Back to Journals » International Journal of General Medicine » Volume 16
Value of Methylation Status of RPRM, SDC2, and TCF4 Genes in Plasma for Gastric Adenocarcinoma Screening
Authors Guo J , Li J, Chang J, Wang L , Xi Y
Received 17 November 2022
Accepted for publication 8 February 2023
Published 22 February 2023 Volume 2023:16 Pages 673—681
DOI https://doi.org/10.2147/IJGM.S395951
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jianghong Guo, Jing Li, Jiang Chang, Li Wang, Yanfeng Xi
Department of Pathology, Cancer Hospital Affiliated to Shanxi Medical University, Shanxi Province Cancer Hospital, Shanxi Hospital Affiliated to Cancer Hospital, Chinese Academy of Medical Sciences, Taiyuan, People’s Republic of China
Correspondence: Yanfeng Xi, Email [email protected]
Objective: To explore the clinical value of the combined screening of the methylation statuses of the RPRM, SDC2, and TCF4 genes in plasma of gastric cancer patients.
Methods: Differential expressed genes (DEGs) were selected from the Gene Expression Omnibus database, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed using DAVID, and a protein-protein interaction network was constructed. Hub genes were obtained using Cytoscape. Screening results combined with literature reports identified three genes (RPRM, SDC2, and TCF4). Further analysis was done using biopsies collected through gastroscopy at Shanxi Cancer Hospital from January 8, 2020 to February 22, 2021. The patients were divided into two groups: gastric adenocarcinoma group, and control group which are not gastric adenocarcinoma according to pathological or gastroscopic results. The methylation statuses of the three genes in peripheral blood plasma were detected by fluorescence polymerase chain reaction, and the relationships between the positive rates of the three combined genes with pathology and/or gastroscopy results were analyzed. The clinical value of the combined detection of the three genes was evaluated according to these indicators. The diagnostic specificity and sensitivity of this detective method were analyzed.
Results: A total of 197 DEGs were identified and 12 hub genes were obtained. The enriched functions and pathways of DEGs include regulation of cell proliferation, extracellular space, cytokine activity, and pathways in cancer. The combination of RPRM, SDC2, and TCF4 gene methylation had a specificity of 93.39% and sensitivity of 80.33%. The combined positive rate of RPRM, SDC2, and TCF4 gene methylation in patients with gastric adenocarcinoma was significantly higher compared with those without gastric adenocarcinoma (χ2=151.179, P< 0.05).
Conclusion: Combined detection of RPRM, SDC2, and TCF4 gene methylation in peripheral blood plasma maybe helpful in screening for gastric adenocarcinoma, and maybe a complementary method to gastroscopy and serum tumor markers.
Keywords: gastric cancer, RPRM, SDC2, TCF4, methylation
Introduction
Gastric adenocarcinoma (GAC) is the fourth most common cause of cancer-related mortality globally, leading more than 750,000 deaths annually.1,2 GAC has become a major health problem in many countries. Early detection and intervention are important factors in the diagnosis and treatment of malignant gastric tumors. Gastroscopy and pathological biopsy are the main methods for GAC screen, but their limitations including invasiveness and high cost, are big challenges in population-based screening. The identification of painless and sensitive biomarkers for GAC screening has a potential role to reduce the pain of patients. It is reported that measuring serum pepsinogen (PG) levels can be a screening method for gastric cancer.3 The serum tumor markers comprising carbohydrate antigen 199 (CA199), carcinoembryonic antigen (CEA), and carbohydrate antigen 724 (CA724) are also helpful for the diagnosis of GAC. However, current methylation markers are few applied in GAC.
Many studies have shown an important role for DNA methylation in carcinogenesis.4,5 DNA methylation can lead to abnormal expression of many tumor suppressor genes and may thus promote the occurrence and development of gastric cancer. Although methylated genes in gastric cancer have been reported in the literatures, few of them have been used clinically in terms of diagnosis, possibly because of low positive rates. The detection of multigene combinations may thus improve the detection rate. RPRM encodes the highly glycosylated protein Reprimo, which is mainly found in the cytoplasm. RPRM is the founding member of the RPRM gene family and has been found to be highly methylated in blood samples from patients with gastric cancer.6 RPRM has been explored as a noninvasive biomarker in gastric cancer.6–8 Syndecan-2 (SDC2), encoded by the SDC2 gene, is a transmembrane protein involved in cell proliferation, cell migration, angiogenesis, and cell-matrix interactions.9 SDC2 is hypermethylated in most patients with colorectal cancer.10 Recent studies also showed that SDC2 played a regulatory role in the migration and invasion of gastric cancer cells.11
Array technology, high-throughput sequencing, and bioinformatics analysis have recently been widely used to identify differentially expressed genes (DEGs) and to understand the functional pathways involved in GAC carcinogenesis. We therefore determined the methylation profiles in the GSE25869,12 GSE28094,13 and GSE3060114 array datasets from the Gene Expression Omnibus (GEO) database. We downloaded the three datasets and analyzed them using GEO2R to identify genes that were significantly different between gastric cancer and normal tissues (P<0.01). Finally, transcription factor 4 (TCF4) is selected. TCF4 is a member of the T cytokine/lymphoid enhancer factor (TCF4/LEF) family, and is also frequently methylated in gastric cancer.
Eventually, through screening and review of the literature, three genes (RPRM, SDC2, and TCF4) were identified. Then we designed this trial to explore the consistency of combined detection of RPRM, SDC2, and TCF4 methylation with pathological biopsy (gold standard). Our study may provide a potential diagnostic strategy for early gastric cancer.
Materials and Methods
Data Screening
GEO (https://www.ncbi.nlm.nih.gov/geo) is a comprehensive public functional genomics data repository, including chips, arrays, and high-throughput sequencing gene expression data. We downloaded three methylation datasets (GSE25869, GSE28094, and GSE30601) from GEO (GPL8490 platform, Illumina HumanMethylation27 BeadChip and GPL9183 platform, Illumina GoldenGate Methylation Cancer Panel I). The selected conditions were series (Entry type) and methylation profiling by array (Study type). GSE25869 contained 11 gastric cancer cell lines and 32 pairs of gastric cancer and normal samples, GSE28094 contained 1628 human samples (we selected 16 stomach cancer and 418 healthy samples), and GSE30601 contained 203 gastric tumor tissues and 94 matched non-malignant gastric tissues.
Identification of DEGs
DEGs between gastric cancer and non-cancerous tissues were screened using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r), which is an online analysis tool for GEO that can be used to compare two or more datasets in GEO sample data to identify DEGs under different experimental conditions. Notably, the tumor group must be defined preferentially when defining the grouping with GEO2R analysis. DEGs were considered significant if the adjusted P-value was <0.01. Overlapping genes among the three datasets were then obtained by the website tool (http://bioinformatics.psb.ugent.be/webtools/Venn).
GO and KEGG Enrichment Analyses of DEGs
The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/) is currently used mainly for functional and pathway enrichment analyses of DEGs. In this study, we analyzed the biological functions of the DEGs using the DAVID online database as follows: on the DAVID home page, click “start analysis” and paste the selected DEGs into the gene list; select identifier (official gene symbol) and select species (Homo sapiens); choose the gene list; and then click “Submit List”. In the new screen, select “Functional Annotation Chart”, the BP (biological process), CC (cell component), and MF (molecular function) charts in GO and KEGG pathways can then be obtained.
PPI Network Construction and Hub Gene Selection
The PPI network was calculated using the Search Tool for the Retrieval of Interacting Genes (http://string-db.org) online database. Functional interactions between proteins were analyzed. It was helpful for us to understand the mechanisms of disease generation. “String_interactions_short.tsv” was downloaded and analyzed using Cytoscape (version 3.10.0), which is an open software platform for visualizing complex networks and integrating these with any type of attribute data. Molecular Complex Detection (MCODE) in Cytoscape is a plug-in app for clustering a given network based on topology, to find densely connected regions.15 In this study, the DEG PPI network was constructed using Cytoscape and the most significant module in the PPI network was identified by MCODE. The criteria for selection were: find clusters = in whole network; degree cutoff = 2; cluster finding = hairout (yes); node density cutoff = 0.1; node score cutoff = 0.2; k-core = 2; and max depth = 100. The hub genes were then selected using the plug-in cytoHubba.
Gene Analysis
Genes screened in the literature and bioinformatics databases were analyzed using gene expression profiling interactive analysis (GEPIA; http://gepia.cancer-pku.cn). We selected single gene analysis: click on “GoPIA”; expression DIY select boxplot, and enter the screened genes separately into the gene name; |Log2FC| cutoff = 1, p-value cutoff = 0.01; for cancer name, select STAD (stomach adenocarcinoma); click “Add”; for log scale, select yes; Jitter Size=0.4; for matched normal data select match TCGA normal and GTEx data; and click “Plot”.
Patient Samples
Biopsy tissues were collected in patients undergoing gastroscopy at Shanxi Cancer Hospital from January 8, 2020 to February 22, 2021. Patients were divided into two groups: an experimental group with GAC confirmed by pathological examination, and a control group excluded GAC confirmed by pathological and/or gastroscopic examination. All participants were over 18 years old. Patients with the following conditions should be excluded: 1) underwent gastrectomy within 1 year, 2) were cured of gastric cancer, 3) pregnant, 4) had a history of blood transfusion within the last month.
Blood Samples
Peripheral blood (5 mL) was collected from each enrolled patient into EDTA anticoagulant tubes and delivered to the laboratory within 2 h. Plasma was centrifuged immediately for 12 min at 1600 × g. If the plasma could not be separated immediately, it was stored at 2–8°C for no more than 4 h, without freezing. Blood sample with severe hemolysis or with extracted plasma less than 2 mL was removed. Samples that were not collected, stored, and/or transported as required, and samples with incomplete information were also removed.
DNA Extraction and Bisulfite Conversion
DNA was extracted from each sample using a nucleic acid extraction kit (lot. no. 21NAE001; Beijing Akron Medical Technology Co, Ltd, China.), according to the manufacturer’s instructions. DNA was quantified using a NanoDrop One ultrafine ultraviolet spectrophotometer (Applied Biosystems, USA) and diluted to 0.8–1.2 ng/µL. Plasma (2 mL, 2 mL quality control substance) free DNA was treated with sodium bisulfite to obtain sulfite-transformed DNA (BisDNA). Samples with low content of DNA (<0.8 ng/µL) were removed.
Polymerase Chain Reaction (PCR)
Real-time PCR was carried out using a diagnostic kit (Beijing Akron Medical Technology Co, Ltd, China.) for methylated genes in gastric cancer, according to the instructions. The parameters of the cycling thermal profile were 98°C for 5min and 95°C for 10s, followed by 45 cycles of 63°C for 5 s and 58°C for 30s, annealing/elongation by 25°C for 10s with acquisition of fluorescent data. The positive maximum cutoff values for RPRM, SDC2, and TCF4 were 38.81, 41.1, and 43.19. ACTB was used as an internal reference; if the amount of DNA was sufficient (ACTB Ct ≤ 35), the PCR results were considered reliable, but if the ACTB Ct was > 35.0, the PCR reaction was considered invalid.
Statistical Analysis
The collected data were analyzed using the SPSS 18.0 program. Differences in pathological features and gene methylation statuses between the two groups were analyzed using χ2 tests. Youden’s index = (Sensitivity+ Specificity-1). All P values were 2-sided and P<0.05 was considered statistically significant.
Results
Identification of DEGs in Gastric Cancer
After standardization of the screening conditions, DEGs (3920 in GSE25869, 458 in GSE28094, and 10,623 in GSE30601) were identified using GEO2R, including 197 overlapping genes among the three datasets (Figure 1A).
 |
Figure 1 Hub gene screening and analysis. (A) Venn diagram (The 3 datasets showed an overlap of 197 genes). (B) *TCF4 were statistically significant. |
GO and KEGG Enrichment Analyses of DEGs
BP, CC, MF, and KEGG pathway enrichment analyses were performed using DAVID to clarify the biological classification of the DEGs (Table 1). GO analysis revealed that in terms of biological process, DEGs were mainly enriched in the regulation of cell proliferation, mitogen-activated protein kinase (MAPK) cascade, kinase activity, transmembrane receptor protein tyrosine kinase signaling pathway, and peptide-tyrosine phosphorylation. As to cellular components, DEGs were mainly enriched in extracellular region, receptor complex, extracellular space, plasma membrane, and integral component of plasma membrane, and in the field of molecular function, DEGs were significantly enriched in transmembrane receptor protein tyrosine kinase activity, protein tyrosine kinase activity, cytokine activity, growth factor activity, and receptor binding. KEGG pathway enrichment analysis showed that the DEGs were significantly enriched in pathways in cancer, Akt signaling pathway, proteoglycans in cancer, and MAPK signaling pathway.
 |
Table 1 GO and KEGG Pathway Enrichment Analysis of DEGs in Stomach Cancer |
PPI Network Construction, Hub Gene Selection, and Analysis
A PPI network of DEGs was constructed using an online analysis tool and the hub genes were identified using Cytoscape. Every hub gene was analyzed with GEPIA. Finally, TCF4 was selected as an important hub gene. (Figure 1B).
Clinicopathological Information
61 samples of GACs and 227 of non-GACs were collected, which are confirmed according to gastroscopic examination and/or pathological examination. The microscopic morphology of GAC showed irregular glandular atypical dysplasia, angulation, polar disorder, fusion into pieces, and nuclear enlargement (Figure 2A). Non-GAC patients mostly had gastritis, with inflammatory cell infiltration in the gastric mucosa, erosion on the surface, infiltration of numerous plasma cells in some areas, and some glandular hyperplasia (Figure 2B).
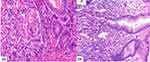 |
Figure 2 Pathological morphology. (A) gastric adenocarcinoma HE X200 (B) gastritis HE X200. |
The basic characteristics of the 288 cases are summarized in Table 2. Age and sex differed significantly between the two groups (P<0.01).
 |
Table 2 Basic Characteristics of Patients and Controls |
Methylation of RPRM, SDC2, and TCF4 in Plasma
With collected 288 plasma samples, the methylation statuses of RPRM, SDC2, and TCF4 were detected using fluorescence PCR analysis (Figure 3). The groupings based on methylation levels of RPRM, SDC2, TCF4, and the three genes combined were significantly related to those based on gastroscopy/tissue biopsy results (P<0.01). We also compared the results for each of the three genes alone and combined, based on the sensitivity, specificity, Youden’s index, positive predictive value, negative predictive value, and kappa score (Table 3). Methylation status of RPRM, SDC2, TCF42 alone picked 37.37, and 41 patients correctly in 61 patients who are diagnosed as gastric cancer by pathologist (sensitivities are 60.66%, 60.66%, and 67.21 respectively). Combined detection could rise the number to 49 patients and 80.33%. Specificity of RPRM, SDC2, TCF42 alone are 98.24%, 98.24%, 95.59% and combined detection is 93.39%. Youden’s index, an indicator for evaluating the authenticity of screening tests, is much higher in the combined detection method, which is 73.72% compared to detection alone methods which are 58.89%, 58.89 and 62.81%. RPRM, SDC2 alone could get the highest ration of positive prediction (both PPV are 0.902). However, when considering ruling out the negative patients, the combined method is better (NPV is 0.946).
 |
Table 3 Results of Gastroscopy/Tissue Biopsy and Methylation of RPRM, SDC2, and TCF4 in Plasma |
 |
Figure 3 (A) methylation of RPRM, SDC2 and TCF4 were positive in GAC. (B) Internal reference ACTB express in gastritis. |
Discussion
Bioinformatics analysis has recently become increasingly important in cancer research. In this study, KEGG pathway enrichment analysis revealed that DEGs between GAC and non-GAC patients were significantly enriched in pathways in cancer. TCF4 was one of the identified DEGs, and GEPIA showed that TCF4 expression was significantly increased in gastric tumors compared with normal tissues. Tumor suppressor genes are crucial negative regulatory factors affecting cell proliferation, differentiation, and apoptosis in vivo. This was consistent with the analyses using DAVID and GEPIA. Gene inactivation caused by hypermethylation of tumor suppressor gene promoters contributes to unlimited cell growth, increased differentiation, and promotion of tumor formation. RPRM, SDC2, and TCF4 are tumor suppressor genes that can be hypermethylated in patients with gastric cancer.6,10,16
The occurrence and development of gastric cancer is closely related to many factors, including the environment, genes, living habits, and Helicobacter pylori infection. In the present study, we found significant differences in sex and age between the two groups (P<0.05). Most of the patients with GAC were male, which may be related to the higher proportion of drinking and smoking in males. The proportion of patients aged over 60 years was also higher in the GAC group, in accordance with age being a risk factor for GAC.
Gastric cancer is a malignant tumor originating from the epithelial cells of the gastric mucosa. Although healthy awareness has increased and regular physical examinations have become relatively common, the morbidity and mortality of gastric cancer in China remain only second to lung cancer.17 The diagnosis of GAC in China currently depends mainly on endoscopy, pathological examination, and the detection of serum tumor markers. However, gastroscopy and pathological examination are invasive, and the sensitivity of serum tumor markers also needs to be improved. In this study, we tested the methylation statuses of RPRM, SDC2, and TCF4 genes in plasma from 288 patients in China (61 GAC, 227 non-GAC controls), and showed that the methylation levels of all three genes were significantly higher in patients with GAC compared with the control group. When the methylation levels of the three genes were tested separately, RPRM and SDC2 had similar specificities of 98.24%, compared with 95.59% for TCF4; and all three genes were satisfactory (over 90%). The sensitivities of RPRM and SDC2 were also similar (but only 60.66%), while that of TCF4 was slightly higher (67.21%). This low sensitivity meant that methylation of any one of these tumor suppressor genes alone was not a credible alternative to gastroscopy for gastric cancer screening. This finding was consistent with clinical practice. However, the combination of RPRM, SDC2, and TCF4 gene methylation had a satisfactory specificity and sensitivity. Youden’s index is a common measure of the veracity of screening tests. In our study, Youden’s index for the combined detection of RPRM, SDC2, and TCF4 methylation was significantly superior to that for methylation of any single gene. The kappa score is an ideal indicator to describe the consistency of diagnosis and is widely used in clinical trials. In this study, the kappa score for the combined detection of triple-gene methylation suggested that the consistency of the combined detection of RPRM, SDC2, and TCF4 methylation and gastroscopy/tissue biopsy was satisfactory.
However, the current study also has some potential limitations. Although the combined detection of RPRM, SDC2, and TCF4 methylation in plasma achieved a satisfactory diagnostic ability in GAC diagnosis, more clinical trials are still needed. More population-based research will be conducted in the future to confirm the value of combined detection of RPRM, SDC2, and TCF4 methylation in the diagnosis of GAC.
In conclusion, the combined detection of RPRM, SDC2, and TCF4 methylation in plasma may helpful in the screening for gastric adenocarcinoma, and maybe a complementary method to gastroscopy and serum tumor markers.
Abbreviations
GAC, Gastric adenocarcinoma; RPRM, Reprimo; SDC2, Syndecan-2; TCF4, Transcription factor 4; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, Differential expressed genes; PPI, protein–protein interaction; DAVID, Visualization and Integrated Discovery; GEPIA, gene expression profiling interactive analysis; BP, biological process; CC, cell component; MF, molecular function.
Data Sharing Statement
The raw data used to support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval and Consent to Participate
The use of human tissues was approved by the Shanxi Provincial Cancer Hospital Ethics Committee (no. 2022G12), and patient consents were obtained.
Patient Consent for Publication
Publication of the clinical datasets in this study does not compromise anonymity or confidentiality, or breach local data protection laws.
Statement of Ethics
All participants gave informed consent before their inclusion in the study. The study protocols were conducted according to the principles of the Declaration of Helsinki and were approved by the Scientific and Medical Ethical Committee of the Shanxi Provincial Cancer Hospital.
Acknowledgments
Thanks to Dr. Wei Cui for helping us revise this manuscript.
Author Contributions
Yanfeng Xi contributed to data collection. Jianghong Guo performed the bioinformatics analyses and wrote the manuscript. Li Wang and Jing Li carried out the experiments of PCR. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
Funded by the medical science and technology innovation team of the Shanxi Provincial Health Commission (No. 2020TD07).
Disclosure
The authors declare that there is no conflict of interest regarding the publication of this article.
References
1. Sung H, Ferlay J, Rebecca L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Etemadi A, Safiri S, Sepanlou SG. The global, regional, and national burden of stomach cancer in 195 countries, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2020;5(1):42–54. doi:10.1016/S2468-1253(19)30328-0
3. Yoshihara M, Sumii K, Haruma K, et al. Correlation of ratio of serum pepsinogen I and II with prevalence of gastric cancer and adenoma in Japanese subjects. Am J Gastroenterol. 1998;93(7):1090–1096. doi:10.1111/j.1572-0241.1998.00335.x
4. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi:10.1038/ng1089
5. Pfeifer GP. Defining driver DNA methylation changes in human cancer. Int J Mol Sci. 2018;19(4):1166. doi:10.3390/ijms19041166
6. Wen JF, Zheng T, Hu KF, et al. Promoter methylation of tumor-related genes as a potential biomarker using blood samples for gastric cancer detection. Oncotarget. 2017;8(44):77783–77793. doi:10.18632/oncotarget.20782
7. Bernal C, Aguayo F, Villarroel C, et al. Reprimo as a potential biomarker for early detection in gastric cancer. Clin Cancer Res. 2008;14(19):6264–6269. doi:10.1158/1078-0432.CCR-07-4522
8. Wang H, Zheng Y, Lai J, Luo Q, Ke H, Chen Q. Methylation-sensitive melt curve analysis of the reprimo gene methylation in gastric cancer. PLoS One. 2016;11(12):e0168635. doi:10.1371/journal.pone.0168635
9. Oh T, Kim N, Moon Y, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15(4):498–507. doi:10.1016/j.jmoldx.2013.03.004
10. Xu F, Yu FF, Han JY, et al. Detection of circulating tumor DNA methylation in diagnosis of colorectal cancer. Clin Transl Gastroenterol. 2021;12(8):e00386. doi:10.14309/ctg.0000000000000386
11. Wang JW, Lu C, Chen JY, et al. Expression of Syndecan-2 in gastric adenocarcinoma and its effect on tumorigenesis in vitro. Transl Cancer Res. 2017;6:658–66.
12. Kwon OH, Park JL, Kim M, et al. Aberrant up-regulation of LAMB3 and LAMC2 by promoter demethylation in gastric cancer. Biochem Biophys Res Commun. 2011;406(4):539–545. PMID: 21345334. doi:10.1016/j.bbrc.2011.02.082
13. Fernandez AF, Assenov Y, Martin-Subero JI, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22(2):407–419. PMID: 21613409. doi:10.1101/gr.119867.110
14. Zouridis H, Deng N, Ivanova T, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med. 2012;4(156):156ra140. PMID: 23076357. doi:10.1126/scitranslmed.3004504
15. Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012;14:83. doi:10.1186/1532-429X-14-83
16. Joo JK, Kim SH, Kim HG, et al. CpG methylation of transcription factor 4 in gastric carcinoma. Ann Surg Oncol. 2010;17(12):3344–3353. doi:10.1245/s10434-010-1131-z
17. CHEN W, ZHENG R, BAADE PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
