Back to Journals » Infection and Drug Resistance » Volume 13
Whole-Genome Analysis of Two Copies of blaNDM-1 Gene Carrying Acinetobacter johnsonii Strain Acsw19 Isolated from Sichuan, China
Authors Tang L, Shen W, Zhang Z, Zhang J, Wang G, Xiang L, She J, Hu X, Zou G , Zhu B , Zhou Y
Received 26 October 2019
Accepted for publication 3 March 2020
Published 23 March 2020 Volume 2020:13 Pages 855—865
DOI https://doi.org/10.2147/IDR.S236200
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Lingtong Tang, 1, 2 Wei Shen, 2, 3 Zhikun Zhang, 2 Jingping Zhang, 2 Guangxi Wang, 2 Li Xiang, 2 Junping She, 2 Xiaoyan Hu, 2 Guoyuan Zou, 4 Baoli Zhu, 5 Yingshun Zhou 2
1Department of Clinical Laboratory, The People’s Hospital of Gao County, Sichuan 644000, People’s Republic of China; 2Department of Pathogenic Biology, School of Basic Medicine, Southwest Medical University, Luzhou, Sichuan, People’s Republic of China; 3Department of Clinical Laboratory, The First People’s Hospital of Yibin, Yibin 644000, Sichuan, People’s Republic of China; 4Institute of Plant Nutrition and Resources, Beijing Academy of Agriculture and Forestry Science, Beijing, People’s Republic of China; 5Key Laboratory of Pathogenic Microbiology & Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, People’s Republic of China
Correspondence: Yingshun Zhou
Department of Pathogenic Biology, School of Basic Medicine, Southwest Medical University, No. 319, Zhongshan Road, Luzhou 646000, Sichuan, People’s Republic of China
Tel +86– 0830– 3160073
Email [email protected]
Guoyuan Zou
Institute of Plant Nutrition and Resources, Beijing Academy of Agriculture and Forestry Science, Beijing, People’s Republic of China
Email [email protected]
Purpose: To characterize the genetic feature of the carbapenems resistant Acinetobacter johnsonii strain Acsw19 isolated from municipal sludge. This strain was found to carry two copies of blaNDM-1, cmlB1-like gene, and blaOXA-211-like gene along with other 8 antimicrobial resistance genes, 3 plasmids, 15 genomic islands and 8 prophages.
Methods: A carbapenem-resistant Acinetobacter johnsonii strain Acsw19 isolated from municipal sludge was subjected to whole-genome sequencing (WGS) via the PacBio and Illumina MiSeq platforms. Thereafter, the characteristic was analyzed by a series of bioinformatics software.
Results: The results showed that the genome of Acsw19 was consisted of a 3,433,749 bp circular chromosome and 3 circular plasmids, pAcsw19-1 (11,161 bp), pAcsw19-2 (351,885 bp) and pAcsw19-3 (38,391bp), respectively. Resistome analysis showed that Acsw19 carried 12 antimicrobial resistance genes, including 6 [cmlB1-like, blaNDM-1, blaOXA-58, aph ( 3ʹ)-VIa, msr(E) and mph(E)] in the plasmid pAcsw19-2 and 6 (blaOXA-211-like, blaNDM-1, aph(3")-Ib, aph(6)-Id, sul2, and floR) in the chromosome genome. Specifically, the cmlB1-like gene shared 86.33%, 71.7% and 71.9% similarities with the cmlB1, cmlA4 and cmlA8 gene, and the blaOXA-211-like gene shared 94.4%, 95.39% and 96.36% similarities with blaOXA-211, blaOXA-643 and blaOXA-652, at the nucleotide level, respectively. Phylogenetic analysis showed that the blaOXA-211-like gene and cmlB1-like gene had the closest evolutionary relationship with blaOXA-643 and cmlB1, respectively. These results indicated that the blaOXA-211-like and cmlB1-like genes identified in the current study should be the novel variant resistance genes.
Conclusion: Carrying of two copies of blaNDM-1, cmlB1-like, blaOXA-211-like and along with other 8 antimicrobial resistance genes, 3 plasmids, 15 genomic islands and 8 prophages Acinetobacter johnsonii strain might increase the possibility of spreading of resistance genes.
Keywords: Acinetobacter johnsonii, blaNDM-1, blaOXA, genomic island
Introduction
Producing of carbapenemases, including the β-lactamases of Ambler classes A, B (metallo-β-lactamases) and D, are the most common mechanism of bacterial carbapenems resistance.1–3 Especially, the New Delhi Metallo-β-lactamase (NDM), Klebsiella pneumoniae carbapenemase (KPC) and some Class D β-lactamases (CHDLs) have been identified worldwide in gram-negative bacterial isolates from clinical, environmental samples, and food animals, especially in Enterobacteriaceae,1,4–9 and also in Pseudomonas aeruginosa and Acinetobacter species.10–12 Carbapenem-resistant Acinetobacter species are mainly associated with the carbapenem-hydrolyzing NDM and CHDLs, such as blaOXA-23-like, blaOXA-24/40-like, blaOXA-51-like, blaOXA-134-like and blaOXA-211-like gene.13–18 Especially, co-carrying of the blaNDM and blaOXA in clinical, food, environmental derived isolates of Acinetobacter species were prevalent in the world.11,19-21 In addition, the genes encoding blaNDM and blaOXA are known to be carried on some mobile genetic elements that inserted into the chromosome or plasmids, and it is suspected the mechanism of horizontal gene transfer (HGT) promotes the exchange of resistance genes among pathogenic microorganisms isolated from the clinical, environmental samples, and food animals.11,22–25
In this study, we mainly characterized the genetic feature of a carbapenems resistant strain Acinetobacter johnsonii Acsw19 isolated from municipal sludge. This strain was found to carry two copies of blaNDM-1, cmlB1-like gene, and blaOXA-211-like gene along with other 8 antimicrobial resistance genes, three plasmids, 15 genomic islands and 8 prophages.
Materials and Methods
Bacterial Isolate, Identification and Antimicrobial Susceptibility Testing
The sample of municipal sewage was obtained from the influx of wastewater-related plant in Luzhou City (Sichuan Province, China) in March 2019. The sewage was 1:10 diluted and an aliquot (10 μL) was streaked onto a CHROM Agar Orientation (CHROMAgar, Paris, France) agar plate containing 2 mg/L meropenem (Solarbio, China) and then incubated at 37 °C overnight. Bacterial species identification was carried out by the Vitek2 system (BioMérieux, France), 16srRNA sequencing and matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry. The minimal inhibitory concentrations (MICs) of 15 antimicrobial agents (Solarbio, China) including meropenem, imipenem, cefepime, cefotaxime, ceftazidime, piperacillin-tazobactam, amoxicillin-clavulanic acid, gentamicin, amikacin, aztreonam, erythromycin, chloramphenicol, sulfadiazine, colistin and ciprofloxacin were determined by broth microdilution method according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI 2013, M100-S23). Escherichia coli strain ATCC 25,922 was used as quality control. Polymerase chain reaction (PCR) amplification and DNA sequencing were performed to identify the key carbapenemase-encoding genes (blaNDM and blaKPC) as previously reference.7
Whole-Genome Sequencing and Analysis
Genomic DNA of the strain A. johnsonii Acsw19 was extracted using the DNA Kit (QIAGEN, Germany). The 10kb sequencing library and a 300 bp paired-end library were constructed using the standard PacBio RS sample and Illumina DNA sample preparation instructions, and then sequenced on Pacific Biosciences RS II and MiSeq systems sequencing platforms (Novogene, China). The reads were de novo assembled using the software Celera Assembler (version 8.0). Gene prediction was performed for the whole genome with Glimmer 3.02 (http://www.cbcb.umd.edu/software/glimmer/).26 And the annotation of the Acsw19 genome was achieved using the NCBI Prokaryotic Genome Annotation Pipeline. Pairwise alignment was performed by BLASTn search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The average nucleotide identity (ANI) analysis was performed by the computer.27 The resistome was identified using ResFinder 2.1 (https://cge.cbs.dtu.dk/services/ResFinder/)28 (minimum threshold for identity, 85%; minimum coverage, 60%) and Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/). The genomic island sequences were predicted based on three different genomic islands (GIs) prediction software (IslandPATH-DIMOB, IslandPick, and SIGI-HMM)29–31 and the Prophage was predicted by using phiSpy.32
Results and Discussion
Bacterial Isolate, Identification and Resistance Gene Detection
A gram-stain-negative, blaNDM-1 and blaOXA producing Acinetobacter johnsonii Acsw19 was isolated and identified by the Vitek2 system (BioMérieux, France), 16srRNA sequencing and matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry in Luzhou City, Southwestern China. Acinetobacter johnsonii strain Acsw19 was resistant to meropenem, imipenem, cefepime, cefotaxime, ceftazidime, piperacillin-tazobactam, amoxicillin-clavulanic acid, gentamicin, amikacin, aztreonam, erythromycin, chloramphenicol and sulfadiazine and it was susceptible to colistin and ciprofloxacin according to CLSI breakpoints (M100-S23) (Supplementary Table 1). To the best of our knowledge, the occurrence of multiple antibiotic genes in the multi-drug resistant bacterial isolates from sewage has been evidenced in numerous studies, with the involvement of numerous species.33,34
Characterization of the Whole Genome of Acinetobacter johnsonii Strain Acsw 19
We got 1,876,876,663 bp data of the whole genome by the WGS. The genome of Acinetobacter johnsonii Acsw19 consisted of a 3,433,749bp circular chromosome and three circular plasmids, pAcsw19-1, pAcsw19-2 and pAcsw19-3 in the size of 11,161bp, 351, 885bp and 38, 391 with the 35.66%, 38.79% and 33.76% G+C content, respectively. The chromosome has a 41.78% G+C content, 21 rRNA operons, 88 tRNAs, 44ncRNAs and 3413 predicted protein coding sequences (CDSs) (Table 1).
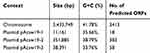 |
Table 1 Characteristic of the Whole Genome of Acinetobacter johnsonii Acsw19 |
Resistome Analysis
A total of 12 drug-resistance genes was detected in the whole genome of Acsw19. Specifically, 6 resistance genes [(blaNDM-1, blaOXA-211-like, aph(3")-Ib (strB), aph(6)-Id (strA), sul2, and floR)] were located in the chromosome genome and 6 [cmlB1-like, mph(E), msr(E), blaOXA-58, blaNDM-1 and aph(3ʹ)-VIa] in plasmid pAcsw19-2 (Table 2). None of the drug-resistance gene was detected in plasmid pAcsw19-1 and pAcsw19-3. The full lengthen of cmlB1-like gene consisted of 1266 nucleotides encoding a protein with 422 amino acids. Sequence analysis showed that the cmlB1-like gene shared 86.33%, 71.7% and 71.9% sequence similarities with the known cmlB1, cmlA4, and cmlA8 genes at nucleotide level and 67.8%, 44.8% and 44.5% at amino acid level, respectively. The full lengthen of blaOXA-211-like gene consisted of 825 nucleotides encoding a protein with 275 amino acids. Sequence analysis showed that the blaOXA-211-like gene shared 94.4%, 95.39%, 95.52%, 96% and 96.36% identity with the known blaOXA-211, blaOXA-643, blaOXA-281, blaOXA-645 and blaOXA-652 gene at nucleotide level and 94.4%, 95.4%, 95.5%, 96% and 96.4% at amino acid level, respectively. Phylogenetic analysis showed that the blaOXA-211-like gene and cmlB1-like gene had the closest evolutionary relationship with blaOXA-643 (Figure 1) and cmlB1 (Figure 2). These results indicated that the blaOXA-211-like and cmlB1-like genes maybe two new allelic variant of the gene blaOXA and cmlB1. To our the knowledge, there are more new allelic variant of the resistance genes have been found from the sewage derived isolates.33,35,36
 |
Table 2 Distribution of the Resistance Genes in Acinetobacter johnsonii Strain Acsw19 |
Characterization of Three Plasmids
Plasmid pAcsw19-1 contained 18 putative coding open reading frames (ORFs). Sequence analysis showed that pAcsw19-1 had 100%, 92%, 68% and 68% query cover and 96.55%, 91.5%, 87.8% and 87.8% sequence similarities with the plasmids p2_010062 (CP033122), p4_010055 (CP032283), p3_010030 (CP029391) and pALWEK1.4 (CP032107) at nucleotide level, and these reported plasmids were all harbored by the Acinetobacter species. None of antimicrobial resistance gene was determined in plasmid pAcsw19-1. Plasmid pAcsw19-1 carried two copies of plasmid replicons, two mobilization proteins (mobL-like), and several hypothetical ORFs. Complete sequence analysis showed that the 351,885 bp plasmid pAcsw19-2 contained 383 putative coding ORFs. Plasmid pAcsw19-2 had 87.34%, 14.76% and 7.06% query cover and 99.28%, 99.78% and 100% sequence similarities with the plasmids pXBB1-9 (CP010350), pACI-df08 (CP026426)37 and pM131-2 (JX101647) at nucleotide level, respectively.11
Plasmid pAcsw19-2 carried 6 resistance genes [cmlB1-like, mph(E), msr(E), blaOXA-58, blaNDM-1, and aph(3ʹ)-VIa] which were distributed in three regions. The first region carried the cmlB1-like gene. The genetic context of this region was ISL3-like transposase-cmlB1-like-LysR (Figure 2). This region was similar to the corresponding area of plasmid pOXA58_010030 (CP029396), pOXA58_010055 (CP032285), and pXBB1-9 (CP010351). The second region carried three resistance genes [mph(E), msr(E), and blaOXA-58]. The first two resistance gene mph(E) and msr(E) were carried by the genetic context which was constituted by IS30 family transposase, helix-turn-helix-ORF, mph(E), msr(E), helix-turn-helix-ORF, plasmid stability associated protein-coding ORFs (addiction module antidote protein, RelE/ParE, brnA/T, and LysE family), ISAba1, and AraC transcriptional regulator gene. The AraC was linked to the third resistance gene blaOXA-58 carrying area which was constituted by ISAba3-blaOXA58-ISAba3 (Figure 3). The context of blaOXA-58 carrying area (ISAba3-blaOXA58-ISAba3) was same to the previous reports.11,38-40
The third region carried 2 resistance genes [plasmid borne blaNDM-1 and aph (3ʹ)-VIa]. Nucleotide sequence analysis revealed that the blaNDM-1 gene was flanked in the upstream region of IS5 transposase-dmc-unknown ORF-umuD-Y-family DNA polymerase-unknown ORF-trpF-bleMBL and downstream by the IS30 family transposase and 6 unknown ORFs carried region, which linked to the resistance gene aph(3ʹ)-Via carrying genetic context [ISAba125-aph(3ʹ)-Via -ISAba125]. This resistance area (ISAba125-aph(3ʹ)-Via-ISAba125) was similar to the corresponding region of the plasmid pAP43-OXA58-NDM1 (CP043053), which was harbored by Acinetobacter pittii.
The plasmid pAcsw19-3 contained 58 putative coding ORFs. Sequence analysis showed that pAcsw19-3 had 72%, 76%, and 83% query cover and 99.90%, 99.96% and 99.84% sequence similarities with the plasmids p3_010055 (CP032282.1), p2_010030 (CP029390), and p4_010060 (CP031712) at the nucleotide level, and these plasmids were all harbored by the Acinetobacter species strains. None of the antimicrobial resistance gene was determined in plasmid pAcsw19-3, either.
Characterization of the Genomic Islands (GI)
Fifteen genomic islands, named GI_Acsw19-1 to GI_Acsw19-15, were identified by the software IslandPATH-DIMOB, IslandPick and SIGI-HMM. Sequence analysis showed that the length of the 15 genomic islands were ranged from ~5.1 kb to ~94.86 kb with the average G+C context of 32.59% to 54.18%, respectively. Moreover, 13 genomic islands (GI_Acsw19-1 to GI_Acsw19-13) were located in the chromosome and 2 genomic islands (GI_Acsw19-14 and GI_Acsw19-15) in the plasmid pAcsw19-2. Among the 15 GIs, two (GI_Acsw19-11 and GI_Acsw19-12) were the resistant genomic islands. Genomic island GI_Acsw19-11 (26,133 bp) carried the aminoglycosides resistance genes strA and strB, sulphonamides resistance gene sul2 and phenicol resistance gene floR. Genomic island GI_Acsw19-12 (12,309 bp) carried the chromosome borne blaNDM-1 (Table 3). Sequence analysis showed that GI_Acsw19-11 had 47%, 37%, and 34% query cover and 100% sequence similarities with the DNA sequence of A. baumannii MRSN15313 chromosome genome (CP033869), A. pittii WCHAP100004 plasmid pOXA58_100004 (CP027249), and uncultured bacterium HHV216 plasmid pHHV216(FJ012880). Based on the specific genetic content, GI_Acsw19-11 could be divided into two regions (regions A and regions B) (Figure 4). The region A was 15,713bp in length which served as the backbone of GI_Acsw19-11. It mainly carried the tyrosine-type recombinase/integrase gene (1137 bp) and an IS1 family tnpA, which might be responsible for encoding the site-specific resolvase and transposition. Additionally, the other genes of region A, including ferrous iron transporter gene feoAB, Cd(II)/Pb(II)-responsive transcriptional regulator encoding gene cadR, hydrolase encoding gene, some of hypothetical protein-encoding genes, were found to be located in the backbone. Sequence of region A was high similar to the corresponding region of other various Acinetobacter species.41 The 4 resistance genes (strA and strB, sul2 and floR) carried region B was 8980 bp in length (Figure 3). These four resistance genes genetic context is ISAba1-sul2-glmM-ISVsa3-LysR-floR-DUF3363-strB-strA-ISAba1. The sul2 and floR carrying area (6814 bp) was similar to the corresponding region of plasmid pOXA58_100004, while the strB and strA carrying area (2832 bp) was similar to corresponding region of plasmid pNDM-AP_882 harbored by the A. pittii AP_882 (CP014478.1).
 |
Table 3 Overall Features of the Acinetobacter johnsonii Strain Acsw19 Genomic Islands |
Sequence analysis showed that GI_Acsw19-12 had 96%, 96%, and 96% query cover and 100%, 99.9%, and 99.9% sequence similarities with the corresponding region of sequences of A. baumannii AR_0083 genome (CP027528), A. pittii ST220 genome (CP029610), and A. baumannii AR_0083 genome (CP027528). GI_Acsw19-12 had 82%, 82%, and 84% query cover and 100%, 100%, and 99.9% sequence similarities with the plasmids including the pNDM1_060092 (CP035935), pNDM-GJ01 (KT965092)42 and pNDM-JVAP01(KM923969)43 at nucleotide level. Moreover, the chromosome borne blaNDM was located in the genetic element (10,023 bp) ISAba125-IS91-family tnpA-groL-groES-cutA-tat-trpF-ble-blaNDM-1-ISAba125 (Figure 5). This blaNDM carrying element was similar equal to the corresponding region of the pNDM1_010045.
Characterization of Prophages
A total of eight prophages, named Pp_Acsw19-1 to Pp_Acsw19-8, was identified by phiSpy (Table 4). Sequence analysis showed that one prophage (Pp_Acsw19-1) was located in the plasmid pAcsw19-2 and 7 prophages (Pp_Acsw19-2 to Pp_Acsw19-8) were located in the chromosome genome. The lengthen of eight prophages was ranged from ~17.22 kb to ~159 kb with average G+C context of 40.03% to 47.12%, respectively. Interestingly, the blaNDM-carrying genomic islands GI_ Acsw19-12 was located in the prophage Pp_Acsw19-7 and the cmlB1-like gene carrying prophage Pp_Acsw19-1 was located in the plasmid pAcsw19-2. These findings suggested that some mobile genetic elements can be excised from chromosome or mobile genetic elements and then integrated into the chromosome or other mobile genetic elements.44–47 These situations may promote the exchange of resistance genes among pathogenic microorganisms.
 |
Table 4 Overall Features of the Acinetobacter johnsonii Strain Acsw19 Prophage |
Conclusions
The current study characterize the complete genome sequence of Acinetobacter Acsw19, which carried two copies of blaNDM-1, cmlB1-like gene, blaOXA-211-like gene along with other 8 antimicrobial resistance genes in 3 plasmids, 15 genomic islands and 8 prophages, which could provide great genetic plasticity for the host dissemination of antimicrobial resistance. The occurrence of multiple antibiotic genes in bacterial isolates from sewage has been evidenced in numerous studies, with the involvement of numerous species, it may be serving as an important reservoir of resistance genes and a hot spot for the transfer of resistance genes and mobile genetic elements. We should be vigilant these isolates or resistance genes transfer to the clinical bacterial.
Nucleotide Sequence Accession Numbers
The genome sequence of Acinetobacter johnsonii Acsw19 is deposited in the NCBI database under accession numbers CP043307 (chromosome) and CP043308 to CP043310 (plasmid pAcsw19-1 to pAcsw19-3).
Funding
This work was supported by the National Natural Science Foundation of China (31500114), and United Funds of Luzhou and Southwest Medical University [2018LZXNYD-ZK51].
Disclosure
The authors declare no conflict of interest.
References
1. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–58, table of contents. doi:10.1128/CMR.00001-07
2. Fu L, Huang M, Zhang X, et al. Frequency of virulence factors in high biofilm formation blaKPC-2 producing Klebsiella pneumoniae strains from hospitals. Microb Pathog. 2018;116:168–172. doi:10.1016/j.micpath.2018.01.030
3. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006.
4. Liu Y, Zhang H, Zhang X, et al. Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagn Microbiol Infect Dis. 2019;93(4):355–361. doi:10.1016/j.diagmicrobio.2018.11.007
5. Fu L, Ang GY, Yu CY, et al. Co-carrying of KPC-2, NDM-5, CTX-M-3 and CTX-M-65 in three plasmids with serotype O89: H10 Escherichia coli strain belonging to the ST2 clone in China. Microb Pathog. 2019;128:1–6. doi:10.1016/j.micpath.2018.12.033
6. Zmarlicka MT, Nailor MD, Nicolau DP. Impact of the New Delhi metallo-beta-lactamase on beta-lactam antibiotics. Infect Drug Resist. 2015;8:297–309. doi:10.2147/IDR.S39186
7. Liu X, Zhang J, Li Y, et al. Diversity and frequency of resistance and virulence genes in blaKPC and blaNDM co-producing Klebsiella pneumoniae strains from China. Infect Drug Resist. 2019;12:2819–2826. doi:10.2147/IDR
8. Kiaei S, Moradi M, Hosseini-nave H, et al. Endemic dissemination of different sequence types of carbapenem-resistant Klebsiella pneumoniae strains harboring blaNDM and 16S rRNA methylase genes in Kerman hospitals, Iran, from 2015 to 2017. Infect Drug Resist. 2019;12:45–54. doi:10.2147/IDR
9. Papagiannitsis CC, Bitar I, Malli E, et al. IncC blaKPC-2-positive plasmid characterised from ST648 Escherichia coli. J Glob Antimicrob Resist. 2019;19:73–77. doi:10.1016/j.jgar.2019.05.001
10. Hu Y, Feng Y, Qin J, Zhang X, Zong Z. Acinetobacter chinensis, a novel Acinetobacter species, carrying blaNDM-1, recovered from hospital sewage. J Microbiol. 2019;57(5):350–355. doi:10.1007/s12275-019-8485-0
11. Feng Y, Yang P, Wang X, et al. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J Antimicrobial Chemother. 2015;71(1):71. doi:10.1093/jac/dkv324
12. Liu WJ, et al. Frequency of antiseptic resistance genes and reduced susceptibility to biocides in carbapenem-resistant Acinetobacter baumannii. J Med Microbiol. 2017;66(1).
13. Waltherrasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrobial Chemother. 2006;57(3):373. doi:10.1093/jac/dki482
14. Hammoudi D, Moubareck CA, Hakime N, et al. Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int J Infect Dis Ijid off Publ Int Soc Infect Dis. 2015;36(C):56–61. doi:10.1016/j.ijid.2015.05.015
15. Higgins PG, Poirel L, Lehmann M, et al. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(12):5035–5038. doi:10.1128/AAC.00856-09
16. Nuno T,A, Lamoureaux TL, Toth M, et al. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother. 2014;58(4):2119–2125. doi:10.1128/AAC.02522-13
17. Yang Q, Rui Y. Two multiplex real-time PCR assays to detect and differentiate acinetobacter baumannii and non- baumannii acinetobacter spp. carrying blaNDM, blaOXA-23-like, blaOXA-40-like, blaOXA-51-like, and blaOXA-58-like genes. PLoS One. 2016;11(7):e0158958. doi:10.1371/journal.pone.0158958
18. Khorsi K, et al. ISAba36 inserted into the outer membrane protein gene carO and associated with the carbapenemase gene blaOXA-24-like in Acinetobacter baumannii. J Glob Antimicrob Resist. 2018;15:107–108. doi:10.1016/j.jgar.2018.08.020
19. Nishida S, Ono Y. Comparative analysis of the pathogenicity between multidrug-resistant Acinetobacter baumannii clinical isolates: isolation of highly pathogenic multidrug-resistant A. baumannii and experimental therapeutics with fourth-generation cephalosporin cefozopran. Infect Drug Resist. 2018;11:1715–1722. doi:10.2147/IDR.S166154
20. Zenati K, Touati A, Bakour S, et al. Characterization of NDM-1- and OXA-23-producing Acinetobacter baumannii isolates from inanimate surfaces in a hospital environment in Algeria. J Hosp Infect. 2016;92(1):19–26. doi:10.1016/j.jhin.2015.09.020
21. Guerra B, Fischer J, Helmuth R. An emerging public health problem: acquired carbapenemase-producing microorganisms are present in food-producing animals, their environment, companion animals and wild birds. Vet Microbiol. 2014;171(3–4):290–297. doi:10.1016/j.vetmic.2014.02.001
22. Sørensen SJ, Bailey M, Hansen LH, et al. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol. 2005;3(9):700–710. doi:10.1038/nrmicro1232
23. Peter JG, Townsend, JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3(9):679–687. doi:10.1038/nrmicro1204
24. Brown-jaque M, Calero-cáceres W, Muniesa M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid. 2015;79(3):1–7. doi:10.1016/j.plasmid.2015.01.001
25. Zhou S, Chen X, Meng X, et al. “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci Rep. 2015;5(1):8976. doi:10.1038/srep08976
26. Delcher AL, Bratke KA, Powers EC, et al. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–679. doi:10.1093/bioinformatics/btm009
27. Goris J, Goris J, Vandamme P, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91. doi:10.1099/ijs.0.64483-0
28. Ea Z, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrobial Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
29. Bertelli C, Brinkman F. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics. 2018;34(13):2161–2167. doi:10.1093/bioinformatics/bty095
30. Langille MG, Hsiao WW, Brinkman FS. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics. 2008;9(1):329. doi:10.1186/1471-2105-9-329
31. Waack S, Keller O, Asper R, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics. 2006;7(1):1–12. doi:10.1186/1471-2105-7-142
32. Langille MGI, Brinkman FSL. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25(5):664–665. doi:10.1093/bioinformatics/btp030
33. Zhao F, Feng Y, Lü X, et al. Remarkable diversity of Escherichia coli Carrying mcr-1 from hospital sewage with the identification of two new mcr-1 variants. Front Microbiol. 2017;8:2094. doi:10.3389/fmicb.2017.02094
34. Hu Y, Feng Y, Zhang X, et al. Acinetobacter defluvii sp. nov., recovered from hospital sewage. Int J Syst Evol Microbiol. 2017;67(6):1709–1713. doi:10.1099/ijsem.0.001847
35. Paiva MC, Reis MP, Costa PS, et al. Identification of new bacteria harboring qnrS and aac(6ʹ)-Ib/cr and mutations possibly involved in fluoroquinolone resistance in raw sewage and activated sludge samples from a full-scale WWTP. Water Res. 2017;110:27–37. doi:10.1016/j.watres.2016.11.056
36. Uyaguari MI, Fichot EB, Scott GI, et al. Characterization and quantitation of a novel beta-lactamase gene found in a wastewater treatment facility and the surrounding coastal ecosystem. Appl Environ Microbiol. 2011;77(23):8226–8233. doi:10.1128/AEM.02732-10
37. Zhang XZ, Lei CW, Zeng JX, et al. An IncX1 plasmid isolated from Salmonella enterica subsp. enterica serovar Pullorum carrying blaTEM-1B, sul2, arsenic resistant operons. Plasmid. 2018.
38. Klotz P, Jacobmeyer L, Leidner U, et al. Acinetobacter pittii from companion animals coharboring blaOXA-58, the tet(39) region, and other resistance genes on a single plasmid. Antimicrob Agents Chemother. 2017;62(1):
39. Ang GY, Yu CY, Cheong YM, et al. Emergence of ST119 Acinetobacter pittii co-harbouring NDM-1 and OXA-58 in Malaysia. Int J Antimicrob Agents. 2015;47(2):168–169. doi:10.1016/j.ijantimicag.2015.11.008
40. Cayô R, Rodrigues-costa F, Pereira Matos A, et al. Old clinical isolates of Acinetobacter seifertii in Brazil producing OXA-58. Antimicrob Agents Chemother. 2016;60(4):
41. Yaligara V, Husain F, Boente R, et al. Deficiency of the ferrous iron transporter FeoAB is linked with metronidazole resistance in Bacteroides fragilis. J Antimicrobial Chemother. 2014;69(10):2634–2643. doi:10.1093/jac/dku219
42. Zou D, Huang Y, Liu W, et al. Complete sequences of two novel blaNDM-1-harbouring plasmids from two Acinetobacter towneri isolates in China associated with the acquisition of Tn125. Sci Rep. 2017;7(1):9405. doi:10.1038/s41598-017-09624-0
43. Paula E, Mosqueda N, Telli M, et al. Identification of NDM-1 in a putatively novel acinetobacter species (“NB14”) closely related to Acinetobacter pittii. Antimicrob Agents Chemother. 2015;59(10):6657–6660. doi:10.1128/AAC.01455-15
44. Frost L, Leplae R, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3(9):722–732.
45. Nicolas C, Matteau D, Luo P, et al. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014;10(10):e1004714. doi:10.1371/journal.pgen.1004714
46. Carraro N, Rivard N, Burrus V, et al. Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob Genet Elements. 2017;7(2):1–6. doi:10.1080/2159256X.2017.1304193
47. Partridge SR, et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4).
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





