Back to Journals » Infection and Drug Resistance » Volume 17
Whole Genome Sequence Analysis of Two Oxacillin-Resistant and mecA-Positive Strains of Staphylococcus haemolyticus Isolated from Ear Swab Samples of Patients with Otitis Media
Authors Liu Z, Wang L, Sun J, Zhang Q, Peng Y, Tang S, Zhang L, Li X , Yu Z, Zhang T
Received 14 January 2024
Accepted for publication 15 March 2024
Published 30 March 2024 Volume 2024:17 Pages 1291—1301
DOI https://doi.org/10.2147/IDR.S455051
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Sandip Patil
Zhao Liu,1,* Ling Wang,2,* Jiabing Sun,1,* Qinghuan Zhang,3 Yue Peng,1 Susu Tang,1 Limei Zhang,4 Xiaobin Li,4,5 Zhijian Yu,1 Tao Zhang6
1Department of Otolaryngology, Zhuhai Hospital Affiliated with Jinan University (Zhuhai People’s Hospital), Zhuhai, People’s Republic of China; 2Department of Obstetrics, Zhuhai Hospital Affiliated with Jinan University (Zhuhai People’s Hospital), Zhuhai, 519000, People’s Republic of China; 3Department of Clinical Laboratory, Zhuhai Hospital Affiliated with Jinan University (Zhuhai People’s Hospital), Zhuhai, 519000, People’s Republic of China; 4Guangdong Provincial Key Laboratory of Tumor Interventional Diagnosis and Treatment, Zhuhai Hospital Affiliated with Jinan University (Zhuhai People’s Hospital), Zhuhai, People’s Republic of China; 5Zhuhai Precision Medical Center, Zhuhai Hospital Affiliated with Jinan University (Zhuhai People’s Hospital), Zhuhai, People’s Republic of China; 6Department of Otolaryngology, The First Affiliated Hospital of Jinan University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tao Zhang; Zhijian Yu, Email [email protected]; [email protected]
Objective: Staphylococcus haemolyticus can cause a series of infections including otitis media (OM), and the oxacillin-resistant S. haemolyticus has become a serious health concern. This study aimed to investigate the genomic characteristics of two strains of oxacillin-resistant and mecA-positive S. haemolyticus isolated from the samples of ear swabs from patients with OM and explore their acquired antibiotic resistance genes (ARGs) and the mobile genetic elements (MGEs).
Methods: Two oxacillin-resistant S. haemolyticus strains, isolated from ear swab samples of patients with OM, underwent antimicrobial susceptibility evaluation, followed by whole-genome sequencing. The acquired ARGs and the MGEs carried by the ARGs, harbored by the genomes of two strains of S. haemolyticus were identified.
Results: The two strains of oxacillin-resistant S. haemolyticus (strain SH1275 and strain SH9361) both carried the genetic contexts of mecA with high similarity with the SCCmec type V(5C2& 5) subtype c. Surprisingly, the chromosomal aminoglycoside resistance gene aac(6’)-aph(2”) harbored by S. haemolyticus strain SH936 was flanked by two copies of IS 256, forming the IS 256-element (IS 256-GNAT-[aac(6’)-aph(2”)]-IS 256), which was widely present in strains of both Staphylococcus and Enterococcus genus. Furthermore, the two strains of oxacillin-resistant and MDR S. haemolyticus were found to harbor antimicrobial resistance plasmids, including one 26.9-kb plasmid (pSH1275-2) containing msr(A)–mph(C)) and qacA, one mobilizable plasmid pSH1275-3 harboring vga(A)LC, one plasmid (pSH9361-1) carrying erm(C), and one plasmid (pSH9361-2) carrying qacJ.
Conclusion: The systematic analysis of whole-genome sequences provided insights into the mobile genetic elements responsible for multi-drug resistance in these two strains of oxacillin-resistant and mecA-positive S. haemolyticus, which will assist clinicians in devising precise, personalized, and clinical therapeutic strategies for treating otitis media caused by multi-drug resistant S. haemolyticus.
Keywords: Staphylococcus haemolyticus, ear swabs, mecA, antimicrobial resistance plasmid, whole genome sequence
Introduction
In hospitals, opportunistic infections caused by coagulase-negative staphylococci (CoNS) have attracted considerable attention, especially among neonates, elderly patients, and immunocompromised patients.1,2 Among all CoNS, Staphylococcus haemolyticus is the second most frequently isolated CoNS in clinical cases (after Staphylococcus epidermidis),3 and S. haemolyticus has been considered as the major species of Staphylococcus in nosocomial foreign device-related infections.4 In addition, S. haemolyticus can cause skin or soft tissue infections, bacteremia, septicemia, peritonitis, otitis media (OM), meningitis, and urinary tract infections.5 Furthermore, S. haemolyticus plays a crucial role in the nosocomial infections caused by multidrug-resistant (MDR) staphylococci,6 which result in limited therapeutic options.7 Infections caused by oxacillin-resistant staphylococci present a major therapeutic challenge to the health of hospitalized patients.8 In staphylococci, methicillin resistance is mainly due to the expression of the mecA gene, which encodes penicillin binding protein 2a (PBP2a), a transpeptidase with a low affinity for β-lactams.9
S. haemolyticus can be a reservoir for antibiotic resistance genes that are shared with other staphylococci, including S. aureus.6 Most of the antibiotic resistance genes (ARGs) were thought to be spread via mobile genetic elements (MGEs) across different staphylococci through horizontal gene transfer.10 The mecA gene is carried by an MGE termed the staphylococcal cassette chromosome mec (SCCmec). To date, 14 types of SCCmec have been identified worldwide,11 of which SCCmec type V is the most prevalent in methicillin-resistant S. haemolyticus.12 Notably, a wide variety of plasmid-borne genes that mediate resistance to antimicrobial agents have been identified in staphylococci of human and animal origin.13 These plasmids have been shown to play a key role as carriers of plasmid-borne resistance genes, as well as being vectors for the dissemination of resistance genes by horizontal gene transfer between members of the same species and between bacteria of different species and genera.14
In this study, we report two strains of oxacillin-resistant and mecA-harboring S. haemolyticus isolated from ear swab samples of patients with OM. The antimicrobial susceptibility testing and whole-genome sequencing (WGS) were performed to identify the antimicrobial resistance profiles and genomic characteristics of the two strains of S. haemolyticus.
Materials and Methods
Identification of Bacterial Strains
The ear swab samples were collected from the patients with otitis media (OM) in 2023 using the sterilized cotton swabs. The present study complies with the Declaration of Helsinki. The samples were inoculated in blood agar medium for 24 h at 35°C. Two strains were isolated from ear swab samples. The strain identification was performed using a fully automatic VITEK 2 COMPACT system (BioMérieux, France) following the manufacturer’s instructions. Identification of the two clinical strains was further confirmed via 16S rRNA gene sequencing using the universal primers 27F and 1492R. The minimum inhibitory concentrations (MICs) of oxacillin, clindamycin, erythromycin, linezolid, penicillin, sulfamethoxazole/trimethoprim, tigecycline, daptomycin, gentamicin, levofloxacin, moxifloxacin, rifampicin, teicoplanin, and vancomycin were determined using the broth microdilution method, and the results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI M100–S32).
Whole Genome Sequencing, Assembly, and Annotation
Whole-genome sequencing of the two strains of S. haemolyticus were performed by Genewiz Biotechnology Co. Ltd. (Suzhou, China) using paired-end sequencing with Novaseq 6000 (Illumina, 2×150 bp paired-end reads) and long sequencing with PacBio Sequel IIe (Pacific Biosciences, 10–15 Kb insert whole-genome shotgun libraries). PacBio reads were assembled using the Hifiasm software15 version 0.13-r308 and Canu16 version 1.7. The genome assembly was then polished using Pilon software version 1.2217 using Illumina reads. The assembled genomes of the two strains of S. haemolyticus were submitted to the National Center for Biotechnology Information (NCBI) GenBank database and annotated using the NCBI Prokaryotic Annotation Pipeline (PGAP).
Bioinformatics Analysis of the Two Strains of S. haemolyticus
The acquired ARGs in the genomes of the two strains of S. haemolyticus were identified using ResFinder 4.1,18 with a minimum coverage of 60% and minimum identity of 90%. Multilocus sequence typing (MLST) of the two strains of S. haemolyticus were performed using MLST 2.0,19 selecting the database as “Staphylococcus haemolyticus”. SCCmecFinder20 was used to determine the SCCmec element type. Replicon types of the plasmids contained by the two strains of S. haemolyticus were determined using PlasmidFinder 2.1,21 with the database of “Gram Positive”, the minimum coverage of 60%, and the minimum identity of 95%. The insertion sequences (ISs) adjacent to ARGs in the genomes of the two strains of S. haemolyticus were identified using ISfinder.22 A sequence similarity search was performed using MegaBLAST scans against the GenBank nonredundant (nr) database. BLAST Ring Image Generator (BRIG) 0.9523 and Easyfig 2.2.524 were used to perform and visualize the sequence comparison.
Nucleotide Sequence Accession Numbers
The genome sequences of S. haemolyticus strain SH1275, which contained one chromosome and three plasmids, was submitted to GenBank under the accession numbers CP123979–CP123982. The genome sequences of S. haemolyticus strain SH9361, which contained a chromosome and two plasmids, was submitted to GenBank under the accession numbers CP123983–CP123985.
Results
Identification and Antimicrobial Susceptibility of Two Strains of S. haemolyticus
Two bacterial strains (SH1275 and SH9361) isolated from ear swab samples of patients with OM were identified as S. haemolyticus using the VITEK 2 COMPACT system, with further confirmation by 16S rRNA gene sequencing. Both of the two strains of S. haemolyticus showed resistance to oxacillin, clindamycin, erythromycin, penicillin, and levofloxacin (Table 1). Moreover, S. haemolyticus SH1275 was shown to be susceptible to sulfamethoxazole/trimethoprim, whereas S. haemolyticus SH9361 showed resistance to these same antibiotics (Table 1). In addition, S. haemolyticus SH1275 showed intermediate-level resistance to moxifloxacin, and S. haemolyticus SH9361 showed resistance to moxifloxacin (Table 1).
 |
Table 1 Minimum Inhibitory Concentration (µg/mL) for the Two Trains of S. haemolyticus |
Genomic Characteristics of the Two Strains of S. haemolyticus
Genome sequencing and analysis revealed that the genome of S. haemolyticus strain SH1275 comprised a 2.55-Mb chromosome and three plasmids with sizes of 37,857 bp (pSH1275-1), 26,934 bp (pSH1275-2), and 6056 bp (pSH1275-3), and the S. haemolyticus strain SH9361 comprised a 2.51-Mb chromosome and two plasmids with sizes of 2473 bp (pSH9361-1) and 2161 bp (pSH9361-2). MLST analysis revealed that strain SH1275 belonged to sequence type (ST) 3 and SH9361 to ST30. Three plasmid replicons, rep20 (pSH1275-1), rep39 (pSH1275-2) and rep5b (pSH1275-3), were identified in the genome of strain SH1275. In the strain SH9361, one of the two plasmids (pSH9361-1) was identified as containing the rep10 replicon, and the other plasmid (pSH9361-2) could not be typed. ResFinder results indicated that S. haemolyticus strain SH1275 carried multiple ARGs located on both chromosomes and two plasmids (pSH1275-2 and pSH1275-3). The chromosome of strain SH1275 was found to carry beta-lactam resistance genes blaZ and mecA. The S. haemolyticus genome contained mecA, which confers resistance to methicillin. Plasmid pSH1275-2 consisted of two macrolide resistance genes (mph(C) and msr(A)) and an antiseptic-resistance gene, qacA. Plasmid pSH1275-3 contained the gene vga(A)LC, which is responsible for significant resistance to both lincosamides and streptogramin A.
ResFinder results indicated that S. haemolyticus strain SH9361 contained six ARGs located on both chromosomes and two plasmids (pSH9361-1 and pSH9361-2). The chromosome of strain SH9361 was found to harbor ARGs conferring resistance to beta-lactam antibiotics (blaZ and mecA), aminoglycosides (aac(6’)-aph(2”)), as well as trimethoprim (dfrG). Plasmid pSH9361-1 carried an erythromycin resistance gene, (erm(C)), and plasmid pSH9361-2 carried an antiseptic-resistance gene, qacJ.
Genetic Contexts of mecA and Other Chromosomal ARG in the Two Strains
With respect to the two mecA-positive isolates of S. haemolyticus in this study, no SCCmec elements were detected based on the results predicted by SCCmecFinder. However, both SH1275 and SH9361 carried chromosomal fragments containing mecA similar to the SCCmec type V(5C2&5) subtype c (GenBank: AB505629) (Figure 1). The methicillin resistance gene mecA was bracketed by two copies of IS431.
Notably, the aminoglycoside resistance gene aac(6’)-aph(2”) and one gene encoding the GNAT family N-acetyltransferase were flanked by two copies of the insertion sequence IS256 in different orientations (Figure 2A) in strain SH9361. Furthermore, we found the IS256-element was widely present in Staphylococcus (mainly S. aureus, S. epidermidis, and S. haemolyticus) and Enterococcus (Enterococcus faecalis and E. faecium), not only on chromosomes but also on plasmids (100.00% coverage and 100% identity) (Figure 2B and Table S1).
Genomic Analysis of the Antibiotic-Resistant Plasmids Carried by the Two Strains
In the case of the plasmid pSH1275-2 of S. haemolyticus strain SH1275, the genes msr(A)–mph(C) and the qacR/qacA system were located on a ~14.4-kb composite transposon, which was bracketed by two copies of the insertion sequence IS1272 in different orientations (Figure 3). Notably, an insertion sequence, IS431, was found to be located adjacent to msr(A)-mph(C) macrolide resistance genes. Based on the results of the BLAST search hit from the nr database of GenBank, the 14.4-kb composite transposon of plasmid pSH1275-2 was nearly identical to that of plasmid pSGAir0252 in S. haemolyticus strain SGAir0252 (100.00% coverage and 99.91% identity; Figure 3). Moreover, the region within the 14.4-kb composite transposon containing msr(A), mph(C), IS431, and qacR/qacA was also found in plasmids from S. aureus, S. epidermidis, and S. hominis (Figure 3). In addition, the BLAST search hit from the nr database of GenBank showed that the structure was present not only in the Staphylococcus plasmids, but also in the chromosomes of Staphylococcus spp. (Figure 4).
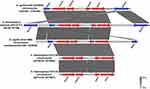 |
Figure 4 The region of plasmid pSH1275-2 containing msr(A)–mph(C), IS431, and qacR/qacA, had high similarity with the chromosomal fragments from strains of Staphylococcus genus. |
With respect to the 6056-bp plasmid pSH1275-3 containing the gene vga(A)LC in S. haemolyticus strain SH1275, the results of oriTfinder indicated that the plasmid harbors the origin of the transfer site (oriT) and relaxase gene in its genome but lacks the genes coding for type IV coupling protein (T4CP) and type IV secretion system (T4SS); thus it was inferred to be a mobilizable plasmid. The 6-kb potential mobilizable plasmid was also found in another S. haemolyticus strain and two S. hominis strains (Figure 5 and Table S2). In addition, two plasmids from S. aureus (pUR4128, 7567 bp; and pUR2355, 7609 bp) also contained the vga(A)LC and a conjugative transfer region (oriT, relaxase), similar to those of plasmid pSH1275-3 (Figure 5 and Table S2).
As for the 2.47-kb plasmid pSH9361-1 harboring erm(C) in S. haemolyticus strain SH9361 only three genes were found in the plasmid genome: the gene encoding replication/maintenance protein RepL, the erythromycin resistance gene erm(C), and the gene encoding the ErmCL peptide. Based on the results of BLAST search hit from the nr database of GenBank, a large number of plasmids with high similarity to the 2.47-kb plasmid (pSH9361-1) were detected in the Staphylococcus genus, especially in S. aureus (coverage ≥ 99% and identity ≥ 99%, Figure 6 and Table S3). In addition, this 2.47-kb plasmid (pSH9361-1) was highly similar to chromosomal fragments from two strains of S. aureus and one strain of S. haemolyticus (Table S3).
The small plasmid pSH9361-2 in S. haemolyticus strain SH9361, comprising one rep gene and one antiseptic-resistance gene qacJ, was found to be highly similar to the 9th plasmid (GenBank: CP027494) of S. aureus strain ST2594 (Figure S1).
Discussion
Among the coagulase-negative staphylococci (CoNS), S. haemolyticus is one of the most common pathogens worldwide and is mainly associated with bloodstream and device-associated infections.25,26 In the present study, we report the whole genome sequences of two strains of oxacillin-resistant and mecA-positive S. haemolyticus (ST3 strain SH1275 and ST30 strain SH9361), which were isolated from ear swab samples of patients with OM. Epidemiological surveillance has shown that S. haemolyticus ST3 is the original strain that has evolved into many other molecular types,26 including the emerging ST42 clone disseminated in the hospital environment.3,27 However, the prevalence of ST3 has continuously decreased since 2013.26 In addition, S. haemolyticus strains belonging to ST30 have been previously detected in different clinical samples of the urinary tract,28 infected eyes,29 nares,3 feces,3 and blood.30
In our study, two strains of oxacillin-resistant S. haemolyticus were found to carry the mecA gene, which confers methicillin resistance and is usually acquired through a SCCmec element in methicillin-resistant strains of staphylococci.31 Although no definite SCCmec elements were detected in either of the two strains, both carried the genetic context of mecA with high similarity to SCCmec type V (5C2&5) subtype c, which was first reported in the clonal complex 398 methicillin-resistant S. aureus strain JCSC6944.32 Genomic analysis showed that the mecA carried by two strains of S. haemolyticus in this study was bracketed by IS431 (IS431-mecA-ΔmecR1-IS431). The most prevalent and widely disseminated mec complex in S. aureus has the structure mecI-mecR1-mecA-IS431 and is designated as the class A mecA gene complex.33 However, in S. haemolyticus, IS431 was found to be associated with the deletion of mecI and mecR1, forming the structure IS431-mecA-ΔmecR1-IS431, designated as the class C mecA gene complex.34 IS431, a well-known mobile genetic element in staphylococci, has been implicated in the transfer of ARFs.35
The chromosomal aminoglycoside resistance gene aac(6’)-aph(2”) harbored by S. haemolyticus strain SH936 is flanked by two copies of IS256. IS256 was first described in strains of S. aureus isolated in Australia in 198736 and was considered the founding member of the IS256 family of insertion sequence elements,37 and was described as a part of the transposon Tn4001, conferring aminoglycoside resistance in S. aureus. IS256 is widely present in MDR staphylococci and enterococci.38 IS256 is frequently associated with the horizontal spread of ARGs,39 including the IS256-element (IS256-GNAT-[aac(6’)-aph(2”)]-IS256) harbored by S. haemolyticus strain SH936, which is widely present in strains of both Staphylococcus and Enterococcus. Furthermore, IS256 has been reported to influence antibiotic resistance either by insertion into regulatory genes40 or by modulating antibiotic resistance gene expression through the formation of strong hybrid promoters resulting from transposition into the neighborhood of antibiotic resistance genes.41 In addition, the presence of IS256 in staphylococci has been reported to be associated with biofilm formation.42
S. haemolyticus can be a reservoir for resistance genes that can be shared with other staphylococci25 via MGEs, including various plasmids.10,13 In the 26.9-kb plasmid pSH1275-2 of S. haemolyticus strain SH1275, the structure containing msr(A)–mph(C), IS431, and qacR/qacA, harbored by the S. haemolyticus plasmid was also present in S. aureus, S. epidermidis, and S. hominis, both in the plasmids and chromosomes. Staphylococcal antimicrobial resistance plasmids may carry mob genes for mobilization, or a tra gene complex for conjugative transfer,13 similar to the mobilizable plasmid pSH1275-3 containing the gene vga(A)LC in S. haemolyticus strain SH1275 in our study. Notably, one 2.47-kb plasmid (pSH9361-1) harboring erm(C) carried by S. haemolyticus strain SH9361 was also widely present in the Staphylococcus genus, especially in S. aureus.
Conclusion
In this study, we describe the genomic characteristics of two strains of oxacillin-resistant and mecA-positive S. haemolyticus (ST3 strain SH1275 and ST30 strain SH9361) isolated from ear swab samples of patients with OM. The two strains of oxacillin-resistant S. haemolyticus both carried the genetic contexts of mecA (IS431-mecA-ΔmecR1-IS431) with high similarity with the SCCmec type V(5C2&5) subtype c. The chromosomal aminoglycoside resistance gene aac(6’)-aph(2”) harbored by S. haemolyticus strain SH936 was flanked by two copies of IS256 in different orientations, forming the IS256-element (IS256-GNAT-[aac(6’)-aph(2”)]-IS256), which was commonly found in strains of both Staphylococcus and Enterococcus genus. Furthermore, the two strains of oxacillin-resistant and mecA-positive S. haemolyticus were found to harbor various antimicrobial resistance plasmids, including one 26.9-kb plasmid (pSH1275-2) containing msr(A)–mph(C)) and qacA, one mobilizable plasmid pSH1275-3 harboring vga(A)LC, one plasmid (pSH9361-1) carrying erm(C), and one plasmid (pSH9361-2) carrying qacJ. These insights can assist clinicians in devising precise, personalized, and clinical therapeutic strategies for treating otitis media caused by multi-drug resistant S. haemolyticus.
Ethical Approval Statement
This study has been approved by the Ethics Committee of Zhuhai People’s Hospital. The present study was a study focusing on bacteria and did not contain any sensitive personal information. Therefore, informed consent was not required according to “Measures for the Ethical Review of Biomedical Research Involving Humans” (https://www.gov.cn/gongbao/content/2017/content_5227817.htm).
Funding
This work was supported financially by the grants from the Science and Technology Projects of Social Development in Zhuhai (Grant No. ZH22036201210111PWC), and the Xiangshan Talent Project of Zhuhai People’s Hospital (Grant No. 2020XSYC-02).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Serra N, Di Carlo P, Andriolo M, et al. Staphylococcus aureus and coagulase-negative Staphylococci from Bloodstream infections: frequency of occurrence and antimicrobial resistance, 2018–2021. Life. 2023;13(6):1356. doi:10.3390/life13061356
2. Becker K, Both A, Weißelberg S, et al. Emergence of coagulase-negative staphylococci. Exp Rev Anti-Infective Ther. 2020;18(4):349–366. doi:10.1080/14787210.2020.1730813
3. Qin M, Chen P, Deng B, et al. The emergence of a multidrug-resistant and pathogenic ST42 lineage of Staphylococcus haemolyticus from a Hospital in China. Microbiol Spectr. 2022;10(3):e0234221. doi:10.1128/spectrum.02342-21
4. Fredheim EG, Klingenberg C, Rohde H, et al. Biofilm formation by Staphylococcus haemolyticus. J Clin Microbiol. 2009;47:1172–1180. doi:10.1128/JCM.01891-08
5. Sabdaningsih A, Cristianawati O, Sibero MT, et al. Screening antibacterial agent from crude extract of marine-derived fungi associated with soft corals against MDR-Staphylococcus haemolyticus.
6. Froggatt JW, Johnston JL, Galetto DW, et al. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1989;33(4):460–466. doi:10.1128/AAC.33.4.460
7. Czekaj T, Ciszewski M, Szewczyk EM. Staphylococcus haemolyticus - an emerging threat in the twilight of the antibiotics age. Microbiology. 2015;161(11):2061–2068. doi:10.1099/mic.0.000178
8. Parvizi J, Azzam K, Ghanem E, et al. Periprosthetic infection due to resistant staphylococci: serious problems on the horizon. Clin Orthopaedics Related Res. 2009;467(7):1732–1739. doi:10.1007/s11999-009-0857-z
9. García-álvarez L, Holden MT, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11(8):595–603. doi:10.1016/S1473-3099(11)70126-8
10. Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 2010;67(18):3057–3071. doi:10.1007/s00018-010-0389-4
11. Uehara Y. Current Status of Staphylococcal Cassette Chromosome mec (SCCmec). Antibiotics. 2022;11. doi:10.3390/antibiotics12010011
12. Bouchami O, Ben Hassen A, de Lencastre H, et al. High prevalence of mec complex C and ccrC is independent of SCCmec type V in Staphylococcus haemolyticus. Eur J Clin Microbiol Infect Dis. 2012;31(4):605–614. doi:10.1007/s10096-011-1354-3
13. Schwarz S, Shen J, Wendlandt S, et al. Plasmid-mediated antimicrobial resistance in Staphylococci and other Firmicutes. Microbiol Spectr. 2014;2(6): doi:10.1128/microbiolspec.PLAS-0020-2014.
14. Zechner EL, Moncalián G, de la Cruz F. Relaxases and plasmid transfer in gram-negative bacteria. In: Type IV Secretion in Gram-Negative and Gram-Positive Bacteria. Springer; 2017:93–113.
15. Cheng H, Concepcion GT, Feng X, et al. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods. 2021;18(2):170–175. doi:10.1038/s41592-020-01056-5
16. Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi:10.1101/gr.215087.116
17. Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi:10.1371/journal.pone.0112963
18. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi:10.1093/jac/dkaa345
19. Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi:10.1128/JCM.06094-11
20. Kaya H, Hasman H, Larsen J, et al. SCCmecFinder, a web-based tool for typing of Staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere. 2018;3. doi:10.1128/mSphere.00612-17
21. Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using plasmid finder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi:10.1128/AAC.02412-14
22. Siguier P, Perochon J, Lestrade L, et al. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(90001):D32–6. doi:10.1093/nar/gkj014
23. Alikhan NF, Petty NK, Ben Zakour NL, et al. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi:10.1186/1471-2164-12-402
24. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi:10.1093/bioinformatics/btr039
25. Montelongo C, Mores CR, Putonti C, et al. Whole-genome sequencing of Staphylococcus aureus and Staphylococcus haemolyticus clinical isolates from Egypt. Microbiol Spectr. 2022;10(4):e0241321. doi:10.1128/spectrum.02413-21
26. Lin LC, Chang SC, Ou YH, et al. Clonal spreading of ST42 Staphylococcus haemolyticus strains occurs possibly due to fusB and tetK resistant genes and capsule-related genes. Int J Mol Sci. 2023:24. doi:10.3390/ijms25010024
27. Lin LC, Liu TP, Chang SC, et al. Characterization of new Staphylococcus haemolyticus ST42 populations in Northern Taiwan. Microb Drug Resist. 2022;28:56–62. doi:10.1089/mdr.2019.0459
28. Phillip S, Mushi MF, Decano AG, et al. Molecular characterizations of the coagulase-negative Staphylococci species causing urinary tract infection in Tanzania: a Laboratory-Based Cross-Sectional Study. Pathogens. 2023;12(2):180. doi:10.3390/pathogens12020180
29. Panda S, Jena S, Sharma S, et al. Identification of novel sequence types among Staphylococcus haemolyticus isolated from variety of infections in India. PLoS One. 2016;11(11):e0166193. doi:10.1371/journal.pone.0166193
30. Sands K, Carvalho MJ, Spiller OB, et al. Characterisation of Staphylococci species from neonatal blood cultures in low- and middle-income countries. BMC Infect Dis. 2022;22(1):593. doi:10.1186/s12879-022-07541-w
31. Schwendener S, Perreten V. The bla and mec families of β-lactam resistance genes in the genera Macrococcus, Mammaliicoccus and Staphylococcus: an in-depth analysis with emphasis on Macrococcus. J Antimicrob Chemother. 2022;77(7):1796–1827. doi:10.1093/jac/dkac107
32. Li S, Skov RL, Han X, et al. Novel types of Staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55(6):3046–3050. doi:10.1128/AAC.01475-10
33. Liu J, Chen D, Peters BM, et al. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathogenesis. 2016;101:56–67. doi:10.1016/j.micpath.2016.10.028
34. Katayama Y, Ito T, Hiramatsu K. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob Agents Chemother. 2001;45:1955–1963. doi:10.1128/AAC.45.7.1955-1963.2001
35. Kobayashi N, Alam MM, Urasawa S. Genomic rearrangement of the mec regulator region mediated by insertion of IS431 in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 2001;45:335–338. doi:10.1128/AAC.45.1.335-338.2001
36. Lyon BR, Gillespie MT, Skurray RA. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J Gen Microbiol. 1987;133(11):3031–3038. doi:10.1099/00221287-133-11-3031
37. Hennig S, Ziebuhr W. Characterization of the transposase encoded by IS256, the prototype of a major family of bacterial insertion sequence elements. J Bacteriol. 2010;192(16):4153–4163. doi:10.1128/JB.00226-10
38. Loessner I, Dietrich K, Dittrich D, et al. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J Bacteriol. 2002;184(17):4709–4714. doi:10.1128/JB.184.17.4709-4714.2002
39. Casagrande Proietti P, Bietta A, Coletti M, et al. Insertion sequence IS256 in canine pyoderma isolates of Staphylococcus pseudintermedius associated with antibiotic resistance. Vet Microbiol. 2012;157(3–4):376–382. doi:10.1016/j.vetmic.2011.12.028
40. Maki H, McCallum N, Bischoff M, et al. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48(6):1953–1959. doi:10.1128/AAC.48.6.1953-1959.2004
41. Yin Y, Chen H, Li S, et al. Daptomycin resistance in methicillin-resistant Staphylococcus aureus is conferred by IS256 insertion in the promoter of mprF along with mutations in mprF and walK. Int J Antimicrob Agents. 2019;54(6):673–680. doi:10.1016/j.ijantimicag.2019.08.021
42. Asante J, Abia ALK, Anokwah D, et al. Phenotypic and genomic insights into biofilm formation in antibiotic-resistant clinical coagulase-negative Staphylococcus species from South Africa. Genes. 2022;14(1):14. doi:10.3390/genes14010014
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.