230. doi:10.1093/jac/dkt516
9. Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the new delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55(12):5952–5954. doi:10.1128/AAC.05108-11
10. Paulshus E, Thorell K, Guzman-Otazo J, et al. repeated isolation of extended-spectrum-β-lactamase-positive Escherichia coli sequence types 648 and 131 from community wastewater indicates that sewage systems are important sources of emerging clones of antibiotic-resistant bacteria. Antimicrob Agents Chemother. 2019;63(9):e00823–19. doi:10.1128/AAC.00823-19
11. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
12. Ye H, Li Y, Li Z, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio. 2016;7(2):e00177. doi:10.1128/mBio.00177-16
13. Wu C, Wang Y, Shi X, et al. Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008–2014. Emerg Microbes Infect. 2018;7(1):30. doi:10.1038/s41426-018-0033-1
14. Wang J, Zeng Z-L, Huang XY. Evolution and comparative genomics of F33:A−:B− plasmids carrying blaCTX-M-55 or blaCTX-M-65 in Escherichia coli and Klebsiella pneumoniae isolated from animals, food products, and humans in China. mSphere. 2018;3(4):e00137–18. doi:10.1128/mSphere.00137-18
15. Yong Z, Xinwei W, Jing Z, et al. High prevalence of CTX-M beta-lactamases in enterobacteriaceae from healthy individuals in Guangzhou, China. Microb Drug Resist. 2015;21(4):398–403. doi:10.1089/mdr.2014.0201
16. Miao M, Wen H, Xu P, et al. Genetic diversity of carbapenem-resistant enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in eastern China. Front Microbiol. 2018;9:3341. doi:10.3389/fmicb.2018.03341
17. Pan YS, Yuan L, Zong ZY, et al. A multidrug-resistance region containing blaCTX-M-65, fosA3 and rmtB on conjugative IncFII plasmids in Escherichia coli ST117 isolates from chicken. J Med Microbiol. 2014;63(3):485–488. doi:10.1099/jmm.0.070664-0
18. Lei CW, Chen YP, Kang ZZ, et al. Characterization of a novel SXT/R391 integrative and conjugative element carrying cfr, blaCTX-M-65, fosA3, and aac(6ʹ)-Ib-cr in proteus mirabilis. Antimicrob Agents Chemother. 2018;62.
19. He L, Partridge SR, Yang X, et al. Complete nucleotide sequence of pHN7A8, an F33:A-:B- type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother. 2013;68(1):46–50. doi:10.1093/jac/dks369
20. Jolley KA, Maiden MC. BIGSdb_scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010;11(1):595. doi:10.1186/1471-2105-11-595
21. Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31(17):2877–2878. doi:10.1093/bioinformatics/btv271
22. Chen Z, Li H, Feng J, et al. NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical. Front Microbiol. 2015;6:294.
23. Bayliss SC, Hunt VL, Yokoyama M, et al. The use of oxford nanopore native barcoding for complete genome assembly. Gigascience. 2017;6(3):1–6. doi:10.1093/gigascience/gix001
24. Li R, Xie M, Dong N, et al. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience. 2018;7(3):1–9. doi:10.1093/gigascience/gix132
25. Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi:10.1186/1471-2164-9-75
26. Overbeek R, Olson R, Pusch GD, et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014;42(D1):D206–D14. doi:10.1093/nar/gkt1226
27. Brettin T, Davis JJ, Disz T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5(1). doi:10.1038/srep08365.
28. Ashok Kumar J, Vinaya Kumar K, Avunje S, et al. Phylogenetic relationship among brackishwater Vibrio species. Evol Bioinform Online. 2020;16:1–8. doi:10.1177/1176934320903288
29. Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi:10.1128/AAC.02412-14
30. Lu Y, Liu W, Liang H, et al. NDM-1 encoded by a pNDM-HN380-like plasmid pNDM-BJ03 in clinical Enterobacter cloacae. Diagn Microbiol Infect Dis. 2018;90(2):153–155. doi:10.1016/j.diagmicrobio.2017.10.022
31. Pak-Leung H. IncX3 epidemic plasmid carrying blaNDM-5 in Escherichia coli from swine in multiple geographic areas in China. Antimicrob Agents Chemother. 2018;62.
32. Wang Q, Sun J, Li J, et al. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome. 2017;5(1):70. doi:10.1186/s40168-017-0288-0
33. Ageevets V, Lazareva I, Mrugova T, et al. IncX4 plasmids harbouring mcr-1 genes: further dissemination. J Glob Antimicrob Resist. 2019;18:166–167. doi:10.1016/j.jgar.2019.07.002
34. Li R, Xie M, Zhang J, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393–401. doi:10.1093/jac/dkw411
35. Li A, Yang Y, Miao M, et al. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing enterobacteriaceae. Antimicrob Agents Chemother. 2016;60(7):4351–4354. doi:10.1128/AAC.00550-16
36. Lu X, Zeng M, Xu J, et al. Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006–2016. EBioMedicine. 2019;42:133–144. doi:10.1016/j.ebiom.2019.03.006
37. Di Pilato V, Arena F, Tascini C, et al. mcr-1.2, a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing klebsiella pneumoniae strain of sequence type 512. Antimicrob Agents Chemother. 2016;60(9):5612–5615. doi:10.1128/AAC.01075-16
38. Sun J, Yang RS, Zhang Q, et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 2016;1(12):16176. doi:10.1038/nmicrobiol.2016.176
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.