Back to Journals » Infection and Drug Resistance » Volume 16
Whole Genome Analysis of a Non-O1, Non-O139 Vibrio cholerae Detected from Human Blood in China
Authors Tang J , Li S, Zhang M, Li F, Tang Y, Yang F
Received 23 May 2023
Accepted for publication 10 August 2023
Published 21 August 2023 Volume 2023:16 Pages 5453—5461
DOI https://doi.org/10.2147/IDR.S420095
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Jinrong Tang,1 Sheng Li,1 Manli Zhang,1 Fengzhi Li,1 Ying Tang,2 Fangfang Yang3
1Department of Clinical Laboratory, Mianyang Hospital of Traditional Chinese Medicine/Mianyang Hospital Affiliated to Chengdu University of Traditional Chinese Medicine, Mianyang, Sichuan, 621000, People’s Republic of China; 2Youxian District Center for Disease Control and Prevention of Mianyang City, Mianyang, Sichuan, 621000, People’s Republic of China; 3Department of Clinical Laboratory, The Third Hospital of Mianyang (Sichuan Mental Health Center), Mianyang, Sichuan, 621000, People’s Republic of China
Correspondence: Fangfang Yang, Department of Clinical Laboratory, The Third Hospital of Mianyang (Sichuan Mental Health Center), Mianyang, Sichuan, 621000, People’s Republic of China, Tel +86-19817592909, Email [email protected]
Abstract: Non-O1, non-O139 Vibrio cholerae (NOVC) can cause cholera-like diarrhea, but it rarely causes extraintestinal infection, so it is easily overlooked. In this report, we present a case of NOVC detected through blood culture in a 58-year-old male patient with cirrhosis, resulting in severe infection. The patient had been diagnosed with cirrhosis seven years prior and was admitted to the hospital due to abdominal distension and gastrointestinal bleeding. Gram-negative bacilli were isolated from blood cultures and identified as V. cholerae using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and average nucleotide identity (ANI). Moreover, the serum agglutination test showed that the strain was non-O1/non-O139. Further whole genome sequencing and analysis of the strain showed that the strain mainly carried virulence genes tox R, RTX, hly A, T3SS/T6SS, but no resistant genes such as sulII, dfrA1, strB were detected. It provides information for the study of the pathogenic mechanism and drug resistance mechanism of V. cholerae. The patient had severe symptoms and a poor prognosis, indicating that although the NOVC strain infected in this patient had few virulence genes, it was not weak in pathogenicity. It may be caused by the effect of some virulence genes, which should be paid attention to.
Keywords: non-O1/non-O139 Vibrio cholerae, whole genome analysis, cirrhosis, virulence factors
Introduction
Vibrio cholerae is a Gram-negative bacterium that can be classified into different serotypes based on the O surface antigen, with O1 and O139 being responsible for causing cholera in humans. Cholera is a highly infectious and severe diarrheal disease that is prevalent worldwide, particularly in Africa, South Asia, and Southeast Asia.1 Non-O1, Non-O139 V. cholerae can also cause cholera-like diarrhea; however, it does not produce the same severity of symptoms as its counterparts due to the absence of cholera toxin.2 NOVC primarily causes intestinal infections but may lead to extraintestinal infections in individuals with weakened immune systems such as those with liver disease or undergoing chemotherapy.3 Studies have shown that NOVC infection is more common in patients with chronic liver disease, but its pathogenesis is still unclear.4,5 It may be related to factors such as increased intestinal permeability, weakened liver detoxification function in cirrhosis, and increased serum iron level.6 These extraintestinal infections have high mortality and should be paid attention to.7
At present, the pathogenic mechanism of NOVC is still unclear. Studies have shown that the main virulence genes of NOVC including hlyA, hapA, rtxA, nanH, stn, ompU, zot, ace, type VI secretion system (T6SS), and type III secretion system (T3SS) gene clusters. The distribution characteristics of virulence genes are closely related to their pathogenicity.8,9
This paper reports a Gram-negative bacterium from the blood culture of a patient with liver cirrhosis. It was identified as V. cholerae by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and average nucleotide identity (ANI). Furthermore, it was reviewed as NOVC by the Centers for Disease Control (CDC).
Materials and Methods
Isolation and Identification of Strain
The suspected strain was isolated from aerobic and anaerobic blood cultures of a patient with liver cirrhosis. The strain was initially observed under the microscope after Gram staining. It was inoculated into a blood plate culture for 24 hours. Then the strain was identified by MALDI-TOF MS (BioMerieux, Germany) with a 99% confidence level (Figure S1).
Antibiotic Susceptibility Test
Antimicrobial susceptibility testing were performed by microdilution method and drug-sensitive slips method, according to the Clinical and Laboratory Standards Institute (CLSI) guideline M45,10 for amikacin, ampicillin, ampicillin/sulbactam, cefazolin, cefepime, ceftazidime, ciprofloxacin, gentamicin, imipenem, levofloxacin, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, meropenem, amoxicillin/clavulanate, cefuroxime, azithromycin, tetracycline, chloramphenicol, polymyxin B, nalidixic acid.
Whole Genome Sequencing and Gene Function Analysis
The DNA of the V. cholerae strain was extracted using the VAMNE Magnetic Pathogen DNA/RNA Kit (Vazyme, CHINA) and sequenced on the Illumina MiSeq platforms. The clean reads are filtered using SOAPnuke software11 (version 1.5.2) and then assembled using SPAdes (version 3.11.0) software.12 The assembled sequences were uploaded to the ribosomal multilocus sequence typing (rMLST) database (https://pubmlst.org/species-id) and the JspeciesWS database (http://jspecies.ribohost.com/jspeciesws) for strain identification and average nucleotide identity (ANI) analysis, respectively. We used Glimmer software (version 3.02) to compare the predicted target gene sequences with sequences in the KEGG, COG, VFDB, and CARD databases to obtain annotation information. The final assembled genome was submitted to the NCBI database (www.ncbi.nlm.nih.gov) with the accession number: JARYMY010000000.
Results
Case Presentation
A 58-year-old man was diagnosed with cirrhosis decompensation and hypersplenism’ 7 years ago. Two hours before admission, the patient had no obvious inducement of nausea, abdominal distension, chills, and general fatigue during sleep. He had hematemesis twice, with an unknown amount, obvious blood clots, clear consciousness, and poor spirit. He was admitted to the hospital for treatment for liver cirrhosis and gastrointestinal bleeding. His temperature was 38.6°C. His heart rate, blood pressure, respiration, and oxygen saturation were within the normal range. Laboratory data showed a white blood cell count of 5.17*109/L, with 78.8% polymorphonuclear cells. Hemoglobin (102g/L) was reduced. Procalcitonin (PCT) (0.17ng/mL), total bilirubin (24.7 µmol/L), direct bilirubin (8.5 µmol/L), total bile acid (25.6 µmol/L), and urea (12.1mmol/L) was increased.
The stool was pasty, and no red blood or white blood cells were found. Besides, the patient tested negative for hepatitis B surface antigen (HBsAg).
The CT (computed tomography) results showed no significant abnormalities in both lungs. There were observed spots with slightly increased density, suggestive of blood accumulation or gastric contents, within the stomach, indicating cirrhosis. Esophageal variceal vein ligation and sclerosis were performed under gastroscopy at the bedside while the patient was under general anesthesia with tracheal intubation. However, after 2 days of admission, there was a worsening of abdominal distension accompanied by dyspnea and decreased oxygen saturation. Physical examination revealed generalized abdominal swelling with multiple areas of tenderness and rebound pain, diminished breath sounds in the right lung, and dullness to percussion over the right chest area. Tracheal intubation was performed followed by ventilator-assisted ventilation. A repeat chest CT scan demonstrated a large pleural effusion in the right hemithorax causing compression and collapse of the right lung as well as mediastinal shift towards the left side. Additionally, ground glass opacities were noted in patches within the left lung along with a small amount of fluid accumulation within the left pleural cavity.
The patient was treated with cefoperazone sodium and tazobactam for anti-infection. Because the patient’s underlying disease was too serious, the patient’s family gave up treatment and asked to be discharged.
Isolation and Identification of Strains
The patient’s blood culture was positive after two days of aerobic and anaerobic bottles. The morphology under the microscope (Lens*100) was Gram-negative bacilli (Figure 1A). Transgenic blood agar plate culture occurred within 24 hours β hemolytic colony with metallic luster (Figure 1B), identified as V. cholerae by MALDI-TOF MS. The local Center for Disease Control and Prevention (CDC) identified the bacteria as non-O1, non-O139 V. cholerae. The strain was named VC1115.
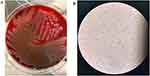 |
Figure 1 Colony characteristics in blood plate cultured for 24 hours (A) and morphological characteristics were observed under oil microscopy (B) of V. cholerae strain VC1115. |
Genome Assembly and Gene Prediction
Quality control, quality assessment, and assembly of VC1115 original reads obtained by sequencing were carried out, and the genomic circle map was mapped by Circos software (Figure 2). It was predicted that the genome size of VC1115 was 4,212,413 bp, the genome encoded 3836 genes, the total length of the encoding genes was 36,344,566 bp, the average length of the encoding genes was 947.49 bp, the gene GC content was 47.49%, and the gene length accounted for 48.36% of the total genome length. The non-coding RNA in the genome of the sequenced strain contained 86 tRNAs, 10 rRNAs, and 72 sRNAs. The assembled sequence was uploaded to the rMLST database of the PubMLST website for strain identification. The results showed that the uploaded sequence was 100% consistent with V. cholerae. The sequence was uploaded to the JspeciesWS database for ANI analysis. The ANIb value was 98.10% and the ANIm value was 98.40%. By comparing with various databases, a total of 3113 (81.15%) genes were annotated, of which 3042 (79.3%) genes were annotated by the COG database, 2502 (65.22%) genes were annotated by the KEGG database, and 370 (9.64%) genes were annotated by the VFDB database.
KEGG Pathway Analysis
By comparing with the KEGG database, 2710 genes related to metabolic pathways were found in the VC1115 genome, which can be divided into metabolism, genetic information processing, organic systems, environmental information processing, cell motility, and pathogenicity (Figure 3). There were a total of 1602 metabolic-related genes, among which there were more genes involved in carbohydrate, amino acid, cofactor, and vitamin metabolism. There were 401 genes related to environmental information processing, mainly involved in membrane transport and signal transduction. A total of 333 genes were related to cellular processes, mainly involved in cell movement, cell growth, and death, as well as transport and metabolism. There are 203 genes involved in genetic information processing, such as transcription, translation, protein folding, sorting, and degradation. Notably, 40 genes were associated with “drug resistances”, including 20 cationic antimicrobial peptide resistance genes, 18 beta-lactam resistance genes, and 6 vancomycin resistance genes. Five of the pathogenicity-related genes were involved in the pathogenesis of V. cholerae (Table 1).
 |
Table 1 Pathogenic KEGG Signaling Pathways in V. cholerae Strain VC1115 |
 |
Figure 3 Distribution map of KEGG functional annotations of V. cholerae VC1115. The ordinate is the annotated entry and the abscissa is the corresponding entry gene number. |
COG Pathway Analysis
In the COG database, genes were assigned to 24 functional categories and divided into four gene functional types: cellular processes and signals, information storage and processing, metabolism, and poorly characterized (Figure 4).
 |
Figure 4 Map of COG functional annotation distribution of V. cholerae VC1115. The ordinate is the annotated entry and the abscissa is the corresponding entry gene number. |
Toxicity Gene Test Results
The VC1115 genome was examined for genes coding for potential virulence factors using the VFDB database. It lacks CTX virulence factor but caries other 140 related factors to virulence, including hlyA virulence factor (Table 2).
 |
Table 2 Prediction of Virulence Factor Genes of the V. cholerae Strain VC1115 |
Resistance Phenotypes and Resistance Genes
The susceptibility test showed that the strain was resistant to cefazolin, imipenem, and polymyxin B, but sensitive to amikacin, ampicillin, ampicillin/sulbactam, cefepime, ceftazidime, ciprofloxacin, gentamicin, levofloxacin, piperacillin/tazobactam, trimethoprim/sulfamethoxazole, meropenem, amoxicillin/clavulanate, cefuroxime, azithromycin, tetracycline, chloramphenicol, nalidixic acid (Table 3).
 |
Table 3 Antimicrobial Resistance Profile of the V. cholerae Strain VC1115 |
Seven drug resistance genes were detected in the genome of strain VC1115, including CRP, catB9, almF, almG, almE, MCR-4.3, and QnrVC5 (Table 4).
 |
Table 4 The Antibiotic Resistance Genes of the Strain VC1115 Annotated in CARD |
Discussion
NOVC has been considered to have limited pathogenicity due to its lack of CTX toxin production. As NOVC-induced infections have been reported, some of them even lead to serious and life-threatening infections, and it has gradually attracted clinical attention.
NOVC is prone to gastrointestinal infections, and extraintestinal infections are rare, but this case is an extraintestinal infection. The patient was a 58-year-old man with cirrhosis. It has been reported that men are more susceptible to NOVC infection than women, and liver disease is also a risk factor for NOVC infection.13,14 NOVC is more likely to cause infection in patients with low immunity, which can cause extraintestinal infection, bloodstream infection, wound infection, and so on.15 Tsuruta et al detected NOVC from blood cultures of patients undergoing chemotherapy.16
The patient had no clear epidemiological history and the cause of infection was unknown. The patient later developed a severe lung infection, which could not be ruled out as being caused by the NOVC strain.
According to gene sequencing of the NOVC strain reported in this paper, the total length of the genome, total length of coding gene, CG content, metabolic pathway, and other aspects of the strain was similar to that of non-toxigenic V. cholerae.17 Compared with the CARD database, the drug resistance genes detected in VC1115 strain were CRP, catB9, almF, almG, almE, MCR-4.3, and QnrVC5. The drug sensitivity test results indicated that the VC1115 strain exhibited susceptibility to sulfonamides, quinolones, aminoglycosides, and other antibiotics while demonstrating resistance to cefazolin, imipenem, and polymyxin B. Wu et al18 reported that the resistance rates of 824 clinical NOVC isolates worldwide were ampicillin (44%), streptomycin (40%), cotrimoxazole (27%), and neomycin (27%). It shows that there are some differences in the drug sensitivity of NOVC strains. Bhandari et al19 reported that among the 56 strains of NOVC isolated from human and animal infections, 17 strains (30%) contained catB9, 13 strains (23%) contained blaCARB-9, and 1 strain (1.8%) carried sulII, strA, and strB. Despite its sensitivity to chloramphenicol, quinolones, and aminoglycoside antibiotics, VC1115 still harbors related resistance genes. Therefore, it is imperative to reinforce the integration of in vitro drug susceptibility testing and drug resistance gene monitoring to facilitate disease treatment.
The main toxin of V. cholerae causing human diarrhea is CT (cholera toxin).20 CT is mainly encoded by ctx AB gene, and the strain carrying ctx AB gene is V. cholerae toxigenic, which can cause cholera.21 V. cholerae O1 and O139, which cause cholera, generally have CTX genes.22 The virulence genes of the strain reported in this paper are tox R, RTX, hlyA, T3SS/T6SS, and lack of CTX gene, ace gene, and zot gene. Compared with O1 or O139, the virulence genes of V. cholerae are less, and the strain does not produce cholera toxin. Therefore, there is no obvious diarrhea in the patient. These results are consistent with the results of previous studies on NOVC.23,24
In this study, VC1115 was found to carry more virulence genes. The clinical symptom in the patient was a severe infection. Possible reasons are as follows: (i) The strain carries genes encoding type III secretory systems (T3SS) and type VI secretory systems (T6SS). Studies have found that T3SS/T6SS is widely distributed in multiple lineages of NOVC strains, which may enhance the virulence of NOVC strains lacking CT and TCP coding genes, thereby enhancing the pathogenic potential of this V. cholerae.25 (ii) This strain carries Hly A but lacks the Tag H gene. Studies26 have shown that Tag H negatively regulates the expression of Hly A at the transcriptional and translational levels. The absence of Tag H enhances the intestinal pathogenicity and extraintestinal invasiveness of V. cholerae, which mainly depends on the expression of Hly A.
Conclusions
The present study ultimately presents findings on the potential pathogenicity of Non-O1, Non-O139 V. cholerae, thereby providing valuable insights into the investigation of both the pathogenic and drug resistance mechanisms associated with V. cholerae. Furthermore, it substantiates that certain NOVCs, under harboring specific virulence genes, can induce severe extraintestinal infections and should be accorded due attention by clinicians. Consequently, regular surveillance of NOVC infections and the development of strategies to address such cases are imperative to potentially save lives.
Abbreviations
NOVC, non-O1 non-O139 V. cholerae; MIC, minimum inhibitory concentration; CT, cholera toxin; MALDI-TOF MS, matrix-assisted laser desorption ionization-time of flight mass spectrometry; ANI, average nucleotide identity; CDC, Centers for Disease Control; CLSI, the Clinical and Laboratory Standards Institute; KEGG, Kyoto Encyclopedia of Genes and Genomes; COG, Clusters of Orthologous Genes; VFDB, Virulence Factor Database; NCBI, National Center of Biotechnology Information; HBsAg, hepatitis B surface antigen; CT, computed tomography; T3SS, type III secretory systems; T6SS, type VI secretory systems.
Data Sharing Statement
Data associated with this study have been deposited in the NCBI database under the accession number: JARYMY010000000.
Compliance with Ethical Standards
The authors declare that they have compliance with ethical standards. This study has been reviewed and approved by the Research Ethics Committee of the Mianyang Traditional Chinese Medicine Hospital (NO. 2023-013). The patient provided informed consent to publish his case details.
Funding
This study was supported by the Health Commission of Mianyang City (202110).
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Al-Tawfiq JA, Chopra H, Dhama K, Sah R, Schlagenhauf P, Memish ZA. The cholera challenge: how should the world respond? New Microbes New Infect. 2023;51:101077. doi:10.1016/j.nmni.2022.101077
2. Baig MZ, Abdullah UH, Shafquat Y, Humayun KN, Zafar A. Non O1, non O139 Vibrio cholerae bacteraemia in an infant; case report and literature review. J Pak Med Assoc. 2018;68(4):650–652.
3. Shanley J, Kanj A, El Zein S, et al. Non-O1, non-O139 Vibrio cholerae bacteremia in an urban academic medical center in the United States. IDCases. 2019;15:e00527. doi:10.1016/j.idcr.2019.e00527
4. Sachu A, Johnson D, Thomas S, Mathew R. Non O1/ O139 Vibrio cholerae septicemia in a patient with hepatocellular carcinoma. Ethiop J Health Sci. 2021;31(6):1303–1306. doi:10.4314/ejhs.v31i6.27
5. Deshayes S, Daurel C, Cattoir V, Parienti JJ, Quilici ML, de La Blanchardière A. Non-O1, non-O139 Vibrio cholerae bacteraemia: case report and literature review. Springerplus. 2015;4:575. doi:10.1186/s40064-015-1346-3
6. Zhang X, Lu Y, Qian H, et al. Non-O1, non-O139 Vibrio cholerae (NOVC) bacteremia: case report and literature review, 2015–-2019. Infect Drug Resist. 2020;13:1009–1016. doi:10.2147/idr.S245806
7. Igere BE, Okoh AI, Nwodo UU. Non-serogroup O1/O139 agglutinable Vibrio cholerae: a phylogenetically and genealogically neglected yet emerging potential pathogen of clinical relevance. Arch Microbiol. 2022;204(6):323. doi:10.1007/s00203-022-02866-1
8. Ceccarelli D, Chen A, Hasan NA, Rashed SM, Huq A, Colwell RR. Non-O1/non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay, Maryland. Appl Environ Microbiol. 2015;81(6):1909–1918. doi:10.1128/aem.03540-14
9. Li F, Du P, Li B, et al. Distribution of virulence-associated genes and genetic relationships in non-O1/O139 Vibrio cholerae aquatic isolates from China. Appl Environ Microbiol. 2014;80(16):4987–4992. doi:10.1128/aem.01021-14
10. Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. In: CLSI Guideline M45.
11. Chen Y, Chen Y, Shi C, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7(1):1–6. doi:10.1093/gigascience/gix120
12. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi:10.1089/cmb.2012.0021
13. Li X, Wu Y, Sun X, et al. Non-O1/non-O139 Vibrio cholerae bacteraemia in mainland China from 2005 to 2019: clinical, epidemiological and genetic characteristics. Epidemiol Infect. 2020;148:e186. doi:10.1017/s0950268820001545
14. Gallardo-Cartagena JA, Chiappe-Gonzalez AJ, Astocondor-Salazar LM, et al. Vibrio cholerae NO-O1/NO-O139 bacteremia in a cirrhotic patient. First case report in Peru and literatura review. Rev Gastroenterol Peru. 2018;38(3):301–305.
15. Hwang S, Kim Y, Jung H, et al. A fatal case of bacteremia caused by Vibrio cholerae non-O1/O139. Infect Chemother. 2021;53(2):384–390. doi:10.3947/ic.2020.0301
16. Tsuruta K, Ueyama T, Watanabe T, Nakano K, Uno K, Fukushima H. Intensive care management of a patient with necrotizing fasciitis due to non-O1/O139 Vibrio cholerae after traveling to Taiwan: a case report. BMC Infect Dis. 2020;20(1):618. doi:10.1186/s12879-020-05343-6
17. Wang H, Yang C, Sun Z, et al. Genomic epidemiology of Vibrio cholerae reveals the regional and global spread of two epidemic non-toxigenic lineages. PLoS Negl Trop Dis. 2020;14(2):e0008046. doi:10.1371/journal.pntd.0008046
18. Wu Q, Vaziri AZ, Omidi N, et al. Antimicrobial resistance among clinical Vibrio cholerae non-O1/non-O139 isolates: systematic review and meta-analysis. Pathog Glob Health. 2023;117(3):235–244. doi:10.1080/20477724.2022.2114620
19. Bhandari M, Rathnayake IU, Huygens F, Jennison AV. Clinical and environmental vibrio cholerae non-O1, non-O139 strains from Australia have similar virulence and antimicrobial resistance gene profiles. Microbiol Spectr. 2023;11(1):e0263122. doi:10.1128/spectrum.02631-22
20. Dutta D, Chowdhury G, Pazhani GP, et al. Vibrio cholerae non-O1, non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg Infect Dis. 2013;19(3):464–467. doi:10.3201/eid1903.121156
21. Ding Y, Hao J, Zeng Z, Jinbo L. Identification and genomic analysis of a Vibrio cholerae strain isolated from a patient with bloodstream infection. Heliyon. 2022;8(11):e11572. doi:10.1016/j.heliyon.2022.e11572
22. Behera DR, Nayak AK, Nayak SR, et al. Genomic diversities of ctxB, tcpA and rstR alleles of Vibrio cholerae O139 strains isolated from Odisha, India. Environ Microbiol Rep. 2022;14(3):376–384. doi:10.1111/1758-2229.13016
23. Lepuschitz S, Baron S, Larvor E, et al. Phenotypic and genotypic antimicrobial resistance traits of Vibrio cholerae non-O1/non-O139 isolated from a large Austrian lake frequently associated with cases of human infection. Front Microbiol. 2019;10:2600. doi:10.3389/fmicb.2019.02600
24. Gao X, Miao Z, Li X, et al. Pathogenicity of non-O1/ O139 Vibrio cholerae and its induced immune response in Macrobrachium rosenbergii. Fish Shellfish Immunol. 2019;92:300–307. doi:10.1016/j.fsi.2019.06.032
25. Arteaga M, Velasco J, Rodriguez S, et al. Genomic characterization of the non-O1/non-O139 Vibrio cholerae strain that caused a gastroenteritis outbreak in Santiago, Chile, 2018. Microb Genom. 2020;6(3). doi:10.1099/mgen.0.000340
26. Wang G, Fan C, Wang H, et al. Type VI secretion system-associated FHA domain protein TagH regulates the hemolytic activity and virulence of Vibrio cholerae. Gut Microbes. 2022;14(1):2055440. doi:10.1080/19490976.2022.2055440
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

