Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
ZNF143 Expression is Associated with COPD and Tumor Microenvironment in Non-Small Cell Lung Cancer
Authors Feng Z, Yin Y, Liu B, Wang L, Chen M, Zhu Y, Zhang H, Sun D, Qin J
Received 22 December 2021
Accepted for publication 24 March 2022
Published 2 April 2022 Volume 2022:17 Pages 685—700
DOI https://doi.org/10.2147/COPD.S352392
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Zhang
Zhenxing Feng,1 Yan Yin,2 Bin Liu,2 Lei Wang,2 Miaomiao Chen,2 Yue Zhu,2 Hong Zhang,1 Daqiang Sun,3 Jianwen Qin2
1Department of Radiology, Tianjin Chest Hospital, Tianjin, 300222, People’s Republic of China; 2Respiratory and Critical Care Medicine, Tianjin Chest Hospital, Tianjin, 300222, People’s Republic of China; 3Department of Thoracic Surgery, Tianjin Chest Hospital, Tianjin, 300222, People’s Republic of China
Correspondence: Jianwen Qin, Respiratory and Critical Care Medicine, Tianjin Chest Hospital, Tianjin, 300222, People’s Republic of China, Email [email protected] Daqiang Sun, Department of Thoracic Surgery, Tianjin Chest Hospital, Tianjin, 300222, People’s Republic of China, Email [email protected]
Background: Chronic obstructive pulmonary disease (COPD) is an inflammatory-related disease highly associated with increased lung cancer risk. Studies have explored the tumor promoting roles for zinc finger protein 143 (ZNF143). However, the role of ZNF143 in COPD and tumor microenvironment of non-small cell lung cancer (NSCLC) has not been fully elucidated.
Methods: COPD-related key genes were identified by differential gene expression evaluation, WGCNA and SVM-RFE analysis using mRNA expression data retrieved from public databases. ROC analysis was conducted to evaluate the diagnostic value of ZNF143. Correlation between ZNF143 and clinic-pathological features, associations with tumor-infiltrating immune cells (TICs) and the relationship with predictors of immunotherapy efficacy were explored. ZNF143 gene expression was validated by qRT-PCR using an independent cohort.
Results: Bioinformatic and machine learning analysis showed that ZNF143 was a COPD-related gene. ZNF143 expression was significantly upregulated in COPD and is a potential diagnostic biomarker in COPD with AUC > 0.85. ZNF143 expression was significantly upregulated in lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). ZNF143 expression levels were significantly higher in LUAD patients with COPD relative to the levels in patients only with LUAD. Upregulation of ZNF143 in patients with comorbidity of NSCLC and COPD was further confirmed by qRT-PCR analysis. High expression of ZNF143 was significantly correlated with advanced TNM stage in LUSC. High ZNF143 expression was associated with activated TICs in both LUAD and LUSC samples. Moreover, ZNF143 expression was significantly correlated with the levels of several known predictors of immunotherapy efficacy, including PD-L1, PD-L2, TMB and TIDE in NSCLC.
Conclusion: ZNF143 is a novel COPD biomarker. High expression level of ZNF143 is associated with immune microenvironment and high risk of progression of COPD to NSCLC.
Keywords: COPD, NSCLC, ZNF143, bioinformatic, tumor microenvironment, PD-L1
Introduction
Lung cancer is the most common cause of cancer-associated deaths globally, with a 5-year survival rate of 19.0%.1 Non-small cell lung cancer (NSCLC) accounts for about 85% of lung cancer cases, with lung squamous cell carcinoma (LUAD) and lung adenocarcinoma (LUSC) being the most prevalent subtypes.2 Tobacco smoking is a lung cancer risk factor and the main cause of chronic obstructive pulmonary disease (COPD).3 COPD is associated with chronic airway inflammation, and patients with COPD have a high risk for lung cancer even after adjusting for smoking status.3 This implies that COPD is an independent lung cancer risk factor, indicating a plausible relationship between lung cancer and COPD.
Tumor microenvironment (TME), particularly the immune microenvironment, plays a key role in NSCLC occurrence and progression.4 Inflammation reprograms the immune microenvironment under pathological conditions such as COPD, thereby increasing risk of NSCLC development.5,6 COPD promotes tumor progression and reduces efficacy of lung cancer therapies.7,8 Immunotherapy, including immune checkpoint inhibitors (ICIs), is an effective modality for lung cancer treatment and is characterized by sustained clinical responses.9,10 However, the overall treatment response rate of immunotherapy in NSCLC is approximately 15–20% and biomarkers that can distinguish between responders and non-responders have not been elucidated.11 Survival rate is higher in ICI recipients among NSCLC patients coexisting with COPD, indicating that COPD-related TME modulates treatment efficacy.12,13 However, the significance of immunomodulatory cues in development and treatment of NSCLC associated with COPD has not been elucidated. Therefore, it is imperative to identify the shared biomarkers of COPD and NSCLC to elucidate the mechanisms underlying transformation from COPD to NSCLC. In addition, these markers will provide a basis for development of personalized therapy for NSCLC coexisting with COPD.
Next-generation sequencing is widely used to study association between various tumors and COPD.14,15 Bioinformatics is currently used in assessment of gene expression profiles to evaluate disease-associated molecular mechanisms and identify disease-specific biomarkers, owing to advances in genomics.16,17 Differential gene expression analysis is used to elucidate the molecular mechanisms involved in genome regulation and evaluation of gene expression levels between diseased and healthy groups.17,18 These gene expression patterns provide a foundation for identification of potential biomarkers for various diseases. In addition, weighted gene co-expression network analysis (WGCNA) is a key approach in elucidating gene functions as well as associations using genome-wide expressions.19 WGCNA is used in evaluation of co-expression modules of highly related genes and clinical trait-associated interested modules.20 WGCNA finding are used to elucidate the roles of co-expression genes and for identification of genes that play vital functions in development of human diseases, such as COPD and cancer.15,21 Moreover, support vector machine-recursive feature elimination (SVM-RFE) is a novel gene selection machine learning approach for identifying features to the model predictive power, and is developed using gene expression profiles.22 Results from differential gene expression analysis, WGCNA and SVM-RFE are pooled to identify differentially expressed genes that can served as candidate markers.
In the present study, COPD-related key genes were identified through differential gene expression analysis, SVM-RFE and WGCNA based on COPD mRNA expression data in GSE76925 dataset. A novel robust diagnostic biomarker, zinc finger protein 143 (ZNF143) was identified in COPD based on GSE76925 and GSE38974 datasets through ROC analysis. The findings showed that that ZNF143 expression was significantly upregulated in LUAD, LUSC and NSCLC coexisting with COPD compared with normal lung tissues from The Cancer Genome Atlas (TCGA) datasets. Notably, ZNF143 expression was significantly correlated with abundance of tumor-infiltrating immune cells (TICs) and signatures of immunotherapy efficacy, including PD-L1, PD-L2, Tumor mutational burden (TMB) and tumor immune dysfunction and exclusion (TIDE) prediction scores in NSCLC. These results imply that ZNF143 is a potential diagnostic marker for COPD. The findings indicates that high expression levels of lung ZNF143 promote cancer development and modulates the immune microenvironment.
Materials and Methods
Analysis of COPD-Related Genes
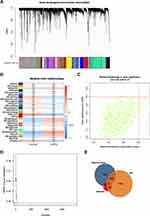
A flowchart showing the design of the study is presented in Figure 1. GSE7692523 and GSE3897424 datasets comprised 111 and 26 lung tissue samples obtained from COPD patients as well as 40 and 9 lung tissue samples obtained from smokers who did not present with COPD. Differential gene expression analysis, WGCNA and SVM-RFE analyses were conducted to explore highly COPD-related genes using GSE76925 dataset. COPD-related key genes were validated using GSE38974 dataset.
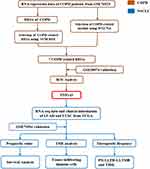 |
Figure 1 Schematic presentation of the flow of this study. |
LUAD and LUSC-Related Genes
Transcriptome profiles, somatic mutation information and the corresponding clinical data of LUAD (tumor samples, 535 cases; normal samples, 59 cases) and LUSC cases (tumor samples, 502 cases; normal samples, 49 cases) were obtained from TCGA database25 using GDC API. Normalized expression profiles for GSE72094 dataset26 comprising gene expression profile for LUAD were retrieved from gene expression omnibus (GEO). GSE72094 dataset consisted of 442 samples from LUAD patients with detailed mRNA expression data and clinical information. GSE72094 dataset was used to verify the role of ZNF143 in LUAD. DNA methylation and copy number variations (CNVs) of ZNF143 in NSCLC were explored using cBioPortal for Cancer Genomics.27 Clinicopathological characteristics of NSCLC patients included in this study were as presented in a previous study.28 COPD was defined as post-bronchodilator forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) ratio < 0.70 based on TCGA datasets.15
Analysis of Differentially Expressed Genes (DEGs) Between Normal Tissues and COPD Tissues
DEGs were evaluated by comparing normal and COPD samples using “limma” package in R software (3.6.3) based on GSE76925 dataset. Genes were considered DEGs if they met the following criteria: 1. adjusted p-value <0.05; 2. absolute value of log2 fold change (FC) > 0.5 (COPD samples vs normal samples).
Establishment of WGCNA Co-Expression Network
WGCNA was conducted to build a co-expression network between normal samples and COPD using “WGCNA” package in R based on gene expression data profiles in GSE76925 dataset. A soft threshold of 8 was used to build a scale-free network. A cluster dendrogram and network heatmap of the genes were generated. The eigengene values of modules were determined and a clustering tree was built.
SVM-RFE Analysis
Significant COPD-related DEGs were identified by SVM-RFE algorithm using “e1071”, “kernlab” and “Caret” packages in R software based on GSE76925 dataset.
RNA Extraction and qRT-PCR Validation
Lung tissue samples were obtained from NSCLC patients who underwent surgical resection in our hospital between October 2019 and October 2020. Patients were randomly selected, and LUAD or LUSC were pathologically confirmed. Histologically normal lung tissues located >5 cm from the edge of the cancerous tissue were used as controls. Lung tissues were stored at −80◦C after surgical resection. Patients with lung function of FEV1/FVC<0.7 were defined as patients with lung cancer coexisting with COPD. Patients with asthma, severe systemic disease, and COPD exacerbation occurring 4 weeks before Surgery were excluded from the study. Patients who had received targeted-therapy, immunotherapy and chemotherapy and radiotherapy before the surgery were also excluded from the study. Characteristics of patients included in the study are presented in Table S1.
Total RNA was extracted from lung tissues (19 NSCLC coexisting with COPD, 28 NSCLC, and 28 controls) using TRIzol reagent according to the manufacturer’s instructions (Thermo Fisher Scientific, USA). Relative expression level of ZNF143 was determined using 2−ΔCt method (ΔCt = CtZNF143–CtGAPDH). Primers used in the study were purchased from Sangon Biotech (Shanghai, China). The primers were as follows 5′-AATCCCATCACCATCTTCCAG-3′; and 5′-CACGAGTCCTTCCACGATACC-3′ used as the forward and reverse primers, respectively, for GAPDH. In addition, 5′-TGAGTCAACAAGTTACAC-3′ and 5′-TATGGAATGCTGCCAAGT-3′ were used as the forward and reverse primers for analysis of ZNF143.
Analysis of Tumor Infiltrating Immune Cells
CIBERSORT tool was used to explore the proportions of TICs in LAUD and LUSC samples retrieved from TCGA database.29 Samples with p < 0.05 were selected for subsequent analyses.
Estimation of TMB and TIDE
TMB and TIDE are biomarkers for assessing responses to immune therapy in lung cancer.30,31 TMB for each LUAD and LUSC sample from TCGA was defined as the number of identified somatic cell mutations, with the exception of silent mutations. Clinical responses of LUAD as well as LUSC were evaluated using TIDE algorithm in TIDE database (http://tide.dfci.harvard.edu/).30 TMB value and TIDE prediction score were calculated as previously described.30,32
Statistical Analysis
The median of gene expression value as cut off value. NSCLC samples were assigned to high- and low- groups based on median ZNF143 mRNA expression levels. Comparison of ZNF143 expression and categorical variables between the two groups was conducted by Kruskal Wallis rank sum test or Wilcoxon rank sum test. Survival analysis was conducted to explore the prognosis value of ZNF143 in LUAD and LUSC patients. Samples with a < 1 month survival time and normal samples were excluded from survival analysis. Survival curves were generated using Kaplan-Meier approach based on log rank test. Receiver operating characteristic (ROC) curve analyses were conducted to evaluated the diagnostic significance of ZNF143 in differentiating of COPD samples from normal samples. GraphPad Prism software (version. 8.0) was used for analyses with p < 0.05 as the significant threshold.
Results
Highly COPD-Related Key Genes Were Identified
A total of 1915 DEGs were identified from COPD tissues, relative to normal tissues (Table S2 and Figure S1A). The results showed that 467 genes were significantly upregulated whereas 1448 genes were downregulated (Table S2 and Figure S1A). The top 50 upregulated and downregulated genes were determined by absolute values of log2 FC (Figure S1B). WGCNA results are shown in Figure 2A–C. A total of 27 modules in the GSE76925 dataset were identified and every module was assigned a different color (Figure 2A). A module-trait relationships heatmap was generated to assess the relationships between every module and the two groups (normal and COPD). The green yellow module (697 genes, Table S3) was highly associated with COPD (Green yellow module: r = 0.390, p<0.0001; Figure 2B and C). A total of 40 COPD-associated DEGs were identified from GSE76925 dataset based on the SVM-RFE algorithm (Figure 2D and Table S4). A Venn diagram was generated and 7 overlapping genes (ARF6, LOC100133770, LOC392285, SLC12A2, UBASH3B, WDR85 and ZNF143) were identified as highly COPD-related key genes for subsequent analysis (Figures S1A and 2E).
ZNF143 is a Potential Biomarker for COPD
Analysis of the seven COPD-related key genes in GSE76925 dataset showed that expression of WDR85 and ZNF143 genes was upregulated, whereas ARF6, LOC100133770, LOC392285, SLC12A2 and UBASH3B genes were downregulated in the COPD samples, relative to normal lung tissue samples (Figure 3A). The results were validated using GSE38974 dataset (Figure 3B). LOC100133770 and LOC392285 expression was not available in GSE38974 dataset, thereby they were not validated. The results showed that only ZNF143 was significantly upregulated in COPD samples, relative to normal lung tissue samples (P=0.001; Figure 3B). Therefore, ZNF143 was used in subsequent studies. ROC analysis was conducted to evaluate the diagnostic value of ZNF143 in COPD based on GSE76925 dataset (Figure 4A) and GSE38974 dataset (Figure 4B). The area under curves (AUCs) of ZNF143 in GSE76925 and GSE38974 datasets were 0.855 (p<0.0001) and 0.930 (p=0.0002) (Table 1). These results showed that ZNF143 had high specificity and sensitivity, therefore, it is a potential diagnostic biomarker for COPD. The findings indicate that ZNF143 is implicated in pathogenesis of COPD and is a promising diagnostic biomarker for COPD.
 |
Table 1 ROC Analysis Results of ZNF143 in GSE76925 and GSE38974 Datasets |
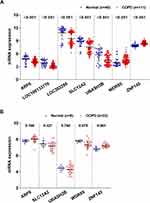 |
Figure 3 Levels of COPD-associated key genes in COPD vs normal based on GSE76925 (A) and GSE38974 (B) datasets. |
 |
Figure 4 ROC curve analysis of ZNF143 expressions for predicting COPD risks for healthy controls in GSE76925 (A) and GSE38974 (B) datasets. |
High Expression of ZNF143 is Associated with LUAD and LUSC
ZNF143 levels in LUAD and LUSC were evaluated to determine the correlation between ZNF143 and NSCLC tumorigenesis. The results showed that ZNF143 was significantly elevated in LUAD (P < 0.0001; Figure 5A) and LUSC (P < 0.0001; Figure 5B) from TCGA cohorts, relative to normal lung tissues. Besides, 25.0% (29/115) of LUAD and 49.0% (41/84) of LUSC patients were found to be coexisting with COPD in TCGA datasets, and there appeared a significant larger percentage of patients coexisting with COPD in LUSC comparing to LUAD (P<0.001; Figure S2). ZNF143 levels were markedly increased in tumor tissues of LUAD (P<0.001; Figure 5C) and LUSC (P=0.010; Figure 5D) coexisting COPD, relative to normal lung tissues. Moreover, ZNF143 levels in LUAD patients with COPD were elevated, relative to LUAD-alone patients (Figure 5C). However, ZNF143 mRNA was not significantly increased patients with LUSC coexisting COPD, relative to LUSC alone patients (P > 0.05; Figure 5D). To further verify the findings, ZNF143 gene expression was detected by qRT-PCR in an independent cohort. As show in Figure S3, the level of ZNF143 was significantly upregulated in tissues of patients with NSCLC coexisting COPD (P<0.001) and NSCLC alone (P<0.001) than controls. Moreover, the level of ZNF143 expression was significantly higher in patients with NSCLC coexisting COPD than that in patients with NSCLC alone (P=0.023). The verification results indicating that this gene may play a crucial role in the tumorigenesis of lung cancer, and the link between the expression of ZNF143 in COPD and NSCLC may be relevant.
Next, clinical information of LUSC and LUAD patients was evaluated to establish the association between ZNF143 and clinic-pathological features. ZNF143 expressions was significantly increased along with the progression of TNM stage in LUSC (P = 0.002; Figure 5F), but not in LUAD (P = 0.212; Figure 5E). These findings imply that ZNF143 has a crucial part in the progress and development of NSCLC. We further performed a survival analysis to determine the potential prognostic significance of ZNF143 in LUSC and LUAD. Results showed that there was no difference in progression-free survival (PFS) and overall survival (OS) between ZNF143 high- and low- groups in LUAD and LUSC based on TCGA and GSE72094 datasets (P>0.05; Figure S4A–E). This result indicated that ZNF143 alone might not be useful as a prognosis predictor for NSCLC.
ZNF143 Expression is Associated with the Proportion of TICs in NSCLC
The relative abundance of 22 kinds of TICs was determined by CIBERSORT algorithm. The relationship between ZNF143 expression and infiltrating levels of these TICs in LUAD and LUSC was evaluated. Differential analyses showed that seven types of TICs exhibited a significant association with ZNF143 levels in LUAD (Figure 6A). Five TICs including memory activated CD4+ T cells, resting mast cells, gamma delta T cells, activated mast cells, eosinophils, as well as neutrophils, exhibited a positive correlation with ZNF143 expression levels. Regulatory T cells (Tregs) exhibited a negative relationship with ZNF143 expression levels. Notably, CD8+ T cells showed a borderline significant association with ZNF143 levels in LUAD (P=0.054; Figure 6A). In addition, eight types of TICs showed a significant correlation with ZNF143 levels in LUSC (Figures 6B). CD8+ T cells, resting dendritic cells, memory activated CD4+ T cells, M1 and M2 macrophages, as well as eosinophils were positive correlated with ZNF143 levels. Naïve B cells and M0 Macrophages exhibited a negative association with ZNF143 expression level in LUSC. These findings indicate that ZNF143 levels exert various immune effects in the TME of LUSC and LUAD.
 |
Figure 6 Associations between TICs abundance with ZNF143 levels in LUAD (A) and LUSC (B) tissues. |
Correlation Between ZNF143 and the Predictors of Immunotherapy Efficacy
The association between ZNF143 levels with known immune check points and predictors of immunotherapy efficacy in NSCLC was evaluated to explore immunotherapeutic responses mediated by ZNF143. Expression of PD-L1 (P < 0.05; Figure 7A) and PD-L2 (P < 0.001; Figure 7B) was significantly upregulated in the ZNF143 high expression group compared with ZNF143 low expression group in LUAD-TCGA dataset. This finding was verified using an independent dataset with large LUAD dataset (GSE72094; P < 0.05; Figure 7C and D). PD-L2 (P = 0.003; Figure 7F) was significantly upregulated in ZNF143 high expression group relative to the ZNF143 low group in LUSC-TCGA dataset. However, there was no significant difference in expression level of PD-L1 between the ZNF143 high expression group and ZNF143 low expression group (P = 0.633; Figure 7E). A higher TMB value was observed in ZNF143 high expression group in LAUD (P < 0.001; Figure 7G) and LUSC compared with the TMB values for ZNF143 low expression groups (P = 0.051; Figure 7H). TIDE algorithm was used for prediction of immunotherapeutic responses and the findings showed that the two ZNF143 groups exhibited significantly different TIDE prediction score in LUAD (P < 0.001; Figure 7I) as well as LUSC (P < 0.001; Figure 7J). These results indicated that LUAD and LUSC patients with high ZNF143 expression levels were more likely to respond to immunotherapy compared with those with low ZNF143 expression levels.
DNA Copy Gain and DNA Methylation Modulate ZNF143 Upregulation in NSCLC
Further analysis was conducted to explore the mechanisms involved in ZNF143 upregulation in LUAD and LUSC from a genetic and epigenetic alterations perspective using TCGA datasets. ZNF143 copy gain was significantly associated with ZNF143 mRNA overexpression in LUSC dataset (P = 0.009; Figure 8A), but not in LUAD dataset (P = 0.613; Figure 8B). Further, the relationship between ZNF143 levels and DNA methylation in NSCLC was explored. Spearman correlation analysis revealed a significant correlation between ZNF143 mRNA levels and the total DNA methylation levels in both LUAD (r=−0.145, P=0.001; Figure 8C) and LUSC datasets (r=−0.254, P<0.001; Figure 8D). These results provide a basis for understanding the regulation mechanism of ZNF143 expression in NSCLC.
Discussion
ZNF143 is the human homolog of the transcriptional activator staf. It is a C2H2-type protein comprising seven zinc finger domains.33 ZNF143 is involved in chromatin loop formation as well as gene regulation. It is a highly expressed in transcriptional activation factor, that regulates expressions of genes involved in DNA replication and cell cycle.33 ZNF143 plays various roles in several cellular as well as pathogenic processes, including cell survival and proliferation.34 Molecular mechanisms underlying ZNF143-associated gene transcriptional regulation have been explored through molecular and bioinformatics approaches. However, only few available studies have explored the expression profile of ZNF143 in COPD and its role in pathogenesis of COPD has not been fully elucidated.
The findings of the present study showed that ZNF143 was significantly upregulated in COPD compared with the expression in normal lung tissues. ZNF143 was identified as a COPD-related key gene using integrated bioinformatics analysis and machine learning methods. ROC analyses as well as AUC statistics have been recommended for objective evaluation of biomarker performance in various diseases.35 It is also used for comparisons of the significance of biomarkers in COPD, whereby biomarkers with AUC >0.85 have a high accuracy while those with AUC between 0.7 and 0.85 have borderline potential for clinical translations, which warrants refinement and validation.35,36 In this study, the AUC of ZNF143 for diagnosis of COPD were 0.855 and 0.930 for the two independent datasets, indicating a high predictive value.
ZNF143 expression was upregulated in LUAD and LUSC samples and high ZNF143 expression was significantly associated with advanced TNM stage in LUSC in the current study. Moreover, ZNF143 mRNA levels were associated with its CNVs and methylation levels in NSCLC. This finding implies that CNVs and methylation account for the significant upregulation of ZNF143 expression in NSCLC. Survival analysis showed that ZNF143 was not a prognostic biomarker in NSCLC. These findings are in tandem with findings from previous studies that reported overexpression of ZNF143 and its possible effects in promoting in development of various cancer types, including lung cancer.37 ZNF143 is present in various solid cancers, and plays a key role in cisplatin resistance since cisplatin-induced ZNF143 binds cisplatin-modified DNA.38 Dysregulation of ZNF143 expression is associated with cell survival, proliferation, migration, as well as invasion. Therefore, novel small molecules can be designed to target ZNF143 as a potential marker and therapeutic target for cancers.38 Kawatsu et al reported that ZNF143 protein is highly expressed in LUAD samples. In addition, high ZNF143 expression levels were significantly associated with pathologically moderate to poor differentiation and invasive characteristics in LUAD. However, ZNF143 expression is not significantly correlated with disease-specific postoperative survival in LUAD.37 YPC-22026 and YPC-21661 have been identified as ZNF143 inhibitors, hence they can be used to target ZNF143 for development of cancer therapeutics.39
Previous studies mainly focused on the functional roles as well as mechanisms of ZNF143 in tumor progression and metastasis of cancers. In the present study, the role of ZNF143 in LUAD and LUSC with COPD comorbidity was evaluated. Comorbidity of COPD and lung cancer is common, and leads to worse prognostic outcomes compared with occurrence of lung cancer alone. Approximately 40–70% of lung cancer patients are diagnosed with COPD.40 The results of the present study showed that 35.0% (70/199) of NSCLC patients had COPD and the frequency of COPD in the LUSC patients was higher compared with that in LUAD patients based on TCGA datasets which is consistent with findings from our previous research.8 ZNF143 was upregulated in COPD patients as well as in LUAD and LUSC patients, indicating ZNF143 is a potentially biomarker for NSCLC with COPD comorbidity. Notably, ZNF143 expression was significantly upregulated in tumor tissues from LUAD patients with coexisting COPD, relative to the expression level in LUAD alone and normal lung tissues. qRT-PCR validation using a different cohort showed upregulation of ZNF143 expression in patients with NSCLC, and the level of gene expression was significantly higher in patients with COPD comorbidity. These results on ZNF143 in COPD and NSCLC analyzed in the current study indicates a relationship between COPD and lung cancer. These results imply that ZNF143 induces COPD progression to lung cancer. However, this postulate should be explored further.
CIBERSORT analysis revealed that ZNF143 high expression was significantly associated with relative abundance of activated immune infiltrating cells in NSCLC. High expression level of ZNF143 was correlated with high infiltration levels of TICs, especially CD8+ T cells and CD4+ T cells, in both LUAD and LUSC patients. This finding indicates that ZNF143 is associated with “hot” TME, which promotes efficacy of ICIs therapy.41,42 Trujillo et al previously reported that abundance of T-cell inflamed TME is correlated with favorable efficacy of PD-1 blockade,43 thus it can promote better responses to anti-PD-1 treatment in NSCLC coexisting with COPD.12,13,44 In the present study, ZNF143 expression in LUAD was positively correlated with levels of immune check points and predictors of immunotherapy efficacy (such as PD-L1/PD-L2 and TMB) and was inversely correlated with TIDE. This implies that patients with high expression level of ZNF143 present a better outcome after ICIs therapy. The findings indicated that expression of ZNF143 in LUSC was significantly correlated with PD-L2 level, but not correlated with PD-L1 level. This difference can be attributed development of LUAD and LUSC in distinct anatomical regions and cells-of-origin with different molecular profiles in TME.45,46 High expression of ZNF143 in COPD may modulate immune microenvironment by regulating PD-L1/PD-L2 thus enhancing lung cancer development. This finding is consistent with previous results that COPD is correlated with better outcome in NSCLC patients who have received ICIs therapy.12,13,44 Upregulated PD-L1 levels have been reported in emphysematous bullae-related NSCLC.47 Future studies should explore immunoregulation roles of ZNF143 in development and progression of COPD and NSCLC.
This is the first study to explore COPD-related genes through bioinformatic and machine learning analysis. Combination of the two approaches can provide highly accurate and reliable results compared with use of single methods. Diagnostic value of ZNF143 was explored by ROC analysis using two independent datasets of COPD. Moreover, we the possible immunoregulation significance of ZNF143 in LUAD and LUSC development was evaluated, which has not been reported previously. The study has certain limitations: (1) The samples in the external validation GSE38974 dataset of COPD were relatively few. Independent datasets of LUSC were not available for validation of the results. Sample sizes for LUAD and LUSC patients presenting with COPD comorbidity retrieved from TCGA were small, therefore, survival analyses were not conducted. Large sample studies should be performed to verify these results. (2) Although the sample size for the validation cohort was small and survival analysis was not conducted due to incomplete follow-up data, the findings showed upregulated expression of ZNF143 gene in NSCLC and COPD comorbidity. (3) Infiltrating levels of TICs and PD-L1 represented markers of immunotherapy efficacy in NSCLC, and the relationship between ZNF143 expression and these predictors was evaluated. However, the role of ZNF143 in regulating the TICs through PD-1/PD-L1 axis was not validated through basic experiments and clinical trials. (4) The role of ZNF143 in COPD was explored through bioinformatics analyses, therefore, in vitro as well as in vivo assays should be conducted to verify these findings. (5) The relationship between the expression level of ZNF143 in COPD and LUAD and LUSC may be relevant but subsequent studies should determine the association between COPD and lung cancer and the role of ZNF143.
Conclusion
ZNF143 was identified as a robust diagnostic biomarker in COPD via integrated bioinformatic and machine learning analysis. ZNF143 was implicated in progression of NSCLC and was correlated with the relative abundance of TICs and predictors of immunotherapy efficacy in NSCLC. Moreover, the level of ZNF143 expression was significantly upregulated in NSCLC coexisting with COPD and NSCLC alone compared with the expression levels in normal lung tissues. These findings indicate that ZNF143 acts as an important link between COPD and NSCLC. Further studies should explore the role of ZNF143 in COPD and in tumorigenesis and development of NSCLC as well as the immunoregulation significance.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Tianjin Chest Hospital and was carried out in accordance with the Declaration of Helsinki. All participants were informed about the study and written informed consent was obtained.
Acknowledgments
We would like to thank Hong Zhang for the financial support and Weishuai Liu for experimental technical support.
Funding
This study was supported by the CAPTRA-Lung Research Funds (No. CAPTRALung2022009) and Tianjin Health Science and Technology Project (Grant No. MS20015 to Hong Zhang).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Faruki H, Mayhew GM, Serody JS, Hayes DN, Perou CM, Lai-Goldman M. Lung adenocarcinoma and squamous cell carcinoma gene expression subtypes demonstrate significant differences in tumor immune landscape. J Thorac Oncol. 2017;12(6):943–953. doi:10.1016/j.jtho.2017.03.010
3. Malhotra J, Malvezzi M, Negri E, La Vecchia C, Boffetta P. Risk factors for lung cancer worldwide. Eur Respir J. 2016;48(3):889–902. doi:10.1183/13993003.00359-2016
4. Liu C, Zhou X, Zeng H, Wu D, Liu L. HILPDA is a prognostic biomarker and correlates with macrophage infiltration in pan-cancer. Front Oncol. 2021;11:597860. doi:10.3389/fonc.2021.597860
5. Treekitkarnmongkol W, Hassane M, Sinjab A, et al. Augmented Lipocalin-2 is associated with chronic obstructive pulmonary disease and counteracts lung adenocarcinoma development. Am J Resp Crit Care. 2021;203(1):90–101. doi:10.1164/rccm.202004-1079OC
6. Liu C, Chen Z, Chen K, et al. Lipopolysaccharide-mediated chronic inflammation promotes tobacco carcinogen-induced lung cancer and determines the efficacy of immunotherapy. Cancer Res. 2020:
7. Eapen MS, Hansbro PM, Larsson Callerfelt A, et al. Chronic obstructive pulmonary disease and lung cancer: underlying pathophysiology and new therapeutic modalities. Drugs. 2018;78(16):1717–1740. doi:10.1007/s40265-018-1001-8
8. Qin J, Li G, Zhou J. Characteristics of elderly patients with COPD and newly diagnosed lung cancer, and factors associated with treatment decision. Int J Chron Obstruct Pulmon Dis. 2016;11:1515–1520. doi:10.2147/COPD.S104670
9. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. New Engl J Med. 2015;373(2):123–135. doi:10.1056/NEJMoa1504627
10. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. New Engl J Med. 2015;373(17):1627–1639. doi:10.1056/NEJMoa1507643
11. Lievense LA, Sterman DH, Cornelissen R, Aerts JG. Checkpoint blockade in lung cancer and mesothelioma. Am J Resp Crit Care. 2017;196(3):274–282. doi:10.1164/rccm.201608-1755CI
12. Biton J, Ouakrim H, Dechartres A, et al. Impaired tumor-infiltrating T cells in patients with chronic obstructive pulmonary disease impact lung cancer response to PD-1 blockade. Am J Resp Crit Care. 2018;198(7):928–940. doi:10.1164/rccm.201706-1110OC
13. Mark NM, Kargl J, Busch SE, et al. Chronic obstructive pulmonary disease alters immune cell composition and immune checkpoint inhibitor efficacy in non–small cell lung cancer. Am J Resp Crit Care. 2018;197(3):325–336. doi:10.1164/rccm.201704-0795OC
14. Zhang L, Chen J, Yang H, et al. Multiple microarray analyses identify key genes associated with the development of non-small cell lung cancer from chronic obstructive pulmonary disease. J Cancer. 2021;12(4):996–1010. doi:10.7150/jca.51264
15. Miao T, Du L, Xiao W, Mao B, Wang Y, Fu J. Identification of survival-associated gene signature in lung cancer coexisting with COPD. Front Oncol. 2021;11. doi:10.3389/fonc.2021.600243
16. Li CY, Cai J, Tsai JJP, Wang CCN. Identification of hub genes associated with development of head and neck squamous cell carcinoma by integrated bioinformatics analysis. Front Oncol. 2020;10. doi:10.3389/fonc.2020.00681
17. Can T. Introduction to bioinformatics. Methods Mol Biol. 2014;1107:51–71. doi:10.1007/978-1-62703-748-8_4
18. San Segundo-Val I, Sanz-Lozano CS. Introduction to the Gene Expression Analysis. New York, NY: Springer New York; 2016:29–43.
19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9(1):559. doi:10.1186/1471-2105-9-559
20. Zhou J, Guo H, Liu L, et al. Construction of co‐expression modules related to survival by WGCNA and identification of potential prognostic biomarkers in glioblastoma. J Cell Mol Med. 2021;25(3):1633–1644. doi:10.1111/jcmm.16264
21. Tian W, Yang X, Yang H, Zhou B. GINS2 functions as a key gene in lung adenocarcinoma by WGCNA Co-Expression network analysis. Onco Targets Ther. 2020;13:6735–6746. doi:10.2147/OTT.S255251
22. López NC, García-Ordás MT, Vitelli-Storelli F, Fernández-Navarro P, Palazuelos C, Alaiz-Rodríguez R. Evaluation of feature selection techniques for breast cancer risk prediction. Int J Env Res Pub He. 2021;18(20):10670. doi:10.3390/ijerph182010670
23. Morrow JD, Zhou X, Lao T, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep-Uk. 2017;7(1). doi:10.1038/srep44232
24. Ezzie ME, Crawford M, Cho J, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi:10.1136/thoraxjnl-2011-200089
25. Feng Z, Zhang J, Zheng Y, Wang Q, Min X, Tian T. Elevated expression of ASF1B correlates with poor prognosis in human lung adenocarcinoma. Pers Med. 2021;18(2):115–127. doi:10.2217/pme-2020-0112
26. Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35(24):3209–3216. doi:10.1038/onc.2015.375
27. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
28. Feng Z, Zhang J, Zheng Y, Liu J, Duan T, Tian T. Overexpression of abnormal spindle-like microcephaly-associated (ASPM) increases tumor aggressiveness and predicts poor outcome in patients with lung adenocarcinoma. Transl Cancer Res. 2021;10(2):983–997. doi:10.21037/tcr-20-2570
29. Zhou R, Zhang J, Zeng D, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I–III colon cancer. Cancer Immunol Immunother. 2019;68(3):433–442. doi:10.1007/s00262-018-2289-7
30. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi:10.1038/s41591-018-0136-1
31. Wang X, Kong C, Xu W, et al. Decoding tumor mutation burden and driver mutations in early stage lung adenocarcinoma using CT‐based radiomics signature. Thorac Cancer. 2019;10(10):1904–1912. doi:10.1111/1759-7714.13163
32. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi:10.1186/s13073-017-0424-2
33. Izumi H, Wakasugi T, Shimajiri S, et al. Role of ZNF143 in tumor growth through transcriptional regulation of DNA replication and cell-cycle-associated genes. Cancer Sci. 2010;101(12):2538–2545. doi:10.1111/j.1349-7006.2010.01725.x
34. Paek AR, Mun JY, Hong K, Lee J, Hong DW, You HJ. Zinc finger protein 143 expression is closely related to tumor malignancy via regulating cell motility in breast cancer. Bmb Rep. 2017;50(12):621–627. doi:10.5483/BMBRep.2017.50.12.177
35. Chen YR, Leung JM, Sin DD. A systematic review of diagnostic biomarkers of COPD exacerbation. PLoS One. 2016;11(7):e0158843. doi:10.1371/journal.pone.0158843
36. Sin DD, Hollander Z, DeMarco ML, McManus BM, Ng RT. Biomarker development for chronic obstructive pulmonary disease. from discovery to clinical implementation. Am J Resp Crit Care. 2015;192(10):1162–1170. doi:10.1164/rccm.201505-0871PP
37. Kawatsu Y, Kitada S, Uramoto H, et al. The combination of strong expression of ZNF143 and high MIB-1 labelling index independently predicts shorter disease-specific survival in lung adenocarcinoma. Brit J Cancer. 2014;110(10):2583–2592. doi:10.1038/bjc.2014.202
38. Ye B, Yang G, Li Y, Zhang C, Wang Q, Yu G. ZNF143 in chromatin looping and gene regulation. Front Genet. 2020;11. doi:10.3389/fgene.2020.00338
39. Haibara H, Yamazaki R, Nishiyama Y, et al. YPC‐21661 and YPC‐22026, novel small molecules, inhibit ZNF143 activity in vitro and in vivo. Cancer Sci. 2017;108(5):1042–1048. doi:10.1111/cas.13199
40. Young RP, Hopkins RJ, Christmas T, Black PN, Metcalf P, Gamble GD. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur Respir J. 2009;34(2):380–386. doi:10.1183/09031936.00144208
41. Chen Y, Zhang Y, Lv J, et al. Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: immunotherapeutic implications. Theranostics. 2017;7(14):3585–3594. doi:10.7150/thno.21471
42. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi:10.1126/science.aaa8172
43. Trujillo JA, Sweis RF, Bao R, Luke JJ. T cell–inflamed versus Non-T cell–inflamed tumors: a conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol Res. 2018;6(9):990–1000. doi:10.1158/2326-6066.CIR-18-0277
44. Takayama Y, Nakamura T, Fukushiro Y, Mishima S, Masuda K, Shoda H. Coexistence of emphysema with non-small-cell lung cancer predicts the therapeutic efficacy of immune checkpoint inhibitors. In Vivo (Brooklyn). 2021;35(1):467–474. doi:10.21873/invivo.12280
45. Kadara H, Scheet P, Wistuba II, Spira AE. Early events in the molecular pathogenesis of lung cancer. Cancer Prev Res. 2016;9(7):518–527. doi:10.1158/1940-6207.CAPR-15-0400
46. Kim A, Lim SM, Kim J, Seo J. Integrative genomic and transcriptomic analyses of tumor suppressor genes and their role on tumor microenvironment and immunity in lung squamous cell carcinoma. Front Immunol. 2021;12. doi:10.3389/fimmu.2021.598671
47. Toyokawa G, Takada K, Okamoto T, et al. High frequency of programmed death-ligand 1 expression in emphysematous bullae-associated lung adenocarcinomas. Clin Lung Cancer. 2017;18(5):504–511. doi:10.1016/j.cllc.2016.11.011
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.