Back to Journals » International Journal of General Medicine » Volume 14
Variations in TAP1 and PSMB9 Genes Involved in Antigen Processing and Presentation Increase the Risk of Vitiligo in the Saudi Community
Authors Mufti AH, AlJahdali IA, Elhawary NA, Ekram SN , Abumansour I, Sindi IA, Naffadi H, Elhawary EN, Alyamani NM, Alghamdi G, Alosaimi W, Rawas G, Alharbi A, Tayeb MT
Received 4 October 2021
Accepted for publication 3 December 2021
Published 19 December 2021 Volume 2021:14 Pages 10031—10044
DOI https://doi.org/10.2147/IJGM.S341079
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Ahmad H Mufti, 1 Imad A AlJahdali, 2 Nasser A Elhawary, 1 Samar N Ekram, 1 Iman Abumansour, 1 Ikhlas A Sindi, 3 Hind Naffadi, 4 Ezzeldin N Elhawary, 5 Najiah M Alyamani, 6 Ghydda Alghamdi, 1 Wafaa Alosaimi, 7 Ghufran Rawas, 8 Amaal Alharbi, 9 Mohammed T Tayeb 1
1Department of Medical Genetics, College of Medicine, Umm Al-Qura University, Mecca, 21955, Saudi Arabia; 2Department of Community Medicine, College of Medicine, Umm Al-Qura University, Mecca, 21955, Saudi Arabia; 3Department of Biotechnology, Faculty of Science, King Abdulaziz University, Jeddah, 21589, Saudi Arabia; 4Common Science, First Year Deanship, Umm Al-Qura University, Mecca, Saudi Arabia; 5MS Genomic Medicine Program, Faculty of Medicine, University of Southampton, Southampton General Hospital, Southampton, UK; 6Department of Biology, College of Science, University of Jeddah, Jeddah, Saudi Arabia; 7Department of Hematology, Maternity and Children Hospital, Mecca, Saudi Arabia; 8King Abdullah Medical City, Mecca, Saudi Arabia; 9Saudi Biobank National ProjectKing Abdullah International Medical Research Center, Jeddah, Saudi Arabia
Correspondence: Nasser A Elhawary
Department of Medical Genetics, Faculty of Medicine, Umm Al-Qura University, P.O. Box 57543, Mecca, 21955, Saudi Arabia
Tel +966 55 369 2180
Email [email protected]
Background: The antigen processing 1 (TAP1) and proteasome 20S subunit beta 9 (PSMB9) genes are associated with strong susceptibility to many autoimmune diseases. Here, we explored whether TAP1/PSMB9 genetic variants, individually or combined, affected susceptibility to the complex, autoimmune-based skin disorder vitiligo.
Methods: Samples of genomic DNA from buccal cells of 172 patients with vitiligo and 129 healthy controls were analyzed using TaqMan™ genotyping assays for the TAP1 rs1135216 (A>G) and PSMB9 rs17587 (A>G) single nucleotide polymorphisms (SNPs). SNPStats software (https://www.snpstats.net) was utilized to choose the best interactive inheritance mode for selected SNPs.
Results: The genotype frequencies for the TAP1 rs1135216 and PSMB9 rs17587 SNPs were in Hardy–Weinberg equilibrium for cases (P= 0.11 and P= 0.10, respectively) but not for controls (P< 0.05). The TAP1 rs1135216 (D637G) and PSMB9 rs17587 (R60H) SNPs increased the risk of vitiligo four-fold and two-fold, respectively (odds ratio [OR]= 4.6; 95% confidence interval [CI], 3.2– 6.5; P< 0.0001 and OR= 2.2; 95% CI, 1.5– 3.1; P< 0.0001). The recessive model (G/G-D/G versus D/D) and the codominant model (R/R versus R/H) were the best models of inheritance for the rs113526 and rs17587 SNPs, respectively (OR= 16.4; 95% CI, 2.0– 138; P= 0.0006 and OR= 1.7; 95% CI, 0.3– 1.8; P= 0.013). Vulgaris, focal vulgaris, and acryl/acrofacial were the most common vitiligo subtypes in our sample (51%, 21%, and 19%, respectively). Heterozygous rs113526 (637D/G) and rs17587 (60R/H) were the most common genotypes in most vitiligo subtypes. The heterozygous 637D/G genotype and the 637G variant allele were significantly more common in patients with active disease than in patients with stable disease (P= 0.000052 and P= 0.0063, respectively).
Conclusion: Our findings suggest a crucial role for TAP1 rs1135216 and PSMB9 rs17587 in the risk and progression of vitiligo in the Saudi community. Genomic analyses are needed to identify more candidate genes and more genetic variants associated with vitiligo.
Keywords: vitiligo, TAP1/PSMB9, single nucleotide polymorphisms, TaqMan genotyping, linkage disequilibrium
Plain Language Summary
Vitiligo is characterized by skin depigmentation due to a lack of melanocytes, which are cells that produce melanin deep inside the skin’s outer layer (ie, the epidermis). Initial patchy loss of skin color occurs on the hands, forearms, faces, and areas around body openings and genitalia. Sometimes hair on the scalp, eyelashes, and eyebrows prematurely gray. Most patients develop symptoms between 10 and 30 years of age, but the first lesions are typically observed at approximately 20 years. Multiple genes involved in the pathogenesis of vitiligo have been identified in different cultures and among people of different ethnicities. Small differences in these genes and how they interact could play a considerable role in complex disorders such as vitiligo. In the current study, the authors studied two genes that are associated with autoimmune disorders to see if small differences in the genes affected risk for vitiligo in the Saudi community.
Introduction
Vitiligo, a rare polygenic skin disorder with a non-Mendelian pattern of inheritance, is characterized by depigmented circumscribed macules or lesions supported in the underlying epidermal layers of the skin.1 Initial patchy loss of skin color targets hands, forearms, faces, and the areas around body openings and genitalia. Sometimes premature graying of hair also occurs on the scalp, eyelashes, and eyebrows. Most patients develop symptoms between 10 and 30 years of age,2 but the first lesions are typically observed at approximately 20 years.3
In-depth surveys have estimated the population prevalence of vitiligo to be 0.5–2.28% in both adults and children worldwide.1,3–9 Some studies have found high rates as high as 5–9% in diverse ethnic populations.10−12 In Saudi Arabia, vitiligo affects about 2.5% of the Saudi population and represents between 5% and 7% of skin disorders seen in the hospital setting.13–15 The prevalence of vitiligo among female school children in the Eastern Saudi province has been identified as 0.4%.16
Oxidative stress, autoimmunity, and neurochemistry have been hypothesized to explain the pathophysiology behind the development of vitiligo.17–20 Thirty percent of vitiligo cases have been strongly categorized as concomitant autoimmune disorders.17,19 Several comorbidities, including autoimmune thyroid disease, systemic lupus erythematosus, type 1 diabetes, Sjögren’s syndrome, juvenile rheumatoid arthritis, multiple sclerosis, celiac disease, and Addison’s disease have also been reported in association with vitiligo.21–28
Selection of candidate genes is always challenging, especially in polygenic and immune-associated disorders in which exposure to environmental risk factors and gene-gene interactions are involved. VitiVar, a compendium of genes and variants associated with vitiligo (http://vitivar.igib.res.in/), included 322 candidate genes and 254 genetic variations reported in 202 research articles as of late 2021. Genes involved in antigen processing and presentation, such as transporter associated with antigen processing 1 (TAP1) and proteasome 20S subunit beta 9 (PSMB9), have been shown to play potential roles in several autoimmune diseases, including vitiligo,29–36 and are included in VitiVar. The membrane-associated protein encoded by the TAP1 gene (MIM #170260) is a member of the superfamily of ATP-binding cassette (ABC) transporters. The TAP1 protein assembles with the TAP2 protein, forming a TAP complex. This protein dimer transports degraded cytosolic peptides from foreign invaders into the endoplasmic reticulum, where the peptides are attached to major histocompatibility complex (MHC) class I proteins. The peptide-bound MHC class I proteins are then moved to the cell’s surface so that T-cells can interact with them and launch an immune response.37 PSMB9 (MIM #177045) is a multicatalytic complex protein that degrades damaged or unneeded proteins by breaking peptide bonds. Zaiss et al38 have reported that immunoproteasome subunits protect against the development of CD8+ T cell-mediated autoimmune diseases. The TAP1 and PSMB9 genes are linked to macromolecular catabolic processes associated with proteasomes responsible for degrading short-lived cytoplasmic proteins into peptides.39,40 Because TAP1 and PSMB9 genes may thus restrict antigen processing and presentation, they are attractive candidate genes for increasing susceptibility to vitiligo.
Although much genetic information is available on vitiligo, most reports are from Caucasian and South Asian populations. Only a small number of research articles and reviews have discussed the role of genetics in vitiligo among Saudi populations.41–51 Although Tayeb51 reported that TAP1 (C>T intron 7) and PSMB8 (G>T intron 6) were not significantly associated with vitiligo in the Saudi community, more in-depth studies are needed at these loci to explore possible associations. Hence, the present study investigated associations between vitiligo and the TAP1 rs1135216 A/G (D637G) and PSMB9 rs17587 A/G (R60H) variant alleles and evaluated the influence of various genotypes on the risk and severity of disease.
Patients and Methods
Study Population
The study population included 172 individuals diagnosed with vitiligo referred from dermatology divisions in governmental hospitals in Saudi Western governorates (eg, Jeddah, Mecca, Al-Hada, and Taif). The clinical diagnosis of vitiligo was based on characteristic skin depigmentation with typical localization as detected under a Wood’s lamp and confirmed by dermatologists. Patients had no other associated autoimmune diseases. The demographic and clinical characteristics collected included gender, age of examination, clinical history of both patients and their relatives, consanguineous status, history of other autoimmune disorders, previous treatment, and types of vitiligo lesions. Vitiligo lesions were classified as follows:1,31 focal (one or more maculae in a non-segmented pattern), vulgaris (a symmetric or asymmetric distribution of maculae in one or more areas), segmental (unilateral depigmented macules that do not cross the midlines), acral/acrofacial (depigmentation on tips of fingers and toes, on anogenital areas, on lips, and around the eyes), and universalis (complete or >80% skin depigmentation). These vitiligo subtypes were classified as active (ie, progressive) or stable. Participants with congenital non-progressive depigmentation or depigmentation due to melanotoxic chemicals; lupus erythematosus; or postinfectious, postinflammatory, or post-traumatic localized depigmentation were excluded from the study. Those exposed to any therapy in the past six months were also excluded. A group of healthy controls (n = 129) with no clinical evidence of vitiligo, no family history of the disease, and no other autoimmune disorders were also included in the study (Figure 1).
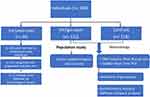 |
Figure 1 Participant eligibility and applied methodology. |
Molecular analyses were performed at the DNA Diagnostic Laboratories, Medical Genetics Department, College of Medicine, Umm Al-Qura University. This study was conducted under the Declaration of Helsinki. The Institutional Biomedical Ethics Committee-Umm Al-Qura University (#HAPO-02-K-012), based on the National Committee of Medical and Bioethics (King Abdulaziz City for Science and Technology-Riyadh), approved the study. All individuals in this study gave their written informed consent before participating in the study.
TaqMan Genotyping Analysis
Genomic DNA samples were isolated from buccal cells by gently scraping the mucosa using an Oragene.DNA-OGR-575 kit (DNA Genotek Inc., Ottawa, ON, Canada) following some modifications.52 We implemented TaqMan real-time PCR (Thermo, Applied Biosystems, USA) to genotype individuals for the TAP1 rs1135216ʹ D637G’ (C____531909_20) and PSMB9 rs17587 “R60H” (C___8849004_1_) single nucleotide polymorphisms (SNPs) using a 7500 Fast-Dx Real-Time PCR System (ThermoFisher Scientific, USA) with the transcript accessions and protein IDs NM_000593.5 (NP_000584.2) and NM_002800.4 (NP_002791.1) on GRCH38. The probe sequences were 5`-CCC TGA CAG CTG GCT CCC AGC CTC G[C/T]C TAC CTC TGC AGA GCA AAG GGC CAA-3` for TAP1 rs1135216 and 5`-GAC AAG CTG TCC CCG CTG CAC GAG C[A/G]C ATC TAC TGT GCA CTC TCT GGT TCA-3` for PSMB9 rs17587. We used VIC/FAM dyes as the reporter/quencher probes. For the sake of genotyping accuracy, all DNA samples and negative controls were included in the assays. The genotypes of all samples were reassessed in duplicate. To confirm our results, some suspected PCR products were purified using the Agencourt AMPure XP kit (Beckman Coulter, Canada) and genotyped with the Genetic Analyzer 3500 (Thermo Fisher Inc., USA).
Bioinformatics Analysis
For the nonsynonymous TAP1 rs1135216 (D637G) and PSMB9 rs17587 (R60H) variants, we used the in silico tools Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping v2 (PolyPhen2), MutationTaster, Functional Analysis through Hidden Markov Models (FATHMM), Mutation Assessor, and loss-of-function tool (LoFtool) to predict the effects of the SNPs on their corresponding functional proteins (Ensemble Variant Effect Predictor; https://www.ensembl.org/vep/).
TAP1-PSMB9 Protein Interaction
The Search Tool for Retrieval of Interacting Genes (STRING) database53 (https://string-db.org) was also used to predict functional interactions between proteins.
Statistical Analysis
Consistency with Hardy-Weinberg equilibrium (HWE) was evaluated at the TAP1 rs1135216 A/G and PSMB9 rs17587 A/G loci by comparing the observed and expected genotypes in cases and controls using chi-squared (χ2) analysis. A P-value < 0.05 was considered a departure from HWE. We considered interactive models of inheritance— codominant, dominant, recessive, overdominant, and additive—using SNPstats software (https://www.snpstats.net). A low Akaike information criterion (AIC) value corresponding to minimum expected entropy was adopted to assess the best inheritance model. Logistic regression of genotype distributions was adjusted by age. Bonferroni correction was performed for multiple tests in genotype analyses of the examined SNPs. The statistical significance of linkage disequilibrium between the polymorphic markers was calculated based on the coefficient of linkage disequilibrium (D`) and the correlation coefficient between pairs of loci (r). Logistic regressions for genotypic distributions of cases and controls were measured in odds ratios (ORs) and 95% confidence intervals (CIs). The paired t-test and one-way ANOVA in chi-square (χ2) values were applied to evaluate demographic and clinical characteristics of cases using MedCalc (https://www.medcalc.org) and Social Science Statistics (https://www.socscistatistics.com/tests/chisquare2/default2.aspx) software. We used G*Power software (https://gpower.software.informer.com/3.1.9.2/) to estimate the required sample sizes of cases and controls to get a power 80% using the priori/post hoc tests in terms of z-test, and 0.05 of α error probability.
Results
Clinico-Epidemiologic Profile of the Population
Among the 172 cases of vitiligo, the female-to-male ratio was 1:1. No significant differences (P < 0.05) were found between cases and controls regarding age or gender. Among vitiligo cases, the mean age at onset was 11.5 years (range, 2–47 years) and mean age at examination was 22 years (range, 4–50 years). When examined by age intervals, the mean age of onset was 8.3 years (standard deviation [SD], 4.7) in children aged 2–10 years, 18.0 years (SD, 5.76) in adolescents aged 11–20 years, and 10.1 years (SD, 3.56) in adults aged > 21 years or older (Figure 2A). Twenty-four (16.3%) of the 172 cases were consanguineous (Figure 2B). In terms of phenotype, more cases of vitiligo were active (ie, progressive) (66.7%; P < 0.0001) than stable (33.3%; P < 0.0001). The patches of depigmentation were sensitive to the sun in 50% of cases (z = 27.1; P < 0.0001). A low percentage of individuals with vitiligo also had diabetes mellitus type 1 (5.8%; z = 0.5, P = 0.63) or hypothyroidism (16.3%; z = 6.8, P < 0.0001).
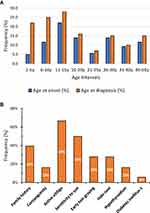 |
Figure 2 Clinico-epidemiologic profile of vitiligo cases. (A) Frequencies of age at onset and age at examination by age interval. (B) Frequencies of demographic and clinical characteristics. |
HWE and Power Analysis
The observed genotype frequencies were consistent with HWE at the rs1135216 and rs17587 genetic loci in cases (χ2 = 2.61; P = 0.11 and χ2 = 2.8; P = 0.10, respectively) but deviated from HWE at rs1135216 (P < 0.05) and rs17587 (P = 0.05) in controls. The statistical priori power analysis found that a power of 80.6% would require 33 cases and 26 controls for the TAP1 rs1135216 SNP, and that a power of 80.0% would require 151 cases and 116 controls for the PSMB9 rs17587 SNP. The post hoc statistical analysis for the TAP1 rs1135216 and rs17587 SNPs revealed actual powers of 99.9% and 84.5%, respectively, among our 301 participants (172 cases and 129 controls).
Allele Frequencies of TAP1 and PSMB9 Loci
Table 1 shows the allele frequencies of the TAP1 rs1135216 and PSMB9 rs17587 polymorphic loci. The TAP1 637G and PSMB9 60H variant alleles were significantly more frequent in cases than controls (73% versus 37% and 39.5% versus 23.3%, respectively). The two SNPs were also associated with four-fold and two-fold increased risks for vitiligo, respectively; the ORs of the allelic variants were 4.6 (95% CI, 3.2–6.5; z = 8.6; P < 0.0001) for TAP1 rs1135216 (D637G) and 2.2 (95% CI, 1.5–3.1; z = 4.2; P < 0.0001) for PSMB9 rs17587 (R60H).
 |
Table 1 Allele Frequencies and Genotype Distributions of TAP1 rs1135216 A>G (D637G) and PSMB9 rs17587 G>A (R60H) Polymorphic Loci in Vitiligo |
Genotypic Distributions of TAP1 and PSMB9 Loci
Using the SNPStats software (https://www.snpstats.net), the recessive model (G/G-D/G versus D/D) and the codominant model (R/R versus R/H) were the best models of inheritance (OR = 16.4; 95% CI, 2.0–138; P = 0.0006 for rs113526 and OR = 1.7; 95% CI, 0.3–1.8; P = 0.013 for PSMB9 rs17587) based on AIC minimum values (Table 1). The 637D/D wild genotype was more common in controls than cases (25.6% versus 2.3%). Also, no controls carried the 637G/G genotype.
Effect of Examined Loci on the Phenotypes of Vitiligo
Vitiligo vulgaris (VV) was the most frequent vitiligo subtype (51%) in our case group, followed by focal vitiligo (FV) (21%) and acral/acrofacial vitiligo (AV) (19%) (Figure 3). Segmented vitiligo (SV) and universalis vitiligo (UV) were the least common (2% and 7%, respectively). Figure 4 presents the frequencies of specific TAP1 (D637G) and PSMB9 (R60H) genotypes in vitiligo patients with different clinical subtypes. Within PSMB9 rs17587 A/G, the heterozygous 60R/H genotype was the most frequent among the subtypes: 59% (VV), 38% (UV), 56% (FV), 50% (AV), and 50% (SV). Within TAP1 rs1135216 A/G, the heterozygous 637D/G genotype was the most frequent: 54% (VV), 67% (UV), 59% (AV), and 75% (SV). The most frequent genotype in any one of the subtype groups was 637G/G within TAP1 rs1135216 A/G, present in more than three-quarters of cases with the FV subtype.
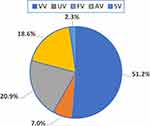 |
Figure 3 Frequencies of vitiligo phenotypes in cases. Abbreviations: AV, acral/acrofacial vitiligo; FV, focal vitiligo; SV, segmental vitiligo; UV, universalis vitiligo; VV, vulgaris vitiligo. |
Association of Genetic Loci with Vitiligo Progression
Table 2 highlights the frequencies of the TAP1 (D637G) and PSMB9 (R60H) genotypes in patients with active versus stable phenotypes. The heterozygous 637D/G genotype was significantly more common in patients with active disease than those with stable disease (64% versus 29%; χ2 = 19.7; P = 0.000052). Hence, the 637G variant allele was also more predominant in cases with active disease than those with stable disease (OR = 2.1; 95% CI, 1.2 3.4; z = 2.7; P = 0.0063). No statistically significant differences were found in the genotype distributions and allele frequencies of the PSMB9 R60H locus in active versus stable phenotypes (χ2 = 0.969 P = 0.616 and OR = 1.0; 95% CI, 0.7–1.6; z = 0.17; P = 0.86).
 |
Table 2 Genotype Distributions and Allelic Frequencies of the Examined TAP1/PSMB9 Genetic Loci for Active versus Stable Vitiligo Phenotypes |
Haplotype Association and Linkage Disequilibrium
Among the four possible haplotypes of the TAP1 rs1135216 (D637G) and PSMB9 rs17587 (R60H) loci, the 637D-60H haplotype had an overall frequency of 8.81% (OR = 0.05; 95% CI, 0.00–1.33; P = 0.0004) (Table 3). The global haplotype analysis also showed that this haplotype was significantly more common than the other haplotypes when adjusted by age (P < 0.0001). In addition, there was strong linkage disequilibrium between the TAP1 rs1135216 A/G (D637G) and PSMB9 rs17587 A/G (R60H) loci (D` = 0.3959; r = ‒0.2476; P = 0.0012).
 |
Table 3 Haplotype Association Analysis of the TAP1 rs1135216 D637G and PSMB9 rs17587 R60H SNPs (Adjusted by Age) |
Bioinformatics and Functional Data
Table 4 shows the predicted functional consequences of the TAP1 rs1135216 (D637G) and PSMB9 rs17587 (R60H) loci (https://www.ensembl.org/vep). The missense TAP1 rs1135216 SNP (c.1978A/G; p.D637G) was found to have damaging effects on the functional protein according to FATHMM (score, −2.37) and LoFtool (score, 0.0865), though it was found to have neutral effects according to Mutation Assessor (score, 1.275) and MutationTaster (score, 1.36×10−36). All the examined tools predicted the missense PSMB9 rs17587 SNP (c.217G/C; p.R60H) to have benign and tolerant effects.
 |
Table 4 In silico Functional Predictions of Non-Synonymous SNPs |
Protein–Protein Interaction Network Analysis
Figure 5 presents the protein-network interaction of the TAP1-PSMB9 protein predicted with STRING software. The TAP1-PSMB9 protein network showed significantly more interactions (P-value = 0.000538) than would be expected for a random set of proteins of the same size and degree of distribution drawn from the genome.
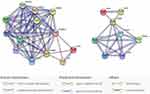 |
Figure 5 Gene-network interactions contained the TAP1 and PSMB9 genes examined in this study (right side) created with STRING (https://string-db.org/), in which there are strong interactions between the TAP1/TAP2 genes and PSMB9 gene. On the Left side: More extended genes contained PSMB and PSMA gene families are shown. The TAP1 gene co-occurred the PSMB10 gene, while the PSMB9 gene considerably interacted and co-expressed with neighboring PSMB and PSMA gene families. Each node represents all the proteins produced by a single, protein-coding gene locus. Colored nodes describe proteins and the first shell of interactors. Edges represent protein–protein associations that are meant to be specific and meaningful, ie, proteins jointly contribute to a shared function; this does not necessarily mean they are physically binding each other. |
Discussion
This case-control study explored common SNPs in the TAP1 rs1135216 A>G (D637G) and PSMB9 rs17587 A>G (R60H) loci as candidate polymorphic biomarkers for vitiligo. Overall, our results showed that the rs1135216 and rs17587 SNPs were associated with four-fold and two-fold increased risks of vitiligo, respectively. In the study, the genotypic distributions were significantly different under recessive and codominant models for the rs1135216 and rs17587 SNPs, respectively.
In Caucasians, Casp et al31 previously reported a significant association between vitiligo and TAP1 rs1135216 (P = 0.0034) but not between vitiligo and PSMB9 rs17587 (P = 0.11). In contrast, a study of 378 cases of vitiligo and 509 healthy controls in an Indian population found no association between TAP1 rs1135216 and either generalized or active vitiligo (P > 0.05 each), but did find associations between PSMB8 rs2071464 and both vitiligo phenotypes (P = 0.019 and P = 0.005, respectively).54 In a more recent study, Dani et al55 found a strongly significant role for the PSMB9 rs17587 polymorphic locus in 1050 generalized and 270 localized cases of vitiligo in Northern Indians.
Our results showed an overrepresentation of VV in the study population (51.2%). This pattern is similar to that reported in most ethnic populations, in which the prevalence of the VV subtype ranges from 50–60%.56–60 It has been reported that VV and other forms of nonsegmental vitiligo (eg, acrofacial, mucosal, generalized, universal) can develop at any age, but it most often occurs in young people between the ages of 10 and 30 years.61–63 In our study, the presence of VV was strongly associated with variant alleles of the rs1135216 and rs17587 SNPs we studied. There were no significant associations between the progression of vitiligo (ie, activity, stability) and the D637G or R60H polymorphic marker (P = 0.83 and P = 0.75, respectively). The 637G variant allele was dominant in cases with progression (P = 0.0063), while the 60H variant allele was not dominant in cases with active patches (P > 0.05).
An earlier study described an association between vitiligo and insulin-dependent type 1 diabetes mellitus (T1DM’), but not between vitiligo and non-insulin-dependent type 2 diabetes mellitus (T2DM).64,65 Among our sample group, the prevalence of cases with T1DM (10/172 cases; 5.8%) was higher than that reported in Caucasians,17 Turkish individuals,66 and Jordanians.67 The higher frequency of co-occurring vitiligo and T1DM in our study might be due to a higher incidence of T1DM in Saudi Arabia or to the interaction of multiple genes affecting both vitiligo and diabetes.
In some instances, individuals with vitiligo develop other autoimmune conditions, particularly those affecting the thyroid.68 The frequency of autoimmune thyroid disease in patients with vitiligo has been estimated to be 14%, and the risk of developing this type of disease has been reported to be 2.5-fold higher for vitiligo patients than for individuals without vitiligo.68 The prevalence of thyroid pathology in our cases (16.3%, P < 0.0001) was slightly higher than that reported in Turkish (4.4%)66 or Chinese (6.8%)69 vitiligo patients. In a Caucasian population, 5.7% of first-degree relatives of individuals with vitiligo were reported as having clinical autoimmune thyroid disease, which was more than twice the population frequency (P < 0.001).17
Several autoimmune disorders, including cutaneous disorders such as vitiligo and halo nevi, have been reported to affect female and male individuals at similar rates.70 No gender differences were seen among vitiligo cases in our study, as vitiligo affected both sexes equally. The equity of vitiligo in male and female individuals could be rationalized in some ethnic, regional populations such as Indians and Northern Chinese.3 Girls and women often seek clinical counseling more frequently and experience more social bullying than boys and men.56 In contrast, among 3280 individuals with vitiligo in a US population, women represented 57.6% of the cases, compared with men representing 42.4%.25
Consanguinity is present in approximately 10% of the global population.71 In the Saudi community, nearly 54% of all marriages are consanguineous (between cousins).72,73 The actual degree of inbreeding is further complicated by the high frequency of marriages occurring within a tribe or caste. Our results showed a 16.3% prevalence of consanguineous relationships (24 of 172 cases) associated with a 39.5% (68/172 cases) positive family history of vitiligo. Another study from the Northern region of Saudi Arabia (Arar city) recorded high rates of consanguinity among vitiligo patients (65%, 45/69 cases).48 Despite the relatively low prevalence of vitiligo in Saudi Arabia, estimated at 0.4–2.5%,13,16,74 the disease is linked to psychosocial burden, as 45–57% of vitiligo patients experience depression.46,75,76
Although age of onset is variable, most patients develop symptoms between 10 and 30 years of age.2 The mean age of onset in our study—11.5 years (range, 2–47 years)—is consistent with a previous report by Alzolibani.77 However, most populations have reported a later onset, with a mean of at least 21 years.17,31,66,67 The age of onset in our Saudi sample group was higher than that reported in American (White and Hispanic/Latino),25 and Northern Indian60 populations. In 47 unrelated Egyptian individuals with vitiligo, ages 2–18 years, our research team also reported an earlier mean age of onset (5.9 years).78 In addition, in a recent cohort study including 220 Egyptian children, El-Husseiny et al79 reported a mean age of onset of 6.18 years (childhood group, < 12 years).
Study Limitations
Our study had several limitations. First, patchy lesions are more obvious in darker-skinned individuals, so age of onset is somewhat lower in these ethnic populations than in our study. Also, the inconsistent prevalence reported in the literature may be due to higher reporting in regions where social and cultural stigma is common.61 Second, our study was limited to only two polymorphic biomarkers whose association with vitiligo risk has been debated in the literature. However, conflicting molecular data regarding candidate genes and some rare variants associated with this autoimmune disease are not uncommon. Because reported inclusion/exclusion criteria have been broad in studies among different ethnic populations, replication of the results of many of these studies would be challenging. Genome-wide linkage analyses in multiplex families have identified several linkage signals for the TAP and PSMB genes as candidate genes.35,80–83 Therefore, we have launched an exome sequencing analysis to identify more candidate genes and rare variants to help us better understand this complex disease.
Conclusion
The associations of the TAP1 rs1135216 D637G and PSMB9 rs17587 R60H polymorphisms with vitiligo support evidence of the defective antigen processing, which potentially influence the immune cells targeting melanocytes. Heterozygosity in TAP1 (637D/G) genotype and the minor allele 637G have increased risk with progressive (active) phenotype than those with stable one in Saudi vitiligo cases. However, further studies need to confirm these functional outcomes. Identifying these new vitiligo susceptibility genes in the Saudi population has opened new avenues for exploring the underlying disease mechanisms for vitiligo. More linkage studies based on a whole-exome approach, rather than a single-gene approach or a focus on a few candidate genes, would be useful for discovering additional genes and variants associated with susceptibility to vitiligo.
Data Sharing Statement
Data sets analyzed during this study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
Written informed consent was obtained from all study participants enrolled in this study, approved by the Institutional Biomedical Ethics Committee of Umm Al-Qura University (reference #HAPO-02-K-012), licensed from the National Committee Medical and Bioethics, KACST (http://bioethics.kacst.edu.sa/About.aspx?lang=en-US).
Consent for Publication
All study participants provided written informed consent to publish the results.
Acknowledgments
The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University, Mecca-Saudi Arabia, for supporting this work through grant 19-Med-1-01-0033. We also thank the dermatology therapists and sympathetic nurses.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Funding
This work was funded by the Deanship of Scientific Research, Umm Al-Qura University, Mecca, Saudi Arabia, under grant 19-MED-1-01-0033.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Krüger C, Schallreuter KU. A review of the worldwide prevalence of vitiligo in children/adolescents and adults. Int J Dermatol. 2012;51(10):1206–1212. doi:10.1111/j.1365-4632.2011.05377.x
2. Mosher DB, Fitzpatrick TB, Ortanne JB. Disorders of pigmentation, hipomelanoses and hypermelanoses. In: Freedberg IM, Eisen AZ, Fitzpatrick TB, eds. Dermatology in General Medicine. New York, NY: McGraw-Hill; 1999:936–945.
3. Alikhan A, Felsten LM, Daly M, Petronic-Rosic V. Vitiligo: a comprehensive overview Part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J Am Acad Dermatol. 2011;65(3):473–491. doi:10.1016/j.jaad.2010.11.061
4. Boisseau-Garsaud AM, Garsaud P, Cales-Quist D, Helenon R, Queneherve C, Claire RC. Epidemiology of vitiligo in the French West Indies (Isle of Martinique). Int J Dermatol. 2000;39(1):18–20. doi:10.1046/j.1365-4362.2000.00880.x
5. Stromberg S, Bjorklund MG, Asplund A, et al. Transcriptional profiling of melanocytes from patients with vitiligo vulgaris. Pigment Cell Melanoma Res. 2008;21(2):162–171. doi:10.1111/j.1755-148X.2007.00429
6. Ezzedine K, Eleftheriadou V, Whitton M, van Geel N. Vitiligo. Lancet. 2015;386(9988):74–84. doi:10.1016/S0140-6736(14)60763-7
7. Zhang Y, Cai Y, Shi M, et al. The prevalence of vitiligo: a meta-analysis. PLoS One. 2016;11(9):e0163806. doi:10.1371/journal.pone.0163806
8. Gianfaldoni S, Wollina U, Tchernev G, Lotti J, Franca K, Lotti T. Vitiligo in children: a review of conventional treatments. Open Access Maced J Med Sci. 2018;6(1):213–217. doi:10.3889/oamjms.2018.054
9. Bergqvist C, Ezzedine K. Vitiligo: a review. Dermatology. 2020;236(6):571–592. doi:10.1159/000506103
10. van den Boorn JG, Konijnenberg D, Dellemijn TA, et al. Autoimmune destruction of skin melanocytes by perilesional T cells from vitiligo patients. J Invest Dermatol. 2009;129(9):2220–2232. doi:10.1038/jid.2009.32
11. Richmond JM, Strassner JP, Zapata L
12. Singh M, Mansuri MS, Jadeja SD, Marfatia YS, Begum R. Association of interleukin 1 receptor antagonist intron 2 variable number of tandem repeats polymorphism with vitiligo susceptibility in Gujarat population. Indian J Dermatol Venereol Leprol. 2018;84(3):285–291. doi:10.4103/ijdvl.IJDVL_1_17
13. Jarallah JS, Al-Sheikh OA, El-Shabrawy M, Al-Wakeel MA. Vitiligo: epidemiology and clinical pattern at King Khalid University Hospital. Ann Saudi Med. 1993;13(4):332–334. doi:10.5144/0256-4947.1993.332
14. Shelleh HH, Al-Hatiti HS. Pattern of skin diseases in a hospital in southwestern Saudi Arabia. Saudi Med J. 2004;25(4):507–510.
15. Alakloby OM. Pattern of skin diseases in Eastern Saudi Arabia. Saudi Med J. 2005;26(10):1607–1610.
16. Al-Saeed WY, Al-Dawood KM, Bukhari IA, Bahnassy AA. Prevalence and pattern of skin disorders among female schoolchildren in Eastern Saudi Arabia. Saudi Med J. 2006;27(2):227–234.
17. Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16(3):208–214. doi:10.1034/j.1600-0749.2003.00032.x
18. Shajil EM, Chatterjee S, Agrawal D, Bagchi T, Begum R. Vitiligo: pathomechanisms and genetic polymorphism of susceptible genes. Ind J Exp Biol. 2006;44:52639.
19. Laddha NC, Dwivedi M, Mansuri MS, et al. Association of neuropeptide Y (NPY), interleukin-1B (IL1B) genetic variants and correlation of IL1B transcript levels with vitiligo susceptibility. PLoS One. 2014;9(9):e107020. doi:10.1371/journal.pone.0107020
20. Praest P, Luteijn RD, Brak-Boer IGJ, et al. The influence of TAP1 and TAP2 gene polymorphisms on TAP function and its inhibition by viral immune evasion proteins. Mol Immunol. 2018;101:55–64. doi:10.1016/j.molimm.2018.05.025
21. Powis SH, Tonks S, Mockridge I, Kelly AP, Bodmer JG, Trowsdale J. Alleles and haplotypes of the MHC-encoded ABC transporters TAP1 and TAP2. Immunogenetics. 1993;37(5):373–380. doi:10.1007/BF00216802
22. Polvi A, Maki M, Partanen J. Celiac patients predominantly inherit HLA-DPB1*0101 positive haplotype from HLA-DQ2 homozygous parent. Hum Immunol. 1997;53(2):156–158. doi:10.1016/S0198-8859(97)00027-X
23. Kostik MM, Klyushina AA, Moskalenko MV, Scheplyagina LA, Larionova VI. Glucocorticoid receptor gene polymorphism and juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2011;9(1):2. doi:10.1186/1546-0096-9-2
24. Sainaghi PP, Collimedaglia L, Alciato F, et al. Growth arrest specific gene 6 protein concentration in cerebrospinal fluid correlates with relapse severity in multiple sclerosis. Mediators Inflamm. 2013;2013:406483. doi:10.1155/2013/406483
25. Sheth VM, Guo Y, Qureshi AA. Comorbidities associated with vitiligo: a ten-year retrospective study. Dermatology. 2013;227(4):311–315. doi:10.1159/000354607
26. Colucci R, Lotti F, Dragoni F, et al. High prevalence of circulating autoantibodies against thyroid hormones in vitiligo and correlation with clinical and historical parameters of patients. Br J Dermatol. 2014;171(4):786–798. doi:10.1111/bjd.13286
27. Kivity S, Arango MT, Ehrenfeld M, et al. Infection and autoimmunity in Sjogren’s syndrome: a clinical study and comprehensive review. J Autoimmun. 2014;51:17–22. doi:10.1016/j.jaut.2014.02.008
28. Kronenberg-Versteeg D, Eichmann M, Russell MA, et al. Molecular pathways for immune recognition of preproinsulin signal peptide in type 1 diabetes. Diabetes. 2018;67(4):687–696. doi:10.2337/db17-0021
29. Djilali-Saiah I, Caillat-Zucman S, Schmitz J, Chaves-Vieira ML, Bach JF. Polymorphism of antigen processing (TAP, LMP) and HLA class II genes in celiac disease. Hum Immunol. 1994;40(1):8–16. doi:10.1016/0198-8859(94)90015-9
30. Teisserenc H, Schmitt W, Blake N, et al. A case of primary immunodeficiency due to a defect of the major histocompatibility gene complex class I processing and presentation pathway. Immunol Lett. 1997;57(1–3):183–187. doi:10.1016/s0165-2478(97)00072-2
31. Casp CB, She JX, McCormack WT. Genes of the LMP/TAP cluster are associated with the human autoimmune disease vitiligo. Genes Immun. 2003;4(7):492–499. doi:10.1038/sj.gene.6364016
32. Kramer U, Illig T, Grune T, Krutmann J, Esser C. Strong associations of psoriasis with antigen processing LMP and transport genes TAP differ by gender and phenotype. Genes Immun. 2007;8(6):513–517. doi:10.1038/sj.gene.6364404
33. Singh A, Sharma P, Kar HK, et al. HLA alleles and amino-acid signatures of the peptide-binding pockets of HLA molecules in vitiligo. J Invest Dermatol. 2012;132(1):124–134. doi:10.1038/jid.2011.240
34. Birlea SA, Ahmad FJ, Uddin RM, et al. Association of generalized vitiligo with MHC class II loci in patients from the Indian subcontinent. J Invest Dermatol. 2013;133(5):1369–1372. doi:10.1038/jid.2012.501
35. Jin Y, Andersen G, Yorgov D, et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet. 2016;48(11):1418–1424. doi:10.1038/ng.3680
36. Dey-Rao R, Sinha AA. Vitiligo blood transcriptomics provides new insights into disease mechanisms and identifies potential novel therapeutic targets. BMC Genomics. 2017;18(1):109. doi:10.1186/s12864-017-3510-3
37. Frisoli ML, Essien K, Harris JE. Vitiligo: mechanisms of pathogenesis and treatment. Annu Rev Immunol. 2020;38:621–648. doi:10.1146/annurev-immunol-100919-023531
38. Zaiss DM, Bekker CP, Grone A, Lie BA, Sijts AJ. Proteasome immunosubunits protect against the development of CD8 T cell-mediated autoimmune diseases. J Immunol. 2011;187(5):2302–2309. doi:10.4049/jimmunol.1101003
39. Kincaid EZ, Che JW, York I, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2011;13(2):129–135. doi:10.1038/ni.2203
40. Basler M, Kirk CJ, Groettrup M. The immunoproteasome in antigen processing and other immunological functions. Curr Opin Immunol. 2013;25(1):74–80. doi:10.1016/j.coi.2012.11.004
41. Shelleh HH, Al-Hatiti HS. Pattern of skin diseases in a hospital in southwestern Saudi Arabia. Saudi Med J. 2004;25(4):507–510.
42. Assiri AA, Yousuf BI, Quantock AJ, Murphy PJ. Incidence and severity of keratoconus in Asir province, Saudi Arabia. Br J Ophthalmol. 2005;89(11):1403–1406. doi:10.1136/bjo.2005.074955
43. Abanmi A, Al Harthi F, Al Baqami R, et al. Association of HLA loci alleles and antigens in Saudi patients with vitiligo. Arch Dermatol Res. 2006;298(7):347–352. doi:10.1007/s00403-006-0699-4
44. Abanmi A, Al Harthi F, Zouman A, et al. Association of Interleukin-10 gene promoter polymorphisms in Saudi patients with vitiligo. Dis Markers. 2008;24(1):51–57. doi:10.1155/2008/210609
45. Alissa A, Al Eisa A, Huma R, Mulekar S. Vitiligo-epidemiological study of 4134 patients at the National Center for Vitiligo and Psoriasis in Central Saudi Arabia. Saudi Med J. 2011;32(12):1291–1296.
46. Al-Shobaili HA. Correlation of clinical efficacy and psychosocial impact on vitiligo patients by excimer laser treatment. Ann Saudi Med. 2014;34(2):115–121. doi:10.5144/0256-4947.2014.115
47. Al-Harthi F, Zouman A, Arfin M, Tariq M, Al-Asmari A. Tumor necrosis factor-alpha and -beta genetic polymorphisms as a risk factor in Saudi patients with vitiligo. Genet Mol Res. 2013;12(3):2196–2204. doi:10.4238/2013.July.8.1
48. Alenizi DA. Consanguinity pattern and heritability of vitiligo in Arar, Saudi Arabia. J Family Community Med. 2014;21(1):13–16. doi:10.4103/2230-8229.128767
49. Al-Shobaili HA, Rasheed Z. Mitochondrial DNA acquires immunogenicity on exposure to nitrosative stress in patients with vitiligo. Hum Immunol. 2014;75(10):1053–1061. doi:10.1016/j.humimm.2014.09.003
50. Alzolibani AA, Al Robaee A, Al-Shobaili H, Al-Saif F, Al-Mekhadab E, Settin AA. Association of CYP2C9 genetic variants with vitiligo. Ann Dermatol. 2014;26(3):343–348. doi:10.5021/ad.2014.26.3.343
51. Tayeb MT. Genetic variations of vitiligo among Saudi patients. Middle East J Sci Res. 2011;7(3):266–271.
52. Sindi IA, Babalghith AO, Tayeb MT, et al. Risk of colorectal carcinoma may predispose to the genetic variants of the GST, CYP450, and TP53 genes among nonsmokers in the Saudi community. Int J Gen Med. 2021;14:1311–1323. doi:10.2147/IJGM.S294802
53. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. doi:10.1093/nar/gku1003
54. Jadeja SD, Mansuri MS, Singh M, Dwivedi M, Laddha NC, Begum R. A case-control study on association of proteasome subunit beta 8 (PSMB8) and transporter associated with antigen processing 1 (TAP1) polymorphisms and their transcript levels in vitiligo from Gujarat. PLoS One. 2017;12(7):e0180958. doi:10.1371/journal.pone.0180958
55. Dani P, Patnaik N, Singh A, et al. Association and expression of the antigen-processing gene PSMB8, coding for low-molecular-mass protease 7, with vitiligo in North India: case-control study. Br J Dermatol. 2018;178(2):482–491. doi:10.1111/bjd.15391
56. McHepange UO, Gao XH, Liu YY, et al. Vitiligo in North-Eastern China: an association between mucosal and acrofacial lesions. Acta Derm Venereol. 2010;90(2):136–140. doi:10.2340/00015555-0799
57. Uncu S, Yayli S, Bahadir S, Okten A, Alpay K. Relevance of autoimmune thyroiditis in children and adolescents with vitiligo. Int J Dermatol. 2011;50(2):175–179. doi:10.1111/j.1365-4632.2010.04665.x
58. Vora RV, Patel BB, Chaudhary AH, Mehta MJ, Pilani AP. A clinical study of vitiligo in a rural set up of Gujarat. Indian J Community Med. 2014;39(3):143–146. doi:10.4103/0970-0218.137150
59. Degboe B, Atadokpede F, Saka B, et al. Vitiligo on black skin: epidemiological and clinical aspects in dermatology, Cotonou (Benin). Int J Dermatol. 2017;56(1):92–96. doi:10.1111/ijd.13366
60. Mahajan VK, Vashist S, Chauhan PS, Mehta KIS, Sharma V, Sharma A. Clinico-epidemiological profile of patients with vitiligo: a retrospective study from a Tertiary Care Center of North India. Indian Dermatol Online J. 2019;10(1):38–44. doi:10.4103/idoj.IDOJ_124_18
61. Sehgal VN, Srivastava G. Vitiligo: compendium of clinico-epidemiological features. Indian J Dermatol Venereol Leprol. 2007;73(3):149–156. doi:10.4103/0378-6323.32708
62. Ezzedine K, Diallo A, Leaute-Labreze C, et al. Pre- vs. post-pubertal onset of vitiligo: multivariate analysis indicates atopic diathesis association in pre-pubertal onset vitiligo. Br J Dermatol. 2012;167(3):490–495. doi:10.1111/j.1365-2133.2012.11002.x
63. Nicolaidou E, Antoniou C, Miniati A, et al. Childhood- and later-onset vitiligo have diverse epidemiologic and clinical characteristics. J Am Acad Dermatol. 2012;66(6):954–958. doi:10.1016/j.jaad.2011.07.010
64. Gould IM, Gray RS, Urbaniak SJ, Elton RA, Duncan LJ. Vitiligo in diabetes mellitus. Br J Dermatol. 1985;113(2):153–155. doi:10.1111/j.1365-2133.1985.tb02057.x
65. Somorin AO, Krahn PM. Vitiligo: a study of 122 cases. Ann Saudi Med. 1997;17(1):125–127. doi:10.5144/0256-4947.1997.125
66. Arýcan O, Koc K, Ersoy L. Clinical characteristics in 113 Turkish vitiligo patients. Acta Dermatovenerol Alp Pannonica Adriat. 2008;17(3):129–132.
67. Alkhateeb A, Al-Dain Marzouka N, Qarqaz F. SMOC2 gene variant and the risk of vitiligo in Jordanian Arabs. Eur J Dermatol. 2010;20(6):701–704. doi:10.1684/ejd.2010.1095
68. Vrijman C, Kroon MW, Limpens J, et al. The prevalence of thyroid disease in patients with vitiligo: a systematic review. Br J Dermatol. 2012;167(6):1224–1235. doi:10.1111/j.1365-2133.2012.11198.x
69. Shong YK, Kim JA. Vitiligo in autoimmune thyroid disease. Thyroidology. 1991;3(2):89–91.
70. De Sanctis V, Khater D. Autoimmune diseases in Turner syndrome: an overview. Acta Biomed. 2019;90(3):341–344. doi:10.23750/abm.v90i3.8737
71. Bittles AH, Black ML. Evolution in health and medicine Sackler colloquium: consanguinity, human evolution, and complex diseases. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1779–1786. doi:10.1073/pnas.0906079106
72. El-Mouzan MI, Al-Herbish AS, Al-Salloum AA, Qurachi MM, Al-Omar AA. Growth charts for Saudi children and adolescents. Saudi Med J. 2007;28(10):1555–1568.
73. Turnpenny P, Ellard S. Emery’s Elements of Medical Genetics. Vol. 269.
74. Al Fahaad HA. Clinico-epidemiological profile of vitiligo patients in Najran region, Saudi Arabia. J Derm Dermatol Surgery. 2015;19(1):31–35. doi:10.1016/j.jdds.2014.05.001
75. AlGhamdi KM. Beliefs and perceptions of Arab vitiligo patients regarding their condition. Int J Dermatol. 2010;49(10):1141–1145. doi:10.1111/j.1365-4632.2010.04514.x
76. Alharbi MA. Identifying patients at higher risk of depression among patients with vitiligo at outpatient setting. Mater Sociomed. 2020;32(2):108–111. doi:10.5455/msm.2020.32.108-111
77. Alzolibani A. Genetic epidemiology and heritability of vitiligo in the Qassim region of Saudi Arabia. Acta Dermatovenerol Alp Pannonica Adriat. 2009;18(3):119–125.
78. Seif Eldin NS, Teama S, Amro K, Farag HM, Nour Eldin SM, Elhawary NA. Polymorphisms of TAP1/LMP7 loci in Egyptian patients with vitiligo. Egy J Med Hum Genet. 2006;7(2):241–249.
79. El-Husseiny R, Abd-Elhaleem A, Salah El-Din W, Abdallah M. Childhood vitiligo in Egypt: clinico-epidemiologic Profile of 483 patients. J Cosmet Dermatol. 2021;20(1):237–242. doi:10.1111/jocd.13451
80. Spritz RA. Modern vitiligo genetics sheds new light on an ancient disease. J Dermatol. 2013;40(5):310–318. doi:10.1111/1346-8138.12147
81. Czajkowski R, Mecinska-Jundzill K. Current aspects of vitiligo genetics. Postepy Dermatol Alergol. 2014;31(4):247–255. doi:10.5114/pdia.2014.43497
82. Shen C, Gao J, Sheng Y, et al. Genetic susceptibility to vitiligo: GWAS approaches for identifying vitiligo susceptibility genes and loci. Front Genet. 2016;7:3. doi:10.3389/fgene.2016.00003
83. Cheng L, Liang B, Tang XF, et al. Validation of susceptibility loci for vitiligo identified by GWAS in the Chinese Han population. Front Genet. 2020;11:542275. doi:10.3389/fgene.2020.542275
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

