Back to Journals » OncoTargets and Therapy » Volume 13
WHSC1 Promotes Cell Proliferation, Migration, and Invasion in Hepatocellular Carcinoma by Activating mTORC1 Signaling
Authors Dai J, Jiang L, Qiu L , Shao Y, Shi P, Li J
Received 6 February 2020
Accepted for publication 5 July 2020
Published 20 July 2020 Volume 2020:13 Pages 7033—7044
DOI https://doi.org/10.2147/OTT.S248570
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Jingjing Dai,1,* Longfeng Jiang,1,* Lei Qiu,2,* Yuyun Shao,1,* Ping Shi,1 Jun Li1
1Department of Infectious Diseases, The First Affiliated Hospital, Nanjing Medical University, Nanjing, People’s Republic of China; 2Department of General Surgery, The Second People’s Hospital of Lianyungang, Lianyungang, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jun Li Email [email protected]
Background: Wolf-Hirschhorn syndrome candidate gene-1 (WHSC1) plays key regulatory roles in cancer development and progression. However, its specific functions and potential mechanisms of action remain to be described in hepatocellular carcinoma (HCC).
Materials and Methods: WHSC1 expression in HCC was evaluated using The Cancer Genome Atlas and verified in HCC tissues and cell lines using qRT-PCR, Western blotting, and immunohistochemistry. Functional assays were performed to explore the role of WHSC1 in HCC progression. Immunoprecipitation-mass spectrometry, co-immunoprecipitation, immunofluorescence, and immunohistochemistry were conducted to evaluate the interaction between WHSC1 and prolyl 4-hydroxylase subunit beta (P4HB). Pathway enrichment was performed using gene set enrichment analysis.
Results: WHSC1 was markedly overexpressed in HCC tissues and cell lines. The level of expression was strongly associated with adverse clinicopathological characteristics. Survival analyses revealed that WHSC1 upregulation predicted poor overall survival and higher recurrence rates in patients with HCC. Functional studies revealed that WHSC1 significantly stimulated HCC proliferation, migration, and invasion in vitro and in vivo. WHSC1 was shown to interact with P4HB to stimulate P4HB expression and subsequently activate mTOR1 signaling.
Conclusion: We determined the oncogenic role of WHSC1 in HCC, via P4HB interaction, which activates mTOR1 signaling, and identified WHSC1 as a promising therapeutic target for HCC.
Keywords: apoptosis, metastasis, P4HB, poor prognosis, hepatocarcinogenesis
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent malignant tumor and the second leading cause of cancer-related deaths worldwide.1–3 A total of 854,000 liver cancer cases and 810,000 deaths were reported worldwide in 2015, with half of them from China.4 Liver resections and transplantations are performed to manage the disease, however, rates of postoperative recurrence and distant metastasis remain high and current therapies do not noticeably improve survival.5–7 Therefore, the molecular mechanisms underlying the initiation and development of HCC need to be investigated further to facilitate the development of novel strategies for the prevention and treatment of this lethal disease.
Wolf-Hirschhorn syndrome candidate 1 (WHSC1) belongs to a family of nuclear receptor SET domain (NSD) proteins, members of which possess a SET domain encoding lysine methyltransferase activity. Although the substrate specificity of the NSD proteins was previously considered as controversial, it was recently narrowed down to the dimethylation of histone H3 lysine 36, a mark commonly associated with active transcription.8 Increasing evidence suggests that the dysregulation of WHSC1 expression is associated with the progression of malignancy, chemoresistance, and poor overall survival.9–11 In prostate cancer, AKT-mediated stabilization of WHSC1 is implicated in metastasis.12 WHSC1 may also activate TWIST1 and contribute to epithelial-mesenchymal transition (EMT) and invasion in prostate cancer.8 It is known to act as an oncogene and its overexpression is associated with poor prognosis in cervical cancer.9 The protein also promotes the progression of squamous cell carcinoma of the head and neck, through regulation of NIMA-related kinase-7.13 Additionally, WHSC1-induced DNA damage repair contributes to chemoresistance.14 It has also been reported that WHSC1 serves as a tumor suppressor through the regulation of cell differentiation.15 Overexpression of WHSC1 is also correlated with a poor prognosis of HCC.16 However, its expression profile and the potential mechanisms of action in HCC remain unclear.
Here, we report a significant overexpression of WHSC1 in HCC tissues and cell lines. Our survival analyses revealed that the upregulation of WHSC1 predicted a poor overall survival and higher recurrence rate in patients with HCC. WHSC1 was observed to promote HCC proliferation, migration, and invasion, possibly through an interaction with prolyl 4-hydroxylase subunit beta (P4HB) and the stimulation of its expression, thus promoting HCC malignancy via P4HB-induced activation of mTOR1 signaling. These findings suggest that WHSC1 may provide a novel therapeutic strategy and diagnostic biomarker for HCC.
Materials and Methods
HCC Tissues and Cell Lines
A total of 70 paired HCC samples were collected through surgical resection at the Department of Hepatobiliary Surgery of the First Affiliated Hospital of Nanjing Medical University (Nanjing, China), between 2017 and 2019. HCC cell lines (SMMC7721, Hep3B, HepG2, Huh7, MHCC97L, and Focus) and the human hepatic cell line (L02) were obtained from the Shanghai Institutes for Biological Sciences (China). All cell lines were cultured in DMEM containing 10% FBS, supplemented with 100 U/mL of penicillin and 100 μg/mL of streptomycin (HyClone, Logan, UT, USA). The cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Lentiviral Transfection
The lentiviral vectors for encoding WHSC1 and P4HB (LV-WHSC1 and LV-P4HB), the short hairpin RNAs (shRNAs) against WHSC1 and P4HB (LV-shWHSC1 and LV-shP4HB), and the empty vectors (LV-vector and LV-shNC) were synthesized by GenePharma Biotech (Shanghai, China). The shRNA sequences of WHSC1 and P4HB are described in Supplementary Data 1. Stable cell lines were obtained using 5 μg/mL puromycin (Sigma-Aldrich).
Cell Viability and Clonogenic Assays
Cell viability was assessed using Cell-Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). A cell counter device (Countstar, Ruiyu Bio-science and Technology, Shanghai, China) was utilized to determine the cell concentration. To summarize the process, SMMC7721 and Hep3B cells were seeded into 96-well plates (500 cells/well), and 10 μL CCK-8 solution was added to each well before incubating the plates for 2 h. Following the incubation, the absorbance was measured at 450 nm. For the clonogenic assay, the SMMC7721 and Hep3B cells were seeded in 6-well plates (1000 cells/well) and incubated for 14 days. Next, proliferating colonies were stained with crystal violet solution (Beyotime, Guangzhou, China), and counted and photographed for statistical analysis. The assay results were confirmed via three independent experiments.
Apoptosis Assay
An apoptosis assay was performed using flow cytometry (FACSCalibur, Becton Dickinson, San Jose, CA, USA). The SMMC7721 and Hep3B cells were collected and incubated after staining with propidium iodide and FITC-conjugated Annexin V (BD Pharmingen), in the absence of light at 37°C for 15 min, before being subjected to flow cytometric analysis. Ten thousand cells were detected in the apoptosis assay to determine the apoptosis index. The results were analyzed with FlowJo software (Tree Star Inc.). The assay results were confirmed through three independent experiments.
Transwell Migration and Invasion Assays
Transwell migration and invasion assays were also performed on SMMC7721 and Hep3B cells. Transwell plates (8-μm pore size), with or without coated Matrigel (BD Biosciences, San Jose, CA, USA), were used to assess the cell invasion and migration, respectively. To summarize the process, 200 μL of serum-free medium was added to the upper chamber, whereas 500 μL of complete culture medium supplemented with 10% fetal bovine serum was added to the lower chamber. After 24 h incubation, the cells were stained with crystal violet solution (Kaigen, Nanjing, China) for 15 min. The HCC cells on the upper side of the membrane were removed with a cotton swab, whereas those on the lower surface were visualized under a light microscope. The assays were confirmed through three independent experiments.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
The target mRNA in the sample cells and tissues was assessed with the qRT-PCR method. The total RNA was extracted using TRIzol® reagent (Thermo Fisher Scientific, USA), according to the manufacturer’s instructions. β-actin was used as the internal control. The qRT-PCR reactions were performed using the SYBR Green One-Step qRT-PCR kit (Thermo Fisher Scientific, USA), according to the manufacturer’s instructions. The primer sequences are detailed in Supplementary Data 1.
Western Blotting
Antibodies against WHSC1 (1:1000, 65127S; Cell Signaling Technology, Beverly, MA, USA), mTOR (1:1000, 2983T; Cell Signaling Technology, Beverly, MA, USA), P70S6K (1:1000, 9204S; Cell Signaling Technology, Beverly, MA, USA), S6 (1:1000, 9204S; Cell Signaling Technology, Beverly, MA, USA), p-mTOR (1:1000, 5536T; Cell Signaling Technology, Beverly, MA, USA), p-P70S6K (1:1000, 9204S; Cell Signaling Technology, Beverly, MA, USA), p-S6 (1:1000, 4858T; Cell Signaling Technology, Beverly, MA, USA), OXCT1 (1:1000, 12,175-1-AP; Proteintech, Wuhan, Hubei, China), RUVBL2 (1:1000, 10,195-1-AP; Proteintech, Wuhan, Hubei, China), TXNRD1 (1:1000, 11,117-1-AP; Proteintech, Wuhan, Hubei, China), HSD17B4 (1:1000, 15,116-1-AP; Proteintech, Wuhan, Hubei, China), PKM (1:1000, 10,078-2-AP; Proteintech, Wuhan, Hubei, China), EIF3A (1:1000, 26,178-1-AP; Proteintech, Wuhan, Hubei, China), TPM4 (1:1000, 13,741-1-AP; Proteintech, Wuhan, Hubei, China), and P4HB (1:1000, ab2792; Abcam, Cambridge, UK) were used in this study. Western blotting was performed as previously described.17
Immunoprecipitation-Mass Spectrometry (IP-MS)
The SMMC7721 cells were harvested for total protein extraction. The protein aliquots were pre-cleared through incubation with protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The pre-cleared samples were then immunoprecipitated with the WHSC1 antibody (1:200, 65127S; Cell Signaling Technology, Beverly, MA, USA), according to the manufacturer’s instructions. The immunoprecipitated proteins were then subjected to a MS analysis to determine the binding protein number in WHSC1.
Co-Immunoprecipitation (Co-IP)
Co-IP assays were utilized to test the interaction between WHSC1 and P4HB, and the complexes were precipitated with protein A/G-agarose beads (Santa Cruz Biotechnology), according to the manufacturer’s protocol. The complexes were then subjected to Western blotting. Antibodies against WHSC1 (1:200, 65127S; Cell Signaling Technology, Beverly, MA, USA) and P4HB (1:100, ab2792; Abcam, Cambridge, UK) were utilized.
Immunohistochemical (IHC) Staining
IHC staining was performed using anti-WHSC1 (1:100, ab223694; Abcam, Cambridge, UK) and anti-P4HB (1:200, ab2792; Abcam, Cambridge, UK) antibodies. The scoring system for IHC staining was applied as previously described.17
Confocal Laser Scanning Microscopy
Cells were seeded on slides, at a density of 2 × 104 cells/well, and incubated overnight. Following incubation, the cells were washed with phosphate buffer saline (PBS), fixed with 4% paraformaldehyde solution, permeabilized with 0.1% Triton X, and blocked with bovine serum albumin (BSA) for 1 h. The cells were then incubated with WHSC1 (1:100, ab223694; Abcam, Cambridge, UK) and P4HB (1:100, ab2792; Abcam, Cambridge, UK) primary antibodies at 4°C overnight. The slides were then washed with PBS and treated with Alexa Fluor® 488 AffiniPure Goat Anti-Rabbit IgG (H+L) (Jackson, Carlsbad, USA), and Alexa Fluor® 594 AffiniPure Goat Anti-Mouse IgG (H+L) (Jackson, Carlsbad, USA) secondary antibodies. The slides were then placed under a confocal laser scanning microscope (ZEISS, Germany) for the immunofluorescence study. The magnification used in our study was 10X and the sampling size was XY. The color Red was generated with a 532 nm laser and LP550 filter; Green with a 488 nm laser and LP505 filter; and Blue with a mercury lamp and BP415-480 filter. We used the Pearson coefficient analysis for our study.
Animal Experiments
Four-week-old male BALB/c-nude mice were purchased from the Animal Core Facility of Nanjing Medical University (Nanjing, China). For tumor growth analysis, a total of 1×106 tumor cells (SMMC7721 and Hep3B) were suspended in 100 μL serum free-DMEM and then subcutaneously injected into the mouse flank (six mice per group). All the mice were monitored once every four days and were euthanized 24 days later. The tumor length and width were monitored and the tumor volume was calculated according to the following equation: Volume (mm3) = 0.5 × Length (mm) × Width2 (mm2). The xenograft tumors were resected, weighted, and dissected. The partial tumor tissues were then fixed in 4% phosphatebuffered neutral formalin and embedded in paraffin for immunostaining analysis.
For the metastasis model, 1×106 tumor cells (SMMC7721 and Hep3B) labeled with RFP-firefly luciferase were injected via the mouse tail vein (eight mice per group). The mice were assessed after six weeks for metastasis via an intraperitoneal injection of D-Luciferin (Caliper LifeSciences). All the animal studies conducted in this study were reviewed and approved by the Institutional Animal Care and Use Committee of Nanjing Medical University.
Bioinformatics Analysis
The Cancer Genome Atlas (TCGA) gene dataset for HCC (with 371 HCC tissues and 50 normal tissues) was downloaded from the UCSC cancer browser (https://xenabrowser.net/datapages/). We obtained the normalized WHSC1 expression values from the TCGA gene dataset. The data employed for gene set enrichment analysis (GSEA) were also acquired from the TCGA dataset, using the GSEA v3.0 software (http://software.broadinstitute.org/gsea/downloads.jsp). A false discovery rate (FDR) < 0.25, and a P value < 0.05 for the normalized enrichment score (NES) were considered statistically significant.
Statistical Analyses
At least three biological replicates were used in each experiment. Data were presented as mean ± standard error of the mean (SEM), and analyzed with unpaired or paired (for matched comparisons) two-tailed Student’s t-test or analysis of variance (ANOVA). A P-value of < 0.05 was considered statistically significant.
Results
Elevated WHSC1 Expression in HCC and Association with Poor Prognosis
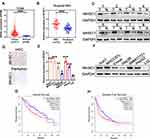
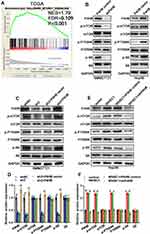
To analyze the WHSC1 expression in HCC, normalized RNA sequencing data of 371 HCC tissues and 50 non-tumor tissues were obtained from TCGA. This showed that WHSC1 was markedly elevated in the tumor tissues compared with the adjacent normal tissues (Figure 1A). We then employed qRT-PCR to assess the WHSC1 expression in the 70 paired samples of HCC-affected and adjacent non-cancerous liver tissue. We discovered that the WHSC1 mRNA levels were markedly elevated in the HCC tissues (Figure 1B). Western blotting and IHC of eight randomly selected paired HCC specimens confirmed that the WHSC1 protein expression was significantly upregulated in tumor tissues (Figure 1C and D). Levels of WHSC1 mRNA and protein were also significantly higher in the six HCC cell lines than in the normal hepatic cell line L02 (Figure 1E and F). As shown in Table 1, WHSC1 expression was distinctly associated with tumor size, microvascular invasion, tumor multiplicity, the tumor-node-metastasis (TNM) stage, and the Edmonson stage. Survival analysis using the GEPIA database (http://gepia2.cancer-pku.cn/#analysis) demonstrated that a higher WHSC1 expression was associated with shorter overall survival and a higher recurrence rate in HCC patients (Figure 1G and H). These results show that WHSC1 expression increases in HCC and may act as a prognostic indicator in patients with the disease.
 |
Table 1 Correlations Between WHSC1 Expression and Clinical Characteristics in HCC Patients (n = 70) |
Increased Proliferation, Migration, and Invasion of HCC
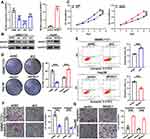
The SMMC7721 cells, which expressed the highest levels of WHSC1 among the six HCC cell lines, were transfected with shRNA to downregulate WHSC1 expression. Based on the qRT-PCR and Western blotting results, sh2 displayed the highest knockdown efficiency and was chosen for subsequent experiments (Figure 2A and B). Low WHSC1-expressing Hep3B cells were transfected with LV-WHSC1 to enhance WHSC1 expression. The upregulation of WHSC1 mRNA and protein levels was verified using qRT-PCR and Western blotting, respectively (Figure 2A and B). CCK-8 and clonogenic assays showed that WHSC1 knockdown markedly inhibited the cell proliferation and colony-forming ability of SMMC7721 cells (Figure 2C and D). Flow cytometric analyses revealed that WHSC1-depleted HCC cells had higher apoptotic rates (Figure 2E), while Transwell assays indicated that WHSC1 knockdown impaired the invasive and migratory abilities of the HCC cells (Figure 2F). Conversely, upregulation of WHSC1 dramatically promoted cell proliferation, inhibited apoptosis, and reinforced the invasive and migratory capabilities of Hep3B cells (Figure 2C–E, G). The results suggest WHSC1 as an oncogenic factor that promotes HCC proliferation, migration, and invasion.
Stimulation of HCC Progression Through P4HB Interaction
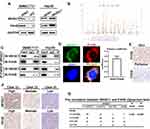
To uncover the deeper mechanism underlying the WHSC1-mediated HCC progression, we performed an immunoprecipitation/mass spectrum (IP/MS) analysis to identify the proteins bound to WHSC1. Combining previous research with the current study, we selected some candidates that could affect the malignant biological behavior of HCC (Supplementary Data 2). Of these, P4HB was downregulated in WHSC1-sh2 SMMC7721 cells and upregulated in LV-WHSC1 Hep3B cells, while the other candidates remained unchanged (Figure 3A, Figure S1A and B). Our results identified P4HB as the binding partner of WHSC1 (Figure 3B). We then explored the possible role of WHSC1/P4HB interaction in the malignancy progression of HCC. P4HB immunoprecipitated with an anti-WHSC1 antibody, and vice versa (Figure 3C), and the two proteins were co-localized in the HCC cells (Figure 3D). Moreover, P4HB expression was higher in HCC tissues than in corresponding non-tumor tissues (Figure 3E). A Spearman analysis revealed that the WHSC1 and P4HB levels were positively correlated (Figure 3F and G).
The expression of P4HB was upregulated in SMMC7721-shWHSC1 cells, to clarify its potential role in WHSC1-induced HCC progression. The overexpression of P4HB reversed decreased proliferation, migration, and invasion of SMMC7721-shWHSC1 cells, and decreased apoptotic rates (Figure S2A–C). The malignant phenotypes discussed earlier were also rescued successfully in Hep3B-WHSC1 cells by downregulating P4HB expression (Figure S2D–F).
Facilitation of HCC Growth and Metastasis in vivo
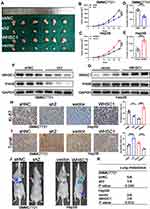
To verify the effects of WHSC1 on the HCC growth in vivo, a murine xenograft HCC model was developed. Downregulation of WHSC1 in the HCC cells decreased tumor size and weight (Figure 4A, B, D), whereas its overexpression had the opposite effect (Figure 4A, C, E). Western blotting revealed that WHSC1 expression was lower in the SMMC7721-shWHSC1 group and higher in the Hep3B-WHSC1 group, compared to the control, and that the WHSC1 and P4HB levels in the xenografts were positively correlated (Figure 4F and G). Additionally, Ki-67 and TUNEL staining revealed that SMMC7721-shWHSC1 xenografts proliferated more rapidly and had a higher apoptosis rate than Hep3B-WHSC1 xenografts (Figure 4H and I). Nude mice injected with luciferase-expressing HCC cells were subjected to bioluminescence imaging, to evaluate the role of WHSC1 in HCC metastasis in vivo. WHSC1 downregulation considerably decreased lung metastasis, whereas its overexpression had the opposite effect (Figure 4J and K). Collectively, our results demonstrate that WHSC1 plays a pivotal role in accelerating HCC growth and metastasis in vivo.
Stimulation of mTORC1 Signaling via P4HB
We performed a GSEA using data from TCGA to assess the downstream signal transduction mechanisms. The WHSC1 and P4HB expression levels were set according to their respective phenotype labels. The activation of the mTORC1 signaling pathway was observed to be positively associated with WHSC1 and P4HB expression levels in HCC tissues (Figure 5A, Figure S3A). To validate the GSEA results, the mTORC1-relevant effectors were detected with Western blotting. Phosphorylation of mTOR (at Ser2448), P70S6K (at Thr389), and S6 (Ser240/244) was significantly downregulated in the SMMC7721-shP4HB cells, and upregulated in the Hep3B-P4HB cells (Figure 5B). Furthermore, downregulation of WHSC1 impaired the mTORC1 signaling, whereas its upregulation activated it (Figure 5C and D). The inhibition of mTORC1 signaling seen in the SMMC7721-shWHSC1 cells was nullified when P4HB was overexpressed. Similarly, the activation of mTORC1 observed in Hep3B-WHSC1 cells was inhibited after downregulation of P4HB expression (Figure 5E and F). These results reveal that WHSC1 positively regulates mTORC1 signaling by interacting with P4HB.
Discussion
The significance of WHSC1 in cancer progression was first emphasized with the identification of t(4;14) translocation in almost 15–20% of multiple myeloma cases.18 This mutation fuses the WHSC1 gene to the immunoglobulin heavy chain enhancer, resulting in significant overexpression of WHSC1. WHSC1 regulates tumor maintenance and chemoresistance, and its expression correlates with overall survival in a variety of malignancies.19,20 However, its functions and associated molecular mechanisms in HCC have not been investigated in detail. In this study, we assessed the WHSC1 expression in HCC using TCGA data, before validating the results in HCC tissues and cell lines. Functional assays showed that WHSC1 stimulates cell proliferation, migration, invasion, and the metastasis of HCC cells while inhibiting their apoptosis. To delineate the mechanisms through which WHSC1 promotes the HCC malignancy, we examined the proteins it interacts with. IP-MS, co-immunoprecipitation, immunofluorescence, and IHC analyses confirmed P4HB as the WHSC1 binding partner. P4HB, which has been observed to be upregulated in various malignancies, including HCC,21 gastric cancer,22 renal carcinoma,23,24 and colon cancer,25 can influence several cancer cell functions, like proliferation, migration, and invasion. Xia et al reported that P4HB promotes HCC tumorigenesis via a downregulation of GRP78 and subsequent upregulation of epithelial-mesenchymal transition.21 A recent study also revealed that P4HB downregulation increased the generation of reactive oxygen species (ROS), thereby promoting cancer cell apoptosis.25 P4HB overexpression is correlated with a poor prognosis in various cancers, including HCC, clear cell renal cell carcinoma, diffuse gliomas, and gastric cancer.24,26,27 We observed that WHSC1 may interact with P4HB to mediate its stimulatory effects on the progression of HCC malignancy.
To further explore the molecular pathways implicated in HCC progression, we performed GSEA and discovered a positive association between the mTORC1 signaling pathway and WHSC1 expression. mTOR (comprising two main complexes, mTORC1 and mTORC2) exhibits a complicated dynamic behavior to influence the fate of cancer cells.28–30 The mTORC1 branch is the central driver of cell progression via protein modulation, lipid synthesis, and energy metabolism in mammals, and is found to be deregulated in various malignancies. After the activation of mTORC1, the phosphorylation and activation of a key downstream effector, namely ribosomal protein S6 kinase (S6K), was also observed. Next, the ribosomal S6 protein was phosphorylated by S6K, which regulates basic cell processes including cell proliferation and metastasis. The influence of mTORC1 signaling on tumor progression, due to its effect on cell proliferation, cell survival, cell cycle, cell death, and metabolic reprogramming, has been widely reported.31–34 In our study, we observed that WHSC1 upregulated the P4HB expression levels and subsequently activated mTORC1 signaling to promote HCC development and progression. However, the precise mechanism (direct or indirect) through which WHSC1 and P4HB regulate mTORC1 signaling remains to be described and requires further investigation.
Conclusion
In conclusion, our study demonstrated that WHSC1 was significantly upregulated in HCC tissues and cell lines, and that high expression levels strongly correlated with adverse clinicopathological parameters, poor overall survival, and higher recurrence rates in patients with HCC. Functional studies confirmed the oncogenic role of WHSC1 in HCC. Mechanistically, WHSC1 increased P4HB expression levels and subsequently activated mTOR1 signaling. Considered together, these findings prove the clinical significance and functional importance of WHSC1 in HCC.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of The First Affiliated Hospital, Nanjing Medical University. All experiments were performed according to the National Institutes of Health guide for the care and use of laboratory animals. Patients or their relatives were informed of the proper usage of human samples, and provided written consent before sample acquisition.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Feng Z, Rong P, Wang W. Hepatocellular carcinoma risk in patients with NASH cirrhosis and diabetes: insufficient for individual management. Hepatology. 2020. doi:10.1002/hep.31100
2. Weiler SME, Lutz T, Bissinger M, et al. TAZ target gene ITGAV regulates invasion and feeds back positively on YAP and TAZ in liver cancer cells. Cancer Lett. 2020;473:164–175. doi:10.1016/j.canlet.2019.12.044
3. Fang G, Zhang P, Liu J, et al. Inhibition of GSK-3beta activity suppresses HCC malignant phenotype by inhibiting glycolysis via activating AMPK/mTOR signaling. Cancer Lett. 2019;463:11–26. doi:10.1016/j.canlet.2019.08.003
4. Hu W, Zheng S, Guo H, et al. PLAGL2-EGFR-HIF-1/2alpha signaling loop promotes HCC progression and erlotinib insensitivity. Hepatology. 2020. doi:10.1002/hep.31293
5. Nault J-C, Martin Y, Caruso S, et al. Clinical impact of genomic diversity from early to advanced hepatocellular carcinoma. Hepatology. 2020;71(1):164–182. doi:10.1002/hep.30811
6. Wu -T-T, Cai J, Tian Y-H, et al. MTF2 induces epithelial-mesenchymal transition and progression of hepatocellular carcinoma by transcriptionally activating snail. Onco Targets Ther. 2019;12:11207–11220. doi:10.2147/OTT.S226119
7. Feng J, Dai W, Mao Y, et al. Simvastatin re-sensitizes hepatocellular carcinoma cells to sorafenib by inhibiting HIF-1alpha/PPAR-gamma/PKM2-mediated glycolysis. J Exp Clin Cancer Res. 2020;39(1):24. doi:10.1186/s13046-020-1528-x
8. Ezponda T, Popovic R, Shah MY, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013;32(23):2882–2890. doi:10.1038/onc.2012.297
9. Wu J, Luo M, Duan Z, et al. WHSC1 acts as a prognostic indicator and functions as an oncogene in cervical cancer. Onco Targets Ther. 2019;12:4683–4690. doi:10.2147/OTT.S204701
10. Yin Z, Sun Y, Ge S, Sun J. Epigenetic activation of WHSC1 functions as an oncogene and is associated with poor prognosis in cervical cancer. Oncol Rep. 2017;37(4):2286–2294. doi:10.3892/or.2017.5463
11. Zhao X, Xie T, Zhao W, Cai W, Su X. Downregulation of MMSET impairs breast cancer proliferation and metastasis through inhibiting Wnt/beta-catenin signaling. Onco Targets Ther. 2019;12:1965–1977. doi:10.2147/OTT.S196430
12. Li N, Xue W, Yuan H, et al. AKT-mediated stabilization of histone methyltransferase WHSC1 promotes prostate cancer metastasis. J Clin Invest. 2017;127(4):1284–1302. doi:10.1172/JCI91144
13. Saloura V, Cho H-S, Kiyotani K, et al. WHSC1 promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol Cancer Res. 2015;13(2):293–304. doi:10.1158/1541-7786.MCR-14-0292-T
14. Shah MY, Martinez-Garcia E, Phillip JM, et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene. 2016;35(45):5905–5915. doi:10.1038/onc.2016.116
15. Yu C, Yao X, Zhao L, et al. Wolf-Hirschhorn Syndrome Candidate 1 (whsc1) functions as a tumor suppressor by governing cell differentiation. Neoplasia. 2017;19(8):606–616. doi:10.1016/j.neo.2017.05.001
16. Zhou P, Wu -L-L, Wu K-M, et al. Overexpression of MMSET is correlation with poor prognosis in hepatocellular carcinoma. Pathol Oncol Res. 2013;19(2):303–309. doi:10.1007/s12253-012-9583-z
17. Li C, Jin Y, Wei S, et al. Hippo signaling controls NLR family pyrin domain containing 3 activation and governs immunoregulation of mesenchymal stem cells in mouse liver injury. Hepatology. 2019;70(5):1714–1731. doi:10.1002/hep.30700
18. Xie Z, Chooi JY, Toh SHM, et al. MMSET I acts as an oncoprotein and regulates GLO1 expression in t(4;14) multiple myeloma cells. Leukemia. 2019;33(3):739–748. doi:10.1038/s41375-018-0300-0
19. Yang P, Guo L, Duan ZJ, et al. Histone methyltransferase NSD2/MMSET mediates constitutive NF-kappaB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol Cell Biol. 2012;32(15):3121–3131. doi:10.1128/MCB.00204-12
20. Hudlebusch HR, Santoni-Rugiu E, Simon R, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17(9):2919–2933. doi:10.1158/1078-0432.CCR-10-1302
21. Xia W, Zhuang J, Wang G, et al. P4HB promotes HCC tumorigenesis through downregulation of GRP78 and subsequent upregulation of epithelial-to-mesenchymal transition. Oncotarget. 2017;8(5):8512–8521. doi:10.18632/oncotarget.14337
22. Zhang J, Guo S, Wu Y, Zheng ZC, Wang Y, Zhao Y. P4HB, a novel hypoxia target gene related to gastric cancer invasion and metastasis. Biomed Res Int. 2019;2019:9749751. doi:10.1155/2019/9749751
23. Xie L, Li H, Zhang L, et al. Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging. 2020;12(2):1828–1842. doi:10.18632/aging.102715
24. Zhu Z, He A, Lv T, et al. Overexpression of P4HB is correlated with poor prognosis in human clear cell renal cell carcinoma. Cancer Biomark. 2019;26(4):431–439. doi:10.3233/CBM-190450
25. Zhou Y, Yang J, Zhang Q, et al. P4HB knockdown induces human HT29 colon cancer cell apoptosis through the generation of reactive oxygen species and inactivation of STAT3 signaling. Mol Med Rep. 2019;19(1):231–237. doi:10.3892/mmr.2018.9660
26. Zhang J, Wu Y, Lin Y-H, et al. Prognostic value of hypoxia-inducible factor-1 alpha and prolyl 4-hydroxylase beta polypeptide overexpression in gastric cancer. World J Gastroenterol. 2018;24(22):2381–2391. doi:10.3748/wjg.v24.i22.2381
27. Zou H, Wen C, Peng Z, et al. P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas. Oncol Rep. 2018;39(2):501–510. doi:10.3892/or.2017.6134
28. Yu VZ, Ko JMY, Ning L, et al. Endoplasmic reticulum-localized ECM1b suppresses tumor growth and regulates MYC and MTORC1 through modulating MTORC2 activation in esophageal squamous cell carcinoma. Cancer Lett. 2019;461:56–64. doi:10.1016/j.canlet.2019.07.005
29. Ruan X-H, Liu X-M, Yang Z-X, et al. INPP4B promotes colorectal cancer cell proliferation by activating mTORC1 signaling and cap-dependent translation. Onco Targets Ther. 2019;12:3109–3117. doi:10.2147/OTT.S186365
30. Wu R, Murali R, Kabe Y, et al. Baicalein targets GTPase-mediated autophagy to eliminate liver tumor-initiating stem cell-like cells resistant to mTORC1 inhibition. Hepatology. 2018;68(5):1726–1740. doi:10.1002/hep.30071
31. He L, Gomes AP, Wang X, et al. mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol Cell. 2018;70(5):949–960 e944. doi:10.1016/j.molcel.2018.04.024
32. Chen J, Ou Y, Yang Y, et al. KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature. 2018;557(7706):585–589. doi:10.1038/s41586-018-0128-9
33. Brandt M, Grazioso TP, Fawal M-A, et al. mTORC1 inactivation promotes colitis-induced colorectal cancer but protects from APC loss-dependent tumorigenesis. Cell Metab. 2018;27(1):118–135 e118. doi:10.1016/j.cmet.2017.11.006
34. Yoon S-O, Shin S, Karreth FA, et al. Focal adhesion- and IGF1R-dependent survival and migratory pathways mediate tumor resistance to mTORC1/2 inhibition. Mol Cell. 2017;67(3):512–527 e514. doi:10.1016/j.molcel.2017.06.033
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.